Introduction
Diabetes mellitus is a chronic disease caused by
metabolic disorders and is frequently accompanied by numerous
complications (1,2); it has become a serious threat to public
health. One of the common complications is gastrointestinal
dysfunction, which usually presents as gastric/colonic dysmotility
(3-5).
However, the underlying molecular mechanisms, which may provide
novel and practical therapeutic strategies, remain largely elusive.
It is known that gastrointestinal motility results from the
coordinated contractions of smooth muscle cells (SMCs) (6). A study by Sun et al (7) has indicated that insulin-like growth
factor-1 (IGF-1) may prevent apoptosis of colonic SMCs and
alleviate colonic dysmotility in diabetic rats. This previous study
suggests a key role of IGF-1 in colonic dysmotility. However, the
upstream regulatory mechanisms of IGF-1 in colonic SMCs and colonic
dysmotility remain to be explored.
microRNAs (miRNAs/miRs) are a group of endogenous,
small non-coding RNAs, which usually have a length of ~22
nucleotides and regulate gene expression at the
post-transcriptional level (8,9). In
general, miRNAs function by binding to the 3'-untranslated regions
(3'-UTRs) of target mRNAs, leading to translational repression or
mRNA degradation. Of note, miRNAs regulate >60% of mammalian
protein-coding genes (10-12).
Therefore, miRNAs are involved in almost all cellular processes,
including proliferation, differentiation and apoptosis (9). Furthermore, miRNAs have pivotal roles
in physiology and pathology (13-16).
miR-155 is one of the miRNAs that is known to regulate
physiological and pathological processes. For instance, miR-155 has
been identified as a tumor-suppressive miRNA in colon cancer
through targeting collagen triple helix repeat containing 1 or
forkhead box O3 (17,18). In addition, miR-155 is able to
mediate endothelial progenitor cell dysfunction caused by high
glucose through targeting patched-1(19) and has been reported to regulate the
inflammatory response in the colonic mucosa (20). However, the role of miR-155 in
colonic SMCs and colonic dysmotility has remained elusive.
In the present study, miR-155 was identified to
directly target IGF-1 to promote apoptosis of colonic SMCs.
Furthermore, miR-155 was identified to aggravate colonic
dysmotility in diabetic mice through targeting IGF-1.
Materials and methods
Cells
Mouse colonic SMCs were purchased from Rochen Pharma
Co., Ltd. (cat. no. RC-RM-0052) and cultured in Dulbecco's modified
Eagle's medium (Thermo Fisher Scientific, Inc.) supplemented with
15% fetal bovine serum (Thermo Fisher Scientific, Inc.) at 37˚C
with 5% CO2.
Protein extraction and western blot
analysis
The colonic tissue samples were frozen in liquid
nitrogen, ground into powder, lysed using radioimmunoprecipitation
assay lysis buffer (Thermo Fisher Scientific, Inc.) containing the
protease inhibitor cocktail (Thermo Fisher Scientific, Inc.) and
incubated on ice for 30 min. Tissue homogenates and cell lysates
were then centrifuged for 10 min at 12,000 x g and 4˚C and the
protein concentration of the supernatant was determined with the
Pierce BCA protein assay kit (Thermo Fisher Scientific, Inc.). The
protein was separated by 15% SDS-PAGE and then transferred onto
Immobilon nitrocellulose membranes (EMD Millipore). Subsequently,
the membranes were blocked in 5% milk for 1 h at room temperature,
and then incubated with the indicated primary antibodies (1:1,000)
at 4˚C overnight. The antibodies were as follows: IGF-1 (cat. no.
ab9572), Caspase-3 (cat. no. ab13847) and GAPDH (cat. no. ab181602)
antibodies were purchased from Abcam. The membranes were then
incubated with the secondary antibody goat anti-rabbit IgG H&L
(HRP) (cat. no. ab97051) for 1 h at room temperature. GAPDH served
as a loading control and protein bands were quantified using ImageJ
software 1.52a (National Institutes of Health).
RNA isolation and reverse
transcription-quantitative (RT-q)PCR
Total RNA was isolated from tissues or cultured
cells using TRIzol reagent (Thermo Fisher Scientific, Inc.) as
described previously and RNA was reverse transcribed to
complementary (c)DNA from 1 µg total RNA by using AMV reverse
transcriptase (Takara Bio Inc.) and a RT primer according to the
manufacturer's protocol. The reaction conditions were as follows:
16˚C for 30 min, 42˚C for 30 min and 85˚C for 5 min. qPCR was
performed by using a Taqman PCR kit on an Applied Biosystems 7300
sequence detection system (Applied Biosystems; Thermo Fisher
Scientific, Inc.), with U6 as the internal control. Taqman probe of
miR-155 (cat. no. A25576) and U6 (cat. no. 4427975) were purchased
from Thermo Fisher Scientific, Inc. The reactions were performed in
a 96-well plate at 95˚C for 10 min, followed by 40 cycles of 95˚C
for 10 sec and 60˚C for 1 min.
To measure the level of IGF-1 mRNA, RNA was
reverse-transcribed to cDNA from 1 µg total RNA using AMV reverse
transcriptase and oligo dT (Takara Bio Inc.). The reaction
conditions were as follows: 42˚C for 60 min and 70˚C for 10 min.
qPCR was performed using SYBR Premix Ex Taq (Takara Bio Inc.) and
the corresponding primers (Nanjing Synthgene Medical Technology
Co., Ltd) on an ABI 7300 sequence detection system (Applied
Biosystems; Thermo Fisher Scientific, Inc.). The reactions were
performed in a 96-well plate at 95˚C for 5 min, followed by 40
cycles of 95˚C for 30 sec, 60˚C for 30 sec and 72˚C for 30 sec.
GAPDH served as the internal control. The PCR primers were used as
follows: IGF-1 sense, 5'-CGTCACCGGGACATTGAGTATTAC-3' and
anti-sense, 5'-AATGCATGGTTAAACCGATGCAAG-3'; GAPDH sense,
5'-GATATTGTTGACATCAATGAC-3' and anti-sense,
5'-TTGATTTTGGAGGGATCTCG-3'.
Overexpression and knockdown of
miR-155
miR-155 mimics (forward,
5'-UUAAUGCUAAUCGUGAUAGGGGU-3' and reverse,
5'-CUCCUACAUAUUAGCAUUAACA-3'); control mimics (forward,
5'-UUCUCCGAACGUGUCACGU-3', and reverse,
(5'-ACGUGACACGUUCGGAGAA-3'); anti-miR-155
(5'-ACCCCUAUCACGAUUAGCAUUAA-3') and anti-miR control
(5'-ACGUGACACGUUCGGAGAA-3') were purchased from Nanjing
Synthgene Medical Technology Co., Ltd. Cells were seeded in
6-well plates at a density of 2x105 per well and
transfected using Lipofectamine 2000 (Thermo Fisher Scientific,
Inc.) with a concentration of 50 nM mimics or inhibitors. on the
following day according to the manufacturer's protocol. Total RNA
and protein were extracted after transfection for 24 and 48 h,
respectively.
The prediction of IGF-1 as a direct target of
miR-155: miRWalk (http://www.umm.uni-heidelberg.de/apps/zmf/mirwalk/)
was used as a tool to predict the direct target of miR-155.
Luciferase reporter assay
The entire 3'-UTR of IGF-1 was inserted into a
luciferase reporter plasmid (Nanjing Synthgene Medical Technology
Co., Ltd.). To examine the binding specificity, the sequences that
interacted with miR-155 were mutated and the mutant IGF-1 3'-UTR
was inserted into an equivalent luciferase reporter plasmid. For
the luciferase reporter assay, cells were seeded in 24-well plates
and each well was transfected with 1 µg luciferase reporter
plasmid, 1 µg β-galactosidase plasmid (internal control) and 100
pmol miR-155 mimics, control mimics, anti-miR-155 or anti-miR
control using Lipofectamine 2000 (Thermo Fisher Scientific, Inc.).
After 48 h, luciferase signals were measured using a luciferase
assay kit (cat. no. E1500) according to the manufacturer's protocol
(Promega Corp.) and β-galactosidase activity was measured using a
microplate reader (Thermo Fisher Scientific, Inc.) at 420 nm, using
a β-galactosidase Assay kit (cat. no. RG0036) according to the
manufacturer's protocol (Beyotime Institute of Biotechnology).
Luciferase activities were normalized to β-galactosidase activities
for each well.
Cell viability assay
Cells were seeded in 96-well plates at a density of
5,000 cells per well. After incubation overnight, cells were
transfected with miR-155 mimics, control mimics, anti-miR-155,
anti-miR control, IGF-1 plasmid or control plasmid using
Lipofectamine 2000 (Thermo Fisher Scientific, Inc.) according to
the manufacturer's protocol. At 2 days after transfection, the cell
viability was then determined using an MTT assay according to the
manufacturer's protocol. The purple MTT-formazan crystals was
dissolved by adding 200 µl dimethyl sulfoxide (Amresco, LLC) to
wells, and the absorbance was record at 490 nm.
Cell apoptosis assay
Cells were seeded in 12-well plates and transfected
with miR-155 mimics, control mimics, anti-miR-155, anti-miR
control, IGF-1 plasmid or control plasmid using Lipofectamine 2000
(Thermo Fisher Scientific, Inc.) according to the manufacturer's
protocol. After 48 h of transfection, the attached and floating
cells were harvested and re-suspended in binding buffer (100 mM
HEPES, pH 7.4, 100 mM NaCl and 25 mM CaCl2) after a wash
with cold PBS. Subsequently, the cells were stained with Annexin
V-FITC/propidium iodide (PI) for 15 min at room temperature in the
dark. Apoptosis was measured using a flow cytometer (FACScalibur;
BD Biosciences) using an Annexin V-FITC/PI staining kit (BD
Biosciences) according to the manufacturer's protocol. Data were
analyzed using FlowJo 7.6 Software (FlowJo LLC).
Animal experiment and histological
analysis
Male BALB/c mice (age, 6 weeks) were purchased from
the Model Animal Research Center of Nanjing University (Nanjing,
China). The mice were maintained, in four mice per cage, at
21-23˚C, with a humidity of 40-60%. The cage was maintained in a 12
h light dark cycle, with chow and water provided ad libitum.
All animal protocols were approved by the Animal Care and Use
Committee of Jiangsu Taizhou People's Hospital. Mice were
administered streptozotocin (40 mg/kg) by a single tail-vein
injection to induce diabetes (7). A
blood glucose meter was used to measure the fasting blood glucose
levels. After 3 days, those mice with blood glucose levels of
>16.7 mmol/l were selected as diabetic mice. The diabetic mice
were treated with a control adenovirus, miR-155 adenovirus, IGF-1
adenovirus or miR-155 adenovirus plus IGF-1 adenovirus via tail
vein injection. The amount of adenovirus was 1011
transforming units. n=15 in each group. After 6 weeks, mice were
sacrificed and colonic smooth muscle tissues were dissected from
the distal and proximal portion of the colon for further analysis.
A total of three tissue samples of the same portion of the colonic
smooth muscle tissues were subjected to H&E staining by Nanjing
Synthgene Medical Technology Co., Ltd. The IGF-1 and Caspase-3
expression levels in the tissues were detected by western blot
analysis.
Statistical analysis
All experiments were performed at least three times.
HE staining was performed three times, and other experiments were
performed six times, as indicating in the figure legends. Values
are expressed as the mean ± standard error of the mean. Statistical
significance was determined using a Student's t-test or a one-way
analysis of variance with post hoc Fisher's least significant
difference test. P<0.05 was considered to indicate a
statistically significant difference.
Results
Prediction of IGF-1 as a target of
miR-155
First, to identify a potential upstream regulator of
IGF-1, miRWalk (21) was used to
search miRNAs that may regulate IGF-1. miR-155, which was
previously reported as a tumor-suppressive miRNA in colon cancer
(17,18) and a regulator of the inflammatory
response in colonic mucosa (20),
was then selected as the candidate miRNA that may target IGF-1 in
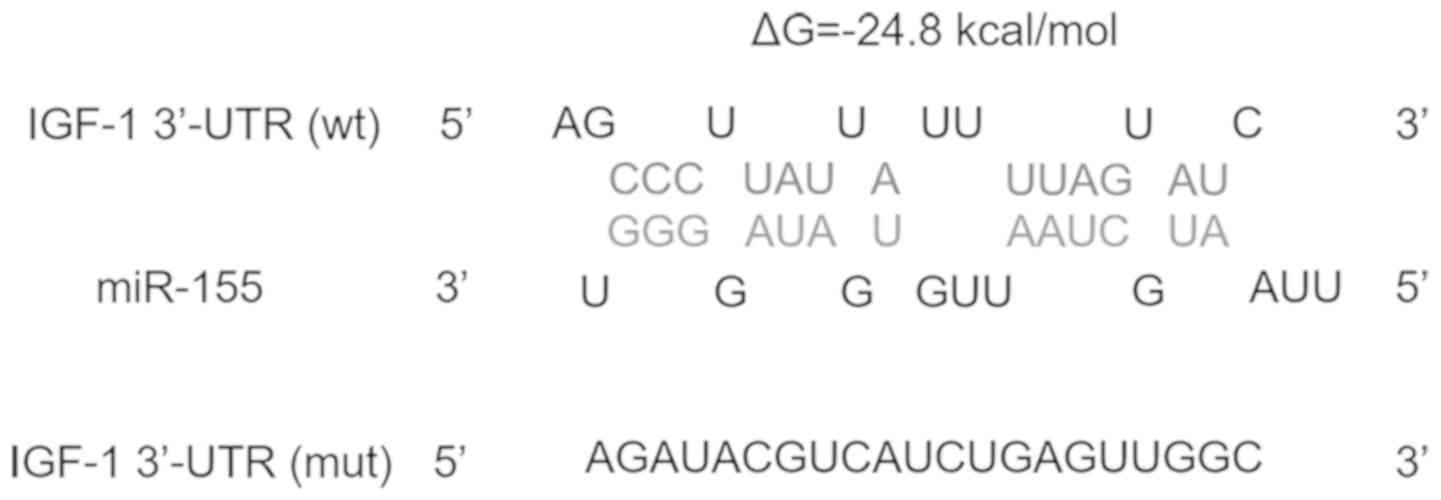
colonic SMCs. Fig. 1 presents the
potential binding between miR-155 and IGF-1. The minimum free
energy value of the hybridization was -24.8 kcal/mol, which is
within the range of genuine miRNA-target interactions.
Validation of IGF-1 as a direct target
of miR-155
Next, to validate whether IGF-1 is a direct target
gene of miR-155, the expression IGF-1 was examined in mouse colonic
SMCs after over-expression or knockdown of miR-155. Overexpression
of miR-155 was achieved by transfection with miR-155 mimics and
knock-down of miR-155 was realized through transfecting cells with
anti-miR-155, which is a chemically synthesized anti-sense
oligonucleotide designed to target mature miR-155. The
over-expression and knockdown efficiencies were confirmed by
measuring the expression levels of miR-155 in colonic SMCs by
RT-qPCR. As presented in Fig. 2A, a
~98-fold increase in miR-155 levels was detected when cells were
transfected with miR-155 mimics, while a ~60% decrease in miR-155
levels was achieved when cells were transfected with anti-miR-155.
The mRNA and protein levels of IGF-1 in these cells were then
determined by RT-qPCR and western blot analysis, respectively. As
indicated in Fig. 2B, the mRNA
levels of IGF-1 remained consistent after over-expression or
knockdown of miR-155. However, the protein levels of IGF-1 were
significantly repressed upon overexpression of miR-155, whereas
knockdown of miR-155 led to significantly increased IGF-1 protein
levels (P<0.05; Fig. 2C and
D). These results suggest that
miR-155 negatively regulates IGF-1 expression at the
post-transcriptional level.
To determine whether the negative regulatory
function of miR-155 on IGF-1 was achieved through the binding of
miR-155 to the predicted recognition site in the 3'-UTR of IGF-1
(Fig. 1), the full-length 3'-UTR of
IGF-1 was inserted into the 3'-UTR of a firefly luciferase plasmid.
The resulting plasmid was then transfected into colonic SMCs and
miR-155 mimics or anti-miR-155 were co-transfected. Measurement of
the luciferase activity in these groups of cells provided clues on
whether miR-155 binds to the 3'-UTR of IGF-1. As presented in
Fig. 3A, overexpression of miR-155
significantly reduced the luciferase signal, while knockdown of
miR-155 significantly increased it (P<0.05). Furthermore,
mutations were introduced into the binding site in the 3'-UTR of
IGF-1 (Fig. 1) and this mutated
3'-UTR was inserted into the reporter vector downstream of the
luciferase gene. The above experiments were re-performed with the
mutant plasmid, revealing that neither overexpression nor knockdown
of miR-155 affected the luciferase signals (Fig. 3B). These results suggest that miR-155
directly binds to the 3'-UTR of IGF-1 to inhibit the translation of
IGF-1 in colonic SMCs.
Role of miR-155 in the apoptosis of
colonic SMCs
Since IGF-1 is able to inhibit the apoptosis of
colonic SMCs (7), it was
hypothesized that miR-155, which negatively regulates IGF-1,
promotes the apoptosis of colonic SMCs through targeting IGF-1. To
test this hypothesis, the viability and apoptosis of colonic SMCs
were evaluated after overexpression or knockdown of miR-155. As
expected, overexpression of miR-155 led to significantly decreased
cell viability (P<0.05; Fig. 4A)
and increased apoptosis (Fig. 4B and
C). By contrast, knockdown of
miR-155 induced the opposite effects (Fig. 4A-C). Furthermore, to confirm that
miR-155 promotes apoptosis via targeting IGF-1, an IGF-1
over-expression plasmid that expresses the full-length open reading
frame of IGF-1 but without the 3'-UTR was constructed and
co-transfected with miR-155 mimics into colonic SMCs. In comparison
with cells transfected with control mimics or control plasmid,
cells overexpressing IGF-1 exhibited significantly increased cell
viability and significantly decreased apoptosis (P<0.05;
Fig. 4D-F), which is consistent with
a previous study (7). However,
co-transfection of miR-155 mimics with IGF-1 overexpression plasmid
led to the reversal of the effect on cell viability and apoptotic
rate (Fig. 4D-F). Taken together,
these results indicate that miR-155 promotes apoptosis of colonic
SMCs through targeting IGF-1.
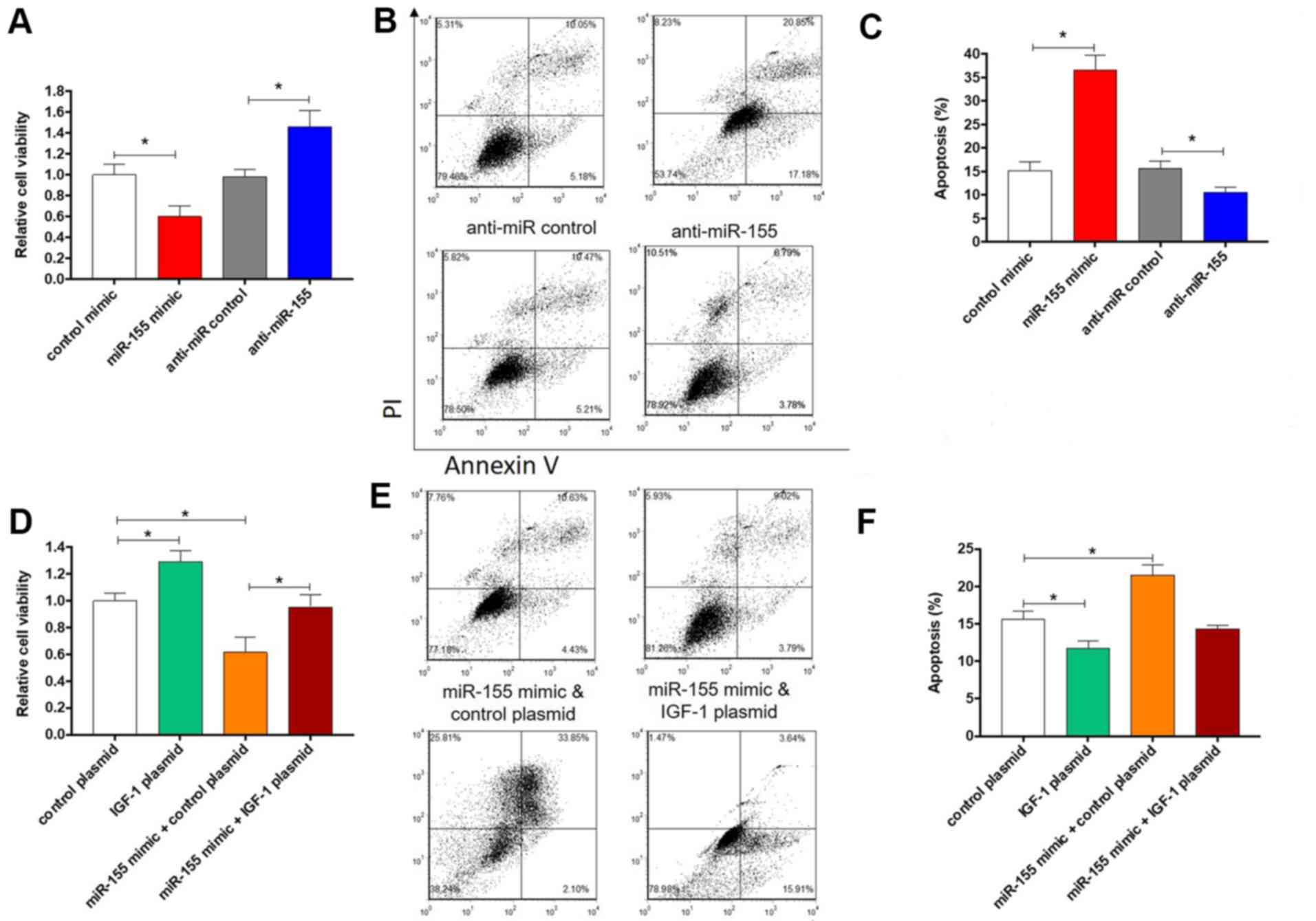 | Figure 4.Effect of miR-155 and IGF-1 on colonic
smooth muscle cells apoptosis. Relative cell viability and
apoptotic rates of colonic smooth muscle cells that were
transfected with (A-C) control mimics, miR-155 mimics, anti-miR
control, anti-miR-155 or (D-F) control plasmid, IGF-1 plasmid,
miR-155 mimics plus control plasmid or miR-155 mimics plus IGF-1
plasmid. (A and D) Cell viability. (B and E) Representative images
and (C and F) quantitative analysis of apoptosis. The columns
represent the mean ± standard error of the mean (n=6).
*P<0.05, one-way analysis of variance with post hoc
Fisher's least significant difference test. IGF, insulin-like
growth factor; UTR, untranslated region; miR, microRNA; PI,
propidium iodide. |
Role of miR-155 in colonic dysmotility
in diabetic mice
Given the evidence that IGF-1 can alleviate colonic
dysmotility, it was then investigated whether miR-155 aggravates
colonic dysmotility. Mice were first treated with streptozotocin to
induce diabetes and colonic dysmotility according to a previous
study (7). The expression levels of
miR-155 in colonic muscle tissues of mice with or without colonic
dysmotility were compared, revealing that miR-155 was significantly
upregulated in colonic muscle tissues of mice with colonic
dysmotility (P<0.001; Fig. 5E).
To overexpress miR-155 or IGF-1, the corresponding adenovirus
expression vectors were constructed. The diabetic mice were then
treated with control adenovirus, miR-155 adenovirus, IGF-1
adenovirus or miR-155 adenovirus plus IGF-1 adenovirus via tail
vein injection. The mice were sacrificed after 6 weeks and the
colonic smooth muscle tissues were dissected, followed by H&E
staining, RT-qPCR analysis of miR-155 levels or western blot
analysis of IGF-1 protein levels. The H&E staining results are
presented in Fig. 5A-D. In comparison with the control, the
thickness of colonic smooth muscle tissues was increased in
diabetic mice treated with IGF-1 adenovirus, which is consistent
with the result of a previous study (7). By contrast, treatment of miR-155
adenovirus led to a decrease in the thickness of colonic smooth
muscle tissues and co-treatment with IGF-1 adenovirus had rescuing
effects (Fig. 5A-D). This result
suggests that miR-155 promotes atrophy of colonic smooth muscle
tissue in diabetic mice. To further validate this result, total RNA
and protein in the isolated colonic smooth muscle tissues were
extracted for RT-qPCR and western blot analysis. As presented in
Fig. 6A, the levels of miR-155 in
the colonic smooth muscle tissues of mice treated with miR-155
adenovirus were significantly increased (P<0.05). However, the
protein levels of IGF-1 were decreased in the colonic smooth muscle
tissues of mice treated with miR-155 adenovirus, but were
upregulated in the colonic smooth muscle tissues of mice treated
with IGF-1 adenovirus (Fig. 6B and
C). The expression of Caspase-3 was
also assessed (Fig. 6B), validating
the function of miR-155 and IGF-1 in colonic dysmotility. Taken
together, these results further demonstrate that miR-155 aggravates
colonic dysmotility through targeting IGF-1.
Discussion
Gastrointestinal dysmotility is a complication of
diabetes and is also accompanied with aging; it has become a
serious threat to public health. Elucidation of the molecular
mechanisms underlying gastrointestinal dysmotility is therefore
urgently required. In general, gastrointestinal motility is
provided by coordinated contractions of SMCs. Identification of the
key molecules and associated molecular events of SMC contraction is
thus essential, but the current knowledge remains insufficient. In
a previous study, IGF-1 was reported to inhibit the apoptosis of
colonic SMCs and alleviate colonic dysmotility (7), suggesting the important role of IGF-1
in colonic dysmotility. However, the upstream regulatory mechanisms
of IGF-1 have remained elusive.
Certain miRNAs are considered to have key roles in
gastrointestinal smooth muscle fibrosis and dysfunction. For
instance, miR-143 and miR-145 were identified to be upregulated and
involved in smooth muscle contractile dysfunction (22). miR-29b was reported to be
downregulated and associated with gastrointestinal fibrosis
(23). However, RNAs associated with
colonic dysmotility as a complication of diabetes remains to be
investigated in detail. In the present study, miR-155 was
identified as an upstream regulator of IGF-1. The role of miR-155
in other diseases has been elucidated in considerable detail. For
instance, miR-155 was previously demonstrated to induce functional
impairment of vascular SMCs through downregulating soluble guanylyl
cyclase (24). miR-155 was also
reported to inhibit the proliferation of tumor cells via targeting
cyclic AMP responsive element binding protein 1(25) and to suppress the proliferation of
fibroblasts during cardiac injury (26). The oncogenic functions of miR-155 in
clear-cell renal cell carcinoma (27), hepatocellular carcinoma (28) and non-small cell lung cancer
(29) have also been demonstrated.
Among colonic diseases, miR-155 was identified as a
tumor-suppressive miRNA in colon cancer (17,18) and
an important regulator of the inflammatory response in colonic
mucosa (20). However, whether
miR-155 participates in colonic dysmotility has remained elusive.
In the present study, miR-155 was demonstrated to negatively
regulate IGF-1 expression, which is achieved through direct binding
of miR-155 to the 3'-UTR of IGF-1. It was also revealed that
miR-155 targets IGF-1 to promote the apoptosis of colonic SMCs and
aggravate colonic dysmotility. Taken together, the present results
suggest a promoting effect of miR-155 in colonic dysmotility.
In conclusion, miR-155 was identified as a direct
negative regulator of IGF-1, which promoted the apoptosis of
colonic SMCs and aggravated colonic dysmotility in diabetic mice.
These results may provide a novel therapeutic target that is
miR-155 for the treatment of colonic dysmotility.
Acknowledgements
Not applicable.
Funding
The present study was supported by a grant from the
National Natural Science Foundation of China (grant no.
81670490).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
XS and ZZ performed the experiment. BY designed the
research. XS and ZZ analyzed the data. XS and BY wrote the
manuscript.
Ethics approval and consent to
participate
All animal protocols were approved by The Animal
Care and Use Committee of Jiangsu Taizhou People's Hospital.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Nathan DM, Cleary PA, Backlund JY, Genuth
SM, Lachin JM, Orchard TJ, Raskin P and Zinman B: DiabetesControl
and Complications Trial/Epidemiology of Diabetes Interventions and
Complications (DCCT/EDIC) Study Research Group: Intensive diabetes
treatment and cardiovascular disease in patients with type 1
diabetes. N Engl J Med. 353:2643–2653. 2005.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Severino P, D'Amato A, Netti L, Pucci M,
De Marchis M, Palmirotta R, Volterrani M, Mancone M and Fedele F:
Diabetes mellitus and ischemic heart disease: The role of ion
channels. Int J Mol Sci. 19(E802)2018.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Bytzer P, Talley NJ, Leemon M, Young LJ,
Jones MP and Horowitz M: Prevalence of gastrointestinal symptoms
associated with diabetes mellitus: A population-based survey of
15,000 adults. Arch Intern Med. 161:1989–1996. 2001.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Samsom M, Vermeijden JR, Smout AJ, Van
Doorn E, Roelofs J, Van Dam PS, Martens EP, Eelkman-Rooda SJ and
Van Berge-Henegouwen GP: Prevalence of delayed gastric emptying in
diabetic patients and relationship to dyspeptic symptoms: A
prospective study in unselected diabetic patients. Diabetes Care.
26:3116–3122. 2003.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Rodrigues ML and Motta ME: Mechanisms and
factors associated with gastrointestinal symptoms in patients with
diabetes mellitus. J Pediatr (Rio J). 88:17–24. 2012.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Sanders KM, Koh SD, Ro S and Ward SM:
Regulation of gastrointestinal motility-insights from smooth muscle
biology. Nat Rev Gastroenterol Hepatol. 9:633–645. 2012.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Sun M, Wang F and Feng P: Insulin-like
growth factor-1 inhibits colonic smooth muscle cell apoptosis in
diabetic rats with colonic dysmotility. Regul Pept. 194-195:41–48.
2014.PubMed/NCBI View Article : Google Scholar
|
|
8
|
He L and Hannon GJ: MicroRNAs: Small RNAs
with a big role in gene regulation. Nat Rev Genet. 5:522–531.
2004.PubMed/NCBI View
Article : Google Scholar
|
|
9
|
Ambros V: The functions of animal
microRNAs. Nature. 431:350–355. 2004.PubMed/NCBI View Article : Google Scholar
|
|
10
|
John B, Enright AJ, Aravin A, Tuschl T,
Sander C and Marks DS: Human MicroRNA targets. PLoS Biol.
2(e363)2004.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Bartel DP: MicroRNAs: Target recognition
and regulatory functions. Cell. 136:215–233. 2009.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Friedman RC, Farh KK, Burge CB and Bartel
DP: Most mammalian mRNAs are conserved targets of microRNAs. Genome
Res. 19:92–105. 2009.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Lujambio A and Lowe SW: The microcosmos of
cancer. Nature. 482:347–355. 2012.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Esquela-Kerscher A and Slack FJ:
Oncomirs-microRNAs with a role in cancer. Nat Rev Cancer.
6:259–269. 2006.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Miao S, Mao X, Pei R, Song K, Lv Y, Jiang
H, Li B, Yang X, Xiu C, Meng H and Sun J: MiR-448 inhibits
laryngeal cancer metastasis by repressing AEG-1 expression. Int J
Clin Exp Med. 11:1587–1596. 2018.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Cao M, Bai L, Wang D, Zhai Q, Li Y, Hai J
and Wang W: miRNA-33 expression and its mechanism in patients and
model rats with type 2 diabetic nephropathy. Int J Clin Exp Med.
11:1661–1668. 2018.
|
|
17
|
Gao Y, Liu Z, Ding Z, Hou S, Li J and
Jiang K: MicroRNA-155 increases colon cancer chemoresistance to
cisplatin by targeting forkhead box O3. Oncol Lett. 15:4781–4788.
2018.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Liu J, Chen Z, Xiang J and Gu X:
MicroRNA-155 acts as a tumor suppressor in colorectal cancer by
targeting CTHRC1 in vitro. Oncol Lett. 15:5561–5568.
2018.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Gao J, Zhao G, Li W, Zhang J, Che Y, Song
M, Gao S, Zeng B and Wang Y: MiR-155 targets PTCH1 to mediate
endothelial progenitor cell dysfunction caused by high glucose. Exp
Cell Res. 366:55–62. 2018.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Pathak S, Grillo AR, Scarpa M, Brun P,
D'Incà R, Nai L, Banerjee A, Cavallo D, Barzon L, Palù G, et al:
MiR-155 modulates the inflammatory phenotype of intestinal
myofibroblasts by targeting SOCS1 in ulcerative colitis. Exp Mol
Med. 47(e164)2015.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Dweep H and Gretz N: miRWalk2.0: a
comprehensive atlas of microRNA-target interactions. Nat Methods.
12(697)2015.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Dahan D, Ekman M, Larsson-Callerfelt AK,
Turczyńska K, Boettger T, Braun T, Swärd K and Albinsson S:
Induction of angiotensin-converting enzyme after miR-143/145
deletion is critical for impaired smooth muscle contractility. Am J
Physiol Cell Physiol. 307:C1093–C1101. 2014.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Nijhuis A, Biancheri P, Lewis A, Bishop
CL, Giuffrida P, Chan C, Feakins R, Poulsom R, Di Sabatino A,
Corazza GR, et al: In Crohn's disease fibrosis-reduced expression
of the miR-29 family enhances collagen expression in intestinal
fibroblasts. Clin Sci (Lond). 127:341–350. 2014.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Park M, Choi S, Kim S, Kim J, Lee DK, Park
W, Kim T, Jung J, Hwang JY, Won MH, et al: NF-kB-responsive miR-155
induces functional impairment of vascular smooth muscle cells by
downregulating soluble guanylyl cyclase. Exp Mol Med.
51(17)2019.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Zhao XS, Han B, Zhao JX, Tao N and Dong
CY: MiR-155-5p affects Wilms' tumor cell proliferation and
apoptosis via targeting CREB1. Eur Rev Med Pharmacol Sci.
23:1030–1037. 2019.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Wang C, Zhang C, Liu L, A X, Chen B, Li Y
and Du J: Macrophage-derived mir-155-containing exosomes suppress
fibroblast proliferation and promote fibroblast inflammation during
cardiac injury. Mol Ther. 25:192–204. 2017.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Ji H, Tian D, Zhang B, Zhang Y, Yan D and
Wu S: Overexpression of miR-155 in clear-cell renal cell carcinoma
and its oncogenic effect through targeting FOXO3a. Exp Ther Med.
13:2286–2292. 2017.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Li DP, Fan J, Wu YJ, Xie YF, Zha JM and
Zhou XM: MiR-155 up-regulated by TGF-β promotes
epithelial-mesenchymal transition, invasion and metastasis of human
hepatocellular carcinoma cells in vitro. Am J Transl Res.
9:2956–2965. 2017.PubMed/NCBI
|
|
29
|
Liu F, Song D, Wu Y, Liu X, Zhu J and Tang
Y: MiR-155 inhibits proliferation and invasion by directly
targeting PDCD4 in non-small cell lung cancer. Thorac Cancer.
8:613–619. 2017.PubMed/NCBI View Article : Google Scholar
|