Introduction
Asthma is a heterogeneous syndrome that is
characterized by inflammation and hyper-responsiveness of the
airway. Although all age groups are affected, the prevalence of
asthma is increasing in many countries, especially among children
(1). The aetiology and pathogenesis
of asthma, which may be associated with genetic, immune and
environmental factors, are incompletely understood and remain under
investigation (2).
The helper T cell family include Th1, Th2,
regulatory T (Treg) and Th17 cells. Recent developments in
immunology and molecular biology have revealed that asthma is not
only associated with the imbalance of Th1/Th2 function (3) but also with Tregs, since imbalances in
forkhead transcription factor P3 (FOXP3)+ Treg/Th17 and
Th2/FOXP3+ Treg cells lead to asthma (4,5). T cells
play a central role in regulating airway inflammation in asthma.
Tregs are a subset of CD4+ T cells that play an
essential role in maintaining peripheral immune tolerance and
controlling allergic diseases, such as asthma. Tregs, together with
effector T cells (Teffs), cytokines, immune antibodies and other
cellular components, play an important role in maintaining immune
balance (6). As important
immunosuppressive cells, CD4+CD25+ Tregs act
in cell-cell contact-dependent inhibition patterns and ultimately
inhibit immune diseases by inhibiting helper T cell activation and
differentiation, and directly inhibiting B cell activation to
produce antibodies (7).
FOXP3 is the most reliable specific molecular marker
of natural Tregs (nTregs) and is associated with the
immunosuppressive function of CD4+CD25+ Tregs
(8). The development and function of
CD4+CD25+ Tregs depend on the expression of
FOXP3 (9). Tregs, specifically
CD4+CD25+FOXP3+ Tregs, tightly
control autoreactive B and T cell responses in the periphery
(10). FOXP3+ Tregs are
the most widely-known type of immune cells and have the strongest
inhibitory function and most extensive inhibitory targets.
FOXP3+ Tregs prevent autoreactive T cell activation,
inhibit autoimmune and allergic disease occurrence, exert
anti-inflammatory functions and maintain autoimmune tolerance
(11,12). In addition, the downregulation of
FOXP3 expression potentially results in the inability of Tregs to
inhibit infection and tumours (13).
FOXP3 is an important transcription factor in the
activation of Tregs, but its expression alone may not be sufficient
to explain all Tregs functions. An additional mechanism is needed
to explain the genes expressed by Tregs and their functional
stability and cell lineage maintenance. One possible mechanism
underlying this phenomenon is epigenetic regulation, which also
provides a new understanding of the interaction between genes and
the environment (14). Epigenetic
inheritance can initiate and maintain FOXP3 expression in nTregs
(15). FOXP3 expression is regulated
by DNA methylation, histone modifications and posttranscriptional
modifications (16). The epigenetic
regulation and methylation of FOXP3 play an important role in its
stable expression (17). Changes in
the methylation level of the FOXP3 gene may affect Treg
differentiation and regulate the occurrence of an immune response.
Thus, detecting the methylation statuses of upstream enhancers of
FOXP3 may help in the diagnosis and subtype classifications of
diseases (18). A comprehensive
study of epigenetic variation will promote our understanding of
complex diseases, especially those in which genetic and
environmental factors interact, such as asthma. Therefore, in order
to further investigate the association between FOXP3 expression and
methylation levels in Tregs and its pathogenesis in childhood
asthma, the percentages of CD4+CD25+ FOXP3
Tregs in CD4+ T lymphocytes in the peripheral blood
mononuclear cells (PBMCs) from children with asthma and healthy
controls were detected by flow cytometry. Furthermore, the mRNA
expression of transcription factor FOXP3 in PBMCs was detected by
RT-qPCR. The mRNA expression of FOXP3 in
CD4+CD25+ FOXP3 Tregs was compared between
asthmatic and healthy control groups and the correlation between
FOXP3 mRNA expression and pulmonary function (FEV1) was analysed in
the asthma group. Additionally, the methylation statuses of 16 CpG
loci (including 7 sites in the exon and 9 sites in the intron
areas) in the FOXP3 gene were compared by bisulfite transformation,
PCR and pyrosequencing.
Patients and methods
Clinical samples
A total of 15 children with asthma, who were
hospitalized in the respiratory department of Jiangxi Children's
Hospital (Jianxi, China) between July 2016 and June 2017, were
included in the asthma group. The group included 10 males and 5
females, aged 5–14 years, and all of these subjects met the
diagnostic criteria for asthma (19). The healthy control group consisted of
15 children, including 9 males and 6 females, aged 5–14 years.
These subjects simultaneously underwent physical examination at the
children's health clinic. The subjects had no family history of
atopic diseases or personal allergy history, no history of other
diseases, and no respiratory tract infections in the past month.
Subjects who failed to complete the study were excluded from both
groups. Independent-Samples t-test confirmed that there were no
significant differences in sex (P=0.702) or age (P=0.338) between
the two groups. All human materials were obtained with informed
consent, and the protocols were approved by the Ethics Review
Committee of Jiangxi Children's Hospital.
Ficoll density gradient separation of
PBMCs
Peripheral venous blood (5 ml) was collected for
anticoagulation and mixed with 5 ml of phosphate-buffered saline
(PBS). The liquid was added slowly against the wall of the tube and
centrifuged at 4°C, 500 × g for 20 min. Following centrifugation,
the liquid in the tube was divided into four layers: Plasma and
PBS; lymphocytes; red blood cells; and granulocytes. A white and
cloudy layer, mainly composed of monocytes, including lymphocytes
and monocytes, appeared at the junction of the upper and middle
layers. A capillary pipette was inserted into the white and cloudy
layer, and the PBMC layer was collected and placed in another
centrifuge tube. Subsequently, 5 ml PBS solution was added, and the
resulting mixture was centrifuged at 4°C, 500 × g for 10 min. After
discarding the supernatant, the cells were washed twice, and the
cell concentration was adjusted to 1×106 cells/ml with
PBS.
Flow cytometry (FCM) analysis
For Treg analysis, cell suspensions were transferred
into tubes and washed with PBS. The cell suspension (100 µl) was
stained with fluorescein isothiocyanate (FITC) anti-human CD4 (20
µl; cat. no. 561005; BD Biosciences), and allophycocyanin (APC)
anti-human CD25 (20 µl; cat. no. 560987; BD Biosciences). IgG1-FITC
(20 µl; cat. no. 556649; BD Biosciences) and IgG1-APC (20 µl; BD
Biosciences; cat. no. 550854) were used as homologous controls.
After shaking and mixing, the solution was incubated for 15 min in
the dark at room temperature (20–25°C), following which 100 µl
Reagent A (cat. no. 641776; BD Biosciences) was added to the tube
and vortexed thoroughly. After a further incubation for 5 min in
the dark at room temperature (20–25°C), the mixture was washed with
1 ml PBS solution and centrifuged under 800–850 × g at 4°C for 5
min. The supernatant was discarded, and 50 µl Reagent B (BD
Biosciences, cat. no. 641776) and 20 µl phycoerythrin (PE)
anti-human FOXP3 (BD Biosciences; cat. no. 560082) were added,
IgG1-PE (20 µl; BD Biosciences; cat. no. 556650) was used as a
homologous control. After shaking and mixing, the cells were
incubated at room temperature (20–25°C) for 15 min, washed with 1
ml of PBS solution and centrifuged under 800–850 × g at 4°C, for 5
min. The supernatant was discarded, and the cells were suspended in
250 µl of PBS solution. All steps were performed using the BD
Instrasure™ Kit according to manufacturer's protocols (Becton,
Dickinson and Company).
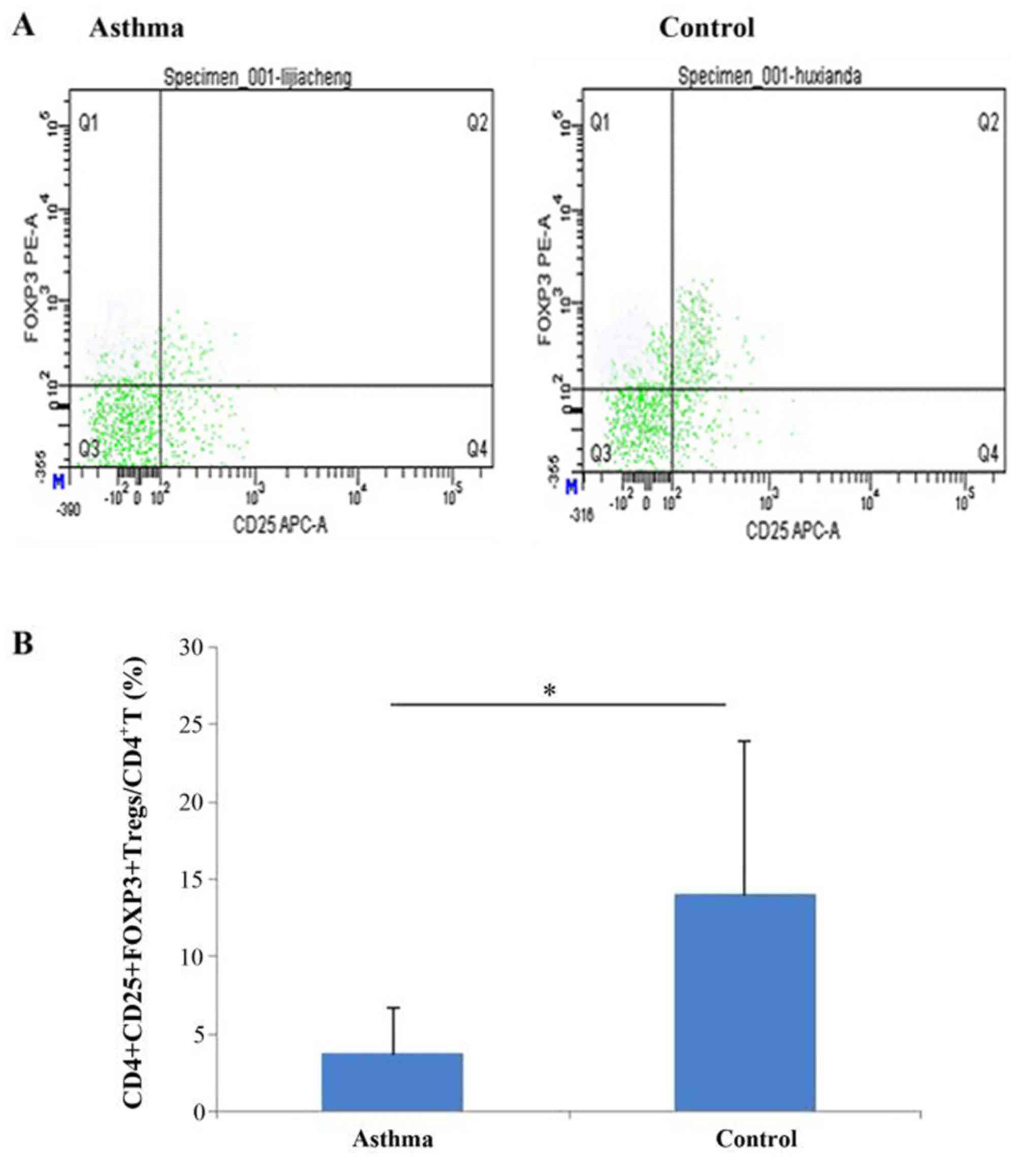
FCM was performed on a BD FACSCanto™ II flow
cytometer (BD, New Jersey, USA) using BD FACSDiVa software v6.1.2
(Becton, Dickinson and Company). CD4+ cells,
CD4+CD25+T cells and
CD4+CD25+FOXP3+ Tregs in the
lymphocyte group were determined by an FSC-SSC scatter plot. The
ratio of CD4+CD25+FOXP3+ Tregs to
CD4+ T cells in children with asthma and healthy
children was compared.
Pulmonary function test
Pulmonary function was measured using a MasterScreen
IOS pulmonary function instrument (Jaeger), and the temperature,
pressure and humidity were corrected before measurement. The
pulmonary function was measured 3 times, and the best value was
taken as the final value. The change in FEV1 was measured as a
percentage of the normal predicted FEV1 (%).
Reverse transcription-quantitative
(RT-q)PCR analysis of FOXP3 mRNA expression
The mRNA sequence of the FOXP3 gene was obtained
from the GenBank database. β-actin (cat. no. ab179467; Abcam) was
used as an internal control. The primers were designed and
synthesized by Shanghai Bioengineering Co., Ltd. The sequences of
the primers are listed in Table
I.
 | Table I.Primer sequences for the FOXP3 and
β-actin genes. |
Table I.
Primer sequences for the FOXP3 and
β-actin genes.
| Name | Direction | Sequence,
5′-3′ | Fragment length,
bp |
|---|
| FOXP3 | F |
CAAGTTCCACAACATGCGAC | 91 |
|
| R |
ATTGAGTGTCCGCTGCTTCT |
|
| β-actin | F |
ATCGTCCACCGCAAATGCTTCTA | 105 |
|
| R |
AGCCATGCCAATCTCATCTTGTT |
|
Total RNA was extracted using TRIzol®
(Invitrogen; Thermo Fisher Scientific, Inc.) and then
reverse-transcribed into cDNA using RevertAid First Strand cDNA
Synthesis Kit (cat. no. K1622; Thermo Fisher Scientific, Inc.)
under the following incubation conditions: 65°C for 5 min, 42°C for
60 min and then 70°C for 5 min according to the manufacturer's
protocol. PCR amplification was carried out using a PCR kit
(Beijing TransGen Biotech Co., Ltd.) under the following reaction
conditions: 35 cycles of denaturing at 94°C for 40 sec; annealing
at 60°C for 40 sec; and extension at 72°C for 60 sec. The PCR
products were subjected to a melting curve analysis to ensure that
a single amplification product was produced. The cycle threshold
(Ct) value of the quantitative results was calculated automatically
and reported by a computer. The relative expression level of FOXP3
was calculated using the quantification cycle (2−ΔΔCq)
method (20).
DNA methylation assays were performed
by bisulfate modification followed by PCR amplification and
pyrosequencing
DNA was from PBMCs using TIANamp Genomic DNA kit
(cat. no. DP304; Tiangen Biotech Co., Ltd.) before hydrogen sulfite
conversion was performed. Subsequently, template DNA (2 µl) was
added to each PCR tube for hydrogen sulfite conversion using a
sodium bisulfite modification kit (EpiTect Fast DNA Bisulfite kit;
Qiagen GmbH), according to the manufacturer's protocols. Each
component of the reaction mixture, along with their respective
volumes, are listed in Table II.
PCR amplification at 20 µl volume per reaction was subsequently
performed using Pyromark PCR Kit (cat. no. 978703; QIAGEN China
Co., Ltd) according to the reaction programme shown in Table III. The primer sequences were
designed and synthesized by KaiJie Transforming Medical Research
Co. Ltd (Table IV). For methylation
analysis, the microspheres were immobilized by the PCR products and
prepared for sequencing on a pyrosequencing apparatus (Pyromark
Q24; QIAGEN China Co., Ltd) that recorded the experimental results.
16 CpG sites from exons −6,210 to −6,334 and introns −2,262 to
−2,376 of the FOXP3 gene were selected in asthma and control
children, respectively to detect the degree of DNA methylation,
which was expressed as the average methylation (Fig. 1).
 | Table II.PCR system. |
Table II.
PCR system.
| Components | Volume/reaction
(µl) |
|---|
| PyroMark PCR Master
Mix, 2X | 12.5 |
| CoralLoad
Concentrate, 10X | 2.5 |
| Forward primer | 0.5 |
| Reverse primer | 0.5 |
| RNase-free
water | 7 |
| Total | 23 |
 | Table III.PCR programme. |
Table III.
PCR programme.
| Step | Process | Temperature,
duration |
|---|
| 1 |
Predenaturation | 95°C, 15 min |
| 2 | Denaturation | 94°C, 30 sec |
| 3 | Anneal | 63°C, 30 sec,
−0.5°C/cycle |
| 4 | Prolongation | 72°C, 30 sec |
|
| Cycle number (steps
2–4) | 10 |
| 5 | Denaturation | 94°C, 30 sec |
| 6 | Anneal | 58°C, 30 sec |
| 7 | Prolongation | 72°C, 30 sec |
|
| Cycle number (steps
5–7) | 40 |
| 8 | Final
extension | 72°C, 5 min |
 | Table IV.PCR primer sequences and
pyrosequencing primer sequences. |
Table IV.
PCR primer sequences and
pyrosequencing primer sequences.
| Name | Sequence,
5′-3′ |
|---|
| FOXP3 F1-2 |
ATTTTTGTGGTGAGGGGAAGAAATTA (Biotin) |
| FOXP3 R1 |
AACCCCAAACCTCTCTCTTCTAATAATCCA |
| FOXP3 Seq1 |
CTCTCTCTTCTAATAATCCAA |
| FOXP3 F2 |
AAATTTGGATTATTAGAAGAGAGAGG |
| FOXP3 R2 |
AACTAACAAAAAAAAATCAACCTAACTTAT
(Biotin) |
| FOXP3 Seq2 |
AGAAGAGAGAGGTTTG |
| FOXP3 F3 |
GGATGTTTTTGGGATATAGATTATGTTT (Biotin) |
| FOXP3 R3 |
ACCTATAAAATAAAATATCTACCCTCTTCT |
| FOXP3 Seq3 |
CCTCTTCTCTTCCTC |
| FOXP3 F4 |
GTTTGTTGTAGGATAGGGTAGT (Biotin) |
| FOXP3 Seq4 |
CCTATTATCACAACCCC |
Statistical analysis
The SPSS 19.0 software (IBM Corp.) was used for the
statistically analysis of the data between the asthma and control
group. The measurement data are expressed as the mean ± standard
deviation. If the two group variances were homogeneous, Student's
t-test was used for comparisons. If the variance was uneven, the
Welch's t-test was used. The correlation between FOXP3 mRNA levels
and FEV1 was analysed by Pearson's linear correlation analysis.
P<0.05 was considered to indicate a statistically significant
difference.
Results
Percentages of peripheral blood
CD4+CD25+FOXP3+ Tregs in
CD4+ T cells in asthma and healthy control groups
The percentage of
CD4+CD25+FOXP3+ Tregs was
significantly lower in the asthma group compared with the healthy
control group (3.75±2.99 vs. 14.01±9.89, respectively; P<0.001;
Fig. 2).
Lung function (FEV1%) is lower in
children with asthma
The percentage change in FEV1 was significantly
lower in the asthma group compared with the healthy control group
(80.32±9.12 vs. 96.40±4.63%, respectively; P<0.001; Table V).
 | Table V.Change in FEV1. |
Table V.
Change in FEV1.
| Group | n | FEV1, % | t | P-value |
|---|
| Asthma | 15 | 80.32±9.12 | −6.091 | P<0.001 |
| Control | 15 | 96.40±4.63 |
|
|
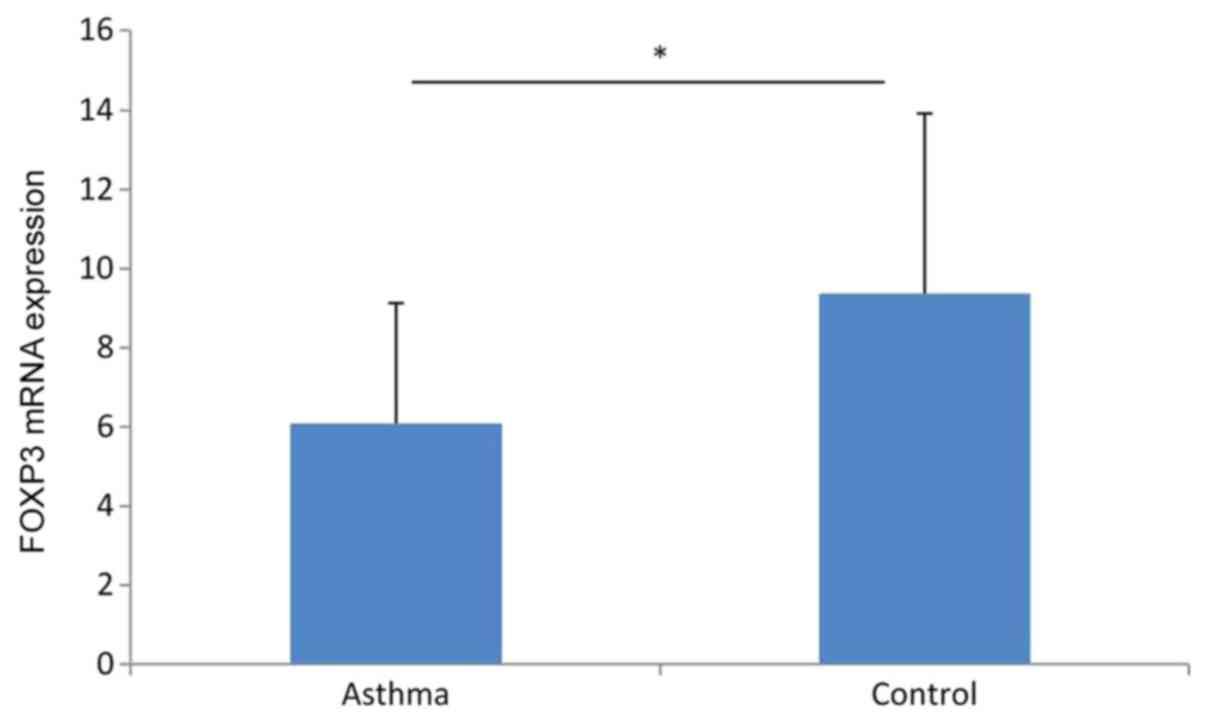
FOXP3 mRNA expression level is lower
in the peripheral blood of children with asthma
The FOXP3 mRNA expression level in the peripheral
blood was 6.09±3.04 in the asthma group compared with 9.38±4.54 in
the healthy control group. The difference between the two groups
was statistically significant (P<0.05; Fig. 3).
Correlation between peripheral blood
FOXP3 mRNA levels and FEV1 in children with asthma
A correlation analysis demonstrated that FOXP3 mRNA
level in the peripheral blood of children with asthma was
positively correlated with FEV1, (P<0.001; r=0.895; Fig. 4).
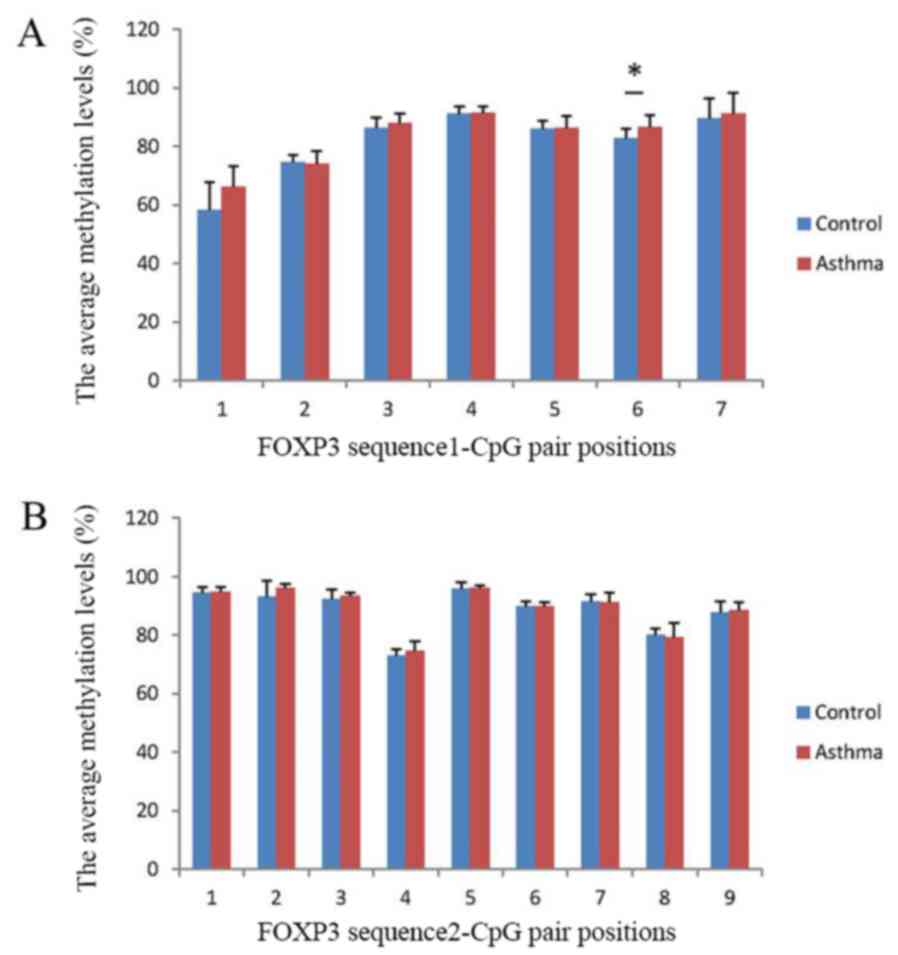
Detection of FOXP3 gene sequence
methylation levels
The average percentage methylation levels of each
FOXP3 CpG loci were compared between the asthma and control groups.
Overall, the average percentage methylation levels of 12 of the 16
FOXP3 CpG loci studied had the tendency to be higher in the asthma
group compared with the control group. However, the methylation
level at only CpG site 6 in exon 1 of sequence 1 was significantly
higher in the asthma group compared with the control group
(P<0.05; Fig. 5).
Discussion
The human immune system maintains a complex balance
of self-defence and tolerance. Tregs differentiate in the human
thymus and express CD4, CD25 and FOXP3, of which
CD4+CD25+ Tregs are a specific subgroup of T
cells that exist mainly in the peripheral blood and spleen of
normal individuals. CD4+CD25+ Tregs have
unique immunomodulatory functions, and FOXP3 is highly expressed in
the cytoplasm. CD4+CD25+ Tregs can be divided
into nTregs and induced CD4+CD25+ Tregs
(iTregs), according to their origin and mechanism of action.
Commonly, CD4+CD25+FOXP3+ Tregs
are referred to as nTregs (21).
Tregs have anti-inflammatory functions and maintain
autoimmune tolerance. The transcription factor FOXP3 can regulate
Treg differentiation, and FOXP3 serves as a key regulator of their
formation and function. FOXP3 expression can be regulated by
multiple epidermal susceptibility enhancers and promoters (22) and is associated with the occurrence
of various immune diseases. Patients with autoimmune hepatitis have
significantly decreased number of
CD3+CD4+CD25+FOXP3+
Tregs and FOXP3 mRNA expression (23). Lower proportions of
CD4+CD25+FOXP3+ Tregs have also
been found in CD4+ T cells harvested from mice with
asthma (24). The percentage of
CD4+CD25+FOXP3+ Tregs in the
peripheral blood was significantly lower in children with allergic
asthma compared with healthy controls (4). Furthermore, the expression of
CD4+CD25+ Tregs and FOXP3 mRNA was lower in
the acute asthma attack group compared with the asthma remission
and normal control groups, whereas the increase in
CD4+CD25+ Tregs and FOXP3 mRNA expression in
the asthma remission group was similar to the control group
(25). Thus, low FOXP3 expression
and insufficient Treg cell function in target cells can lead to the
occurrence of asthma.
Asthma can be both inherited and affected by
environmental factors. Environmental exposure can induce DNA
methylations (26), and hereditary
epigenetic markers may play an important role in the pathogenesis
of asthma (27,28). In order to explore the molecular
mechanism of asthma in the present study, children with asthma and
healthy controls were selected to compare the expression and
methylation status of FOXP3 and for correlation analysis between
FOXP3 mRNA expression and asthma severity. FCM and RT-qPCR were
used to detect the percentage of CD4+ T cells and FOXP3
mRNA expression in PBMCs of children with asthma. The number of
CD4+CD25+FOXP3+ Tregs and the
percentage of CD4+CD25+FOXP3+
Tregs were significantly lower in the asthma group compared with
the healthy control group; these results were consistent with those
of previous studies (4,24).
Children over five years of age were selected for
the present study. The results showed that lung function (FEV1) was
significantly lower in children with asthma than in healthy
controls. FEV1 is the volume of maximal exhalation in the first
second after a maximal deep inhale. The clinical measurement of
FEV1 is often used to judge the severity of asthma. Since
expiratory dyspnoea is most common in patients with asthma, FEV1
levels may be decreased. Some data showed that the Tregs and FOXP3
mRNA levels were decreased in patients with asthma, and that the
FOXP3 mRNA expression levels were positively correlated with FEV1
(29); these results were consistent
with the findings of the present study. The FOXP3 mRNA levels and
FEV1 were positively correlated in the asthma group. Lower FOXP3
mRNA expression was correlated with a more significant decrease in
FEV1 in the same period, which suggested that the FOXP3 gene had an
antagonistic protective effect on lung function injury (30).
The CpG methylation statuses of the FOXP3 gene in
Tregs were compared between children with asthma and healthy
children, which revealed a tendency of hypermethylation in children
with asthma. The methylation degree at the sixth CpG site of exon 1
was significantly higher in children with asthma than that of
healthy children. Epigenetics, including DNA methylation, histone
acetylation, chromatin recombination and nucleosome remodelling, is
a popular topic in modern life sciences. Epigenetic mechanisms have
been shown to regulate many genes, including those involved in
inflammation and immune responses, and to ensure the stability of
phenotypic inheritance and cell differentiation (31). DNA methylation has attracted
increasing attention in the field of epigenetics, and it is of
interest that a previous study has reported that DNA methylation
serves an important role in the development of asthma (32). Changes in DNA methylation can inhibit
gene expression and lead to the differentiation and reactivity of T
cells, where the hypermethylation of DNA CpG frequently leads to
gene silencing and the overall reduction in gene expression
(33). CpG methylation in specific
DNA regions controls the expression of various key transcription
factors, thus contributing to the differentiation of helper T cells
(34). Moreover, DNA CpG
hypermethylation usually results in gene silencing and overall
decrease in gene expression (33),
which can cause Tregs to differentiate into more Th2-type cells
(35). Studies have found that the
interleukin (IL)-4, IL-13 and runt-related transcription factor 3
genes were hypomethylated in patients with asthma (36), whereas the FOXP3 and IL-10 genes were
hypermethylated (26). The
methylation levels of the FOXP3 gene in children predisposed to
risk factors of asthma and/or early, short wheezing were
significantly increased (37). The
methylation of CpG island in the FOXP3 gene could inhibit the
decrease in mRNA expression, affecting DNA binding to transcription
factors and the transcription of proteins, thus weakening the
immunosuppressive effects of CD4+CD25+Tregs
(38). The epigenetic mechanisms
responsible for regulating FOXP3 expression were the key components
of Treg suppressive activity. FOXP3 hypermethylation could impair
the differentiation and function of Tregs, thus increasing the
incidence of asthma and the severity of the disease (38).
The present study had a number of limitations, and
further improvements are required. In the future, larger sample
size for the correlation analysis between the expression and
methylation levels of FOXP3 is required.
In conclusion, the present study demonstrated
decreased proportion of
CD4+CD25+FOXP3+ Tregs and
decreased expression of FOXP3 mRNA, accompanied by increased
methylation level of FOXP3 in children with asthma. Moreover, a
positive correlation was observed between FOXP3 mRNA expression
level and the change in FEV1 in children with asthma. Thus, the
epigenetic modification of FOXP3 could regulate the distribution of
CD4+CD25+FOXP3+ Tregs and affect
the expression of FOXP3. The decreased number of
CD4+CD25+FOXP3+ Tregs and the low
expression and hypermethylation of FOXP3 in the peripheral blood
may be associated with the risk of developing asthma, and impact
the pathogenesis and severity of asthma in children. Therefore,
data from the present study suggest that the upregulation of FOXP3
expression, by suppressing its methylation, can potentially have
immunosuppressive effect in asthma. Further epigenetic studies can
provide new scientific evidence to aid and improve the clinical
diagnosis and treatment of childhood asthma.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Technology
and Science Foundation of Jiangxi Province (grant no.
20142BEG70104).
Availability of data and materials
All data generated or analysed during this study are
included in this published article.
Authors' contributions
XHZ was responsible for project design, clinical
data collection, RT-qPCR, statistical analyses and wrote the
article. QC was involved in the design of the study. ZQL, LL and YZ
performed FCM analysis and pulmonary function test. WH, ZL and DL
performed RT-qPCR and DNA methylation assays. All authors have read
and approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Medical Ethics
Committee of Jiangxi Children's Hospital (approval no. 2015006).
All human materials were obtained with informed consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Global Initiative for Asthma (GINA):
Global Strategy for Asthma Management and Prevention. GINA, 2019.
http://www.ginasthma.org
|
|
2
|
Bergmann KC: Bronchial asthma-many types,
different therapies. Dtsch Med Wochenschr. 141:687–692.
2016.PubMed/NCBI
|
|
3
|
Deng Y, Chen W, Zang N, Li S, Luo Y, Ni K,
Wang L, Xie X, Liu W, Yang X, et al: The antiasthma effect of
neonatal BCG vaccination does not depend on the Th17/Th1 but
IL-17/IFN-γ balance in a BALB/c mouse asthma model. J Clin Immunol.
31:419–429. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Agarwal A, Singh M, Chatterjee BP, Chauhan
A and Chakraborti A: Interplay of T Helper 17 cells with
CD4(+)CD25(high) FOXP3(+) tregs in regulation of allergic asthma in
pediatric patients. Int J Pediatr. 2014:6362382014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kim HJ, Lee HJ, Jeong SJ, Lee HJ, Kim SH
and Park EJ: Cortex Mori Radicis extract exerts antiasthmatic
effects via enhancement of CD4(+)CD25(+)Foxp3(+) regulatory T cells
and inhibition of Th2 cytokines in a mouse asthma model. J
Ethnopharmacol. 138:40–46. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Swamy RS, Reshamwala N, Hunter T,
Vissamsetti S, Santos CB, Baroody FM, Hwang PH, Hoyte EG, Garcia MA
and Nadeau KC: Epigenetic modifications and improved regulatory
T-cell function in subjects undergoing dual sublingual
immunotherapy. J Allergy Clin Immunol. 130:215–224.e7. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Yeh H, Moore DJ, Markmann JF and Kim JI:
Mechanisms of regulatory T cell counter-regulation by innate
immunity. Transplant Rev (Orlando). 27:61–64. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Janson PC, Winerdal ME, Marits P, Thorn M,
Ohlsson R and Winqvist O: FOXP3 promoter demethylation reveals the
committed Treg population in humans. PLoS One. 3:e16122008.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Walker LS: Treg and CTLA-4: Two
intertwining pathways to immune tolerance. J Autoimmun. 45:49–57.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Khan U and Ghazanfar H: T Lymphocytes and
Autoimmunity. Int Rev Cell Mol Biol. 341:125–168. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ray A, Khare A, Krishnamoorthy N, Qi Z and
Ray P: Regulatory T cells in many flavors control asthma. Mucosal
Immunol. 3:216–229. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Sakaguchi S, Yamaguchi T, Nomura T and Ono
M: Regulatory T cells and immune tolerance. Cell. 133:775–787.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Beyer M and Schultze JL: Plasticity of
T(reg) cells: Is reprogramming of T(reg) cells possible in the
presence of FOXP3? Int Immunopharmacol. 11:555–560. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kuo CH, Hsieh CC, Lee MS, Chang KT, Kuo HF
and Hung CH: Epigenetic regulation in allergic diseases and related
studies. Asia Pac Allergy. 4:14–18. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Morikawa H and Sakaguchi S: Genetic and
epigenetic basis of Treg cell development and function: From a
FoxP3-centered view to an epigenome-defined view of natural Treg
cells. Immunol Rev. 259:192–205. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Liu Y, Peng B, Wu S and Xu N: Epigenetic
regulation of regulatory T cells in Kidney disease and
transplantation. Curr Gene Ther. 17:461–468. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wieczorek G, Asemissen A, Model F,
Turbachova I, Floess S, Liebenberg V, Baron U, Stauch D, Kotsch K,
Pratschke J, et al: Quantitative DNA methylation analysis of FOXP3
as a new method for counting regulatory T cells in peripheral blood
and solid tissue. Cancer Res. 69:599–608. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Yang J, Yuan X, Lv C, Bai R, Zhang L,
Ruang L, Zhang C and Quan XQ: Methylation of the FOXP3 upstream
enhancer as a clinical indicator of defective regulatory T cells in
patients with acute coronary syndrome. Am J Transl Res.
8:5298–5308. 2016.PubMed/NCBI
|
|
19
|
CMA RG: Guidelines for diagnosis and
prevention of bronchial Asthma in Children. Chin J Pediat.
54:167–181. 2016.
|
|
20
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Nishimoto T and Kuwana M: CD4+CD25+Foxp3+
regulatory T cells in the pathophysiology of immune
thrombocytopenia. Semin Hematol. 50 (Suppl):S43–S49. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Spreafico R, Rossetti M, van den Broek T,
Jansen NJ, Zhang H, Moshref M, Prakken B, van Loosdregt J, van Wijk
F and Albani S: A sensitive protocol for FOXP3 epigenetic analysis
in scarce human samples. Eur J Immunol. 44:3141–3143. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Liang M, Liwen Z, Yun Z, Yanbo D and
Jianping C: The imbalance between Foxp3+Tregs and
Th1/Th17/Th22 cells in patients with Newly diagnosed autoimmune
hepatitis. J Immunol Res. 2018:37530812018. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Li Y, Fan XS, Yu JH, Xu L and Wang SS:
CD4+CD25+FOXP3+ T cells, Foxp3
gene and protein expression contribute to antiasthmatic effects of
San'ao decoction in mice model of asthma. Phytomedicine.
21:656–662. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Paust S, Lu L, McCarty N and Cantor H:
Engagement of B7 on effector T cells by regulatory T cells prevents
autoimmune disease. Proc Natl Acad Sci USA. 101:10398–10403. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Prunicki M, Stell L, Dinakarpandian D, de
Planell-Saguer M, Lucas RW, Hammond SK, Balmes JR, Zhou X, Paglino
T, Sabatti C, et al: Exposure to NO2, CO, and
PM2.5 is linked to regional DNA methylation differences
in asthma. Clin Epigenetics. 10:22018. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Yang IV and Schwartz DA: Epigenetic
mechanisms and the development of asthma. J Allergy Clin Immunol.
130:1243–1255. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Marques CR, Costa RS, Costa GNO, da Silva
TM, Teixeira TO, de Andrade EMM, Galvão AA, Carneiro VL and
Figueiredo CA: Genetic and epigenetic studies of FOXP3 in asthma
and allergy. Asthma Res Pract. 1:102015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Provoost S, Maes T, van Durme YM, Gevaert
P, Bachert C, Schmidt-Weber CB, Brusselle GG, Joos GF and Tournoy
KG: Decreased FOXP3 protein expression in patients with asthma.
Allergy. 64:1539–1546. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Lu Y, Malmhall C, Sjöstrand M, Rådinger M,
O'Neil SE, Lötvall J and Bossios A: Expansion of CD4(+) CD25(+) and
CD25(−) T-Bet, GATA-3, Foxp3 and RORγt cells in allergic
inflammation, local lung distribution and chemokine gene
expression. PLoS One. 6:e198892011. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Suárez-Álvarez B, Baragaño Raneros A,
Ortega F and Lòpez-Larrea C: Epigenetic modulation of the immune
function: A potential target for tolerance. Epigenetics. 8:694–702.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wysocki K, Conley Y and Wenzel S:
Epigenome variation in severe asthma. Biol Res Nurs. 17:263–269.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Gaffin JM, Raby BA, Petty CR, Hoffman EB,
Baccarelli AA, Gold DR and Phipatanakul W: β-2 adrenergic receptor
gene methylation is associated with decreased asthma severity in
Inner-City Schoolchildren: Asthma and rhinitis. Clin Exp Allergy.
44:681–689. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Polansky JK, Schreiber L, Thelemann C,
Ludwig L, Krüger M, Baumgrass R, Cording S, Floess S, Hamann A and
Huehn J: Methylation matters: Binding of Ets-1 to the demethylated
Foxp3 gene contributes to the stabilization of Foxp3 expression in
regulatory T cells. J Mol Med (Berl). 88:1029–1040. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Zhao M, Liang GP, Tang MN, Luo SY, Zhang
J, Cheng WJ, Chan TM and Lu QJ: Total glucosides of paeony induces
regulatory CD4(+)CD25(+) T cells by increasing Foxp3 demethylation
in lupus CD4(+) T cells. Clin Immunol. 143:180–187. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Yang IV, Pedersen BS, Liu A, O'Connor GT,
Teach SJ, Kattan M, Misiak RT, Gruchalla R, Steinbach SF, Szefler
SJ, et al: DNA methylation and childhood asthma in the inner city.
J Allergy Clin Immunol. 136:69–80. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Brunst KJ, Leung YK, Ryan PH, Khurana
Hershey GK, Levin L, Ji H, Lemasters GK and Ho SM: Forkhead box
protein 3 (FOXP3) hypermethylation is associated with diesel
exhaust exposure and risk for childhood asthma. J Allergy Clin
Immunol. 131:592–594.e1-3. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Nadeau K, McDonald-Hyman C, Noth EM, Pratt
B, Hammond SK, Balmes J and Tager I: Ambient air pollution impairs
regulatory T-cell function in asthma. J Allergy Clin Immunol.
126:845–852.e10. 2010. View Article : Google Scholar : PubMed/NCBI
|