Introduction
Lung cancer is one of the most rapidly growing types
of cancer that is associated with a high rate of morbidity
worldwide (1,2). Non-small cell lung cancer (NSCLC)
constitutes ~80% of lung cancer cases, inclusive of adenocarcinomas
as well as squamous cell carcinomas (1). The prognosis is poor owing to
inefficient therapeutic approaches during the early and advanced
stages of the disease, as well as the frequency of relapses, making
the 5-year survival rate <15% (3). As such, it is necessary to investigate
the mechanisms of action behind the pathogenesis of NSCLC to
identify new therapeutic approaches.
MicroRNAs (miRNAs/miRs) are a set of small,
non-coding RNAs that are 20-24 nucleotides in length and are highly
evolutionarily conserved (4). miRNAs
bind to the 3'-untranslated regions (UTRs) of target mRNAs to
either degrade the target, or prevent further expression (5). Alterations in miRNA expression levels
are linked to the initiation and development of several types of
cancer (6-10).
While the involvement of many miRNAs has been reported in the
development of NSCLC (11,12), the use of these molecules for therapy
is limited (13), with their exact
functions and mechanisms of action in tumorigenesis remaining
unclear. For example, miR-449 shows atypical levels in many cancer
types with variations between different tumors. miR-449 plays an
important role in gynecological clear cell carcinoma (14), hepatocellular cancer (15), papillary thyroid carcinoma (16) and breast cancer (17). Despite this evidence, the mechanism
of action in NSCLC remains to be established.
The current study reported decreased levels of
miR-449b-3p in NSCLC tumor tissues compared to adjacent normal
tissues, with a negative association with tumor stage and
metastasis to lymph nodes. miR-449b-3p overexpression caused the
inhibition of migration, metastasis and NSCLC cell
epithelial-mesenchymal transition (EMT) in vitro.
Additionally, the targeting of interleukin (IL)-6 mRNA, as well as
downregulation of the Janus kinase 2 (JAK2)/STAT3 pathway by
miR-449b-3p, was shown in the cell lines. This work highlighted the
ability of miR-449b-3p to suppress NSCLC and identified one of its
molecular targets as IL-6.
Materials and methods
Patients and tissue details
NSCLC tumor tissue and neighboring adjacent normal
tissue (61 pairs) were collected from surgical lung resection
samples, subject to patients presenting clinicopathological
characteristics of NSCLC, with no prior radiotherapy or
chemotherapy. Samples were collected from the lesion edges, which
were confirmed by a minimum of two experienced pathologists, at The
First Affiliated Hospital of Guangxi University of Traditional
Chinese Medicine between January 2011 and December 2013. Liquid
nitrogen was used to freeze the samples, followed by -80˚C storage
for the analysis. The ethics committee of the First Affiliated
Hospital of Guangxi University of Traditional Chinese Medicine
(Nanning, China) issued approval of the collection of patient
samples. The ethics approval number is GKG-1407014.
Cell culture and transfection
The NSCLC cell lines A549, H1299, CALU-1, H520 and
H1703, in addition to the 293T and Beas-2b normal human bronchial
epithelial cell lines, were supplied by The Cell Bank of Type
Culture Collection of the Chinese Academy of Sciences. RPMI-1640
(Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% FBS
(Gibco; Thermo Fisher Scientific, Inc.), 100 µg/ml streptomycin and
100 U/ml penicillin (Gibco; Thermo Fisher Scientific, Inc.) was
used for cell-culture. Unless otherwise stated, cells were
incubated at 37˚C and 5% CO2 in a humidified atmosphere.
miR-449b-3p mimics, miR-449b-3p inhibitors and the corresponding
miR-control (scrambled/non-coding control) were purchased from
Sigma-Aldrich; Merck KGaA. The sequences were as follows:
miR-449b-3p mimic, 5'-CAGCCACAACUACCCUGCCACU-3'; miR-449b-3p mimic
control, 5'-UUCUCCGAACGUGUCACGUTT-3'; miR-449b-3p inhibitor,
5'-ACAAAGGACAGUCUGCUCCUG-3'; and miR-449b-3p inhibitor control,
5'-CAGUACUUUUGUGUAGUACAA-3'.
To study the roles of miR-449b-3p in the progression
of NSCLC in vitro, A549 and H1299 cells (2x103
cells/ml) were seeded into a 96-well plates and transfected using
Lipofectamine® 2000 (Thermo Fisher Scientific, Inc.)
with the miR-449b-3p mimic (50 nM), miR-449b-3p inhibitor (50 nM)
or corresponding controls (50 nM) for 48 h, according to
manufacturer's protocols. In addition, an overexpression vector
encoding IL-6 (termed IL-6; Shanghai GenePharma Co., Ltd.) and
corresponding control vector (termed pcDNA-NC; Shanghai GenePharma
Co., Ltd.) was transfected into A549 cells according to
manufacturer's protocol. A co-transfection group was also created
for A549 cells, which were co-transfected with IL-6 (100 nM) and
miR-449b-3p mimic (50 nM; termed IL-6 + miR-449b-3p mimic).
Subsequent experimentation was performed at 48 h after
transfection.
Reverse transcription-quantitative PCR
(RT-qPCR)
TRIzol® reagent from Invitrogen; Thermo
Fisher Scientific, Inc. was employed for isolating total RNA from
samples, in accordance with the manufacturer's protocols. Total RNA
was converted into first-strand cDNA using a Mir-X™
miRNA First-Strand Synthesis kit (Takara Bio, Inc.) for miRNAs or
PrimeScript™ RT Reagent kit (Takara Bio, Inc.) for mRNAs
per the prescribed protocols of the manufacturer. The temperature
conditions for reverse transcription were as follows: 37˚C for 15
min followed by 85˚C for 5 sec. Subsequent qPCR was performed using
SYBR® Premix Ex Taq II (Takara Bio, Inc.) and a ViiA 7
real-time PCR system from Applied Biosystems; Thermo Fisher
Scientific, Inc. in accordance with the prescribed protocols of the
manufacturer. The levels of miR-449b-3p were normalized to U6,
whereas the levels of IL-6 were normalized to that of β-actin and
quantified using the 2-∆∆Cq method (18). U6 or β-actin was measured as an
internal control. The thermocycling conditions were as follows:
Initial denaturation at 95˚C for 10 min, followed by 40 cycles of
95˚C for 20 sec and 60˚C for 45 sec. The RT-qPCR primers were as
follows: miR-449b-3p forward, 5'-CCTGGACGGCACGAAA-3' and reverse,
from the aforementioned kit; U6 forward, 5'-CTCGCTTCGGCAGCACA-3'
and reverse, 5'-AACGCTTCACGAATTTGCGT-3'; IL-6 forward,
5'-CCTAATTAGGCATCGGAA-3' and reverse, 5'-ACGACACAGCGAACACAACGTC-3';
and β-actin forward, 5'-TGACGTGGACATCCGCAAAG-3' and reverse,
5'-CTGGAAGGTGGACAGCGAGG-3'.
In vitro cell assays for migration and
invasion
The ability of cells to migrate and invade was
assessed using Transwell assays and wound healing assays. The
Transwell migration assay involved placing 2x104 cells
in RPMI-1640 (Gibco; Thermo Fisher Scientific, Inc.) medium without
serum into the upper chamber of a well with an 8-µm pore insert
(Corning, Inc.). The Transwell invasion assay involved seeding
4x104 cells in medium without serum into the upper
chamber, precoated with Matrigel® (Corning, Inc.) for 48
h at 37˚C. For both the Transwell migration and invasion assays,
the lower chamber contained medium with 20% FBS. Following 24 h
incubation at 37˚C, cotton swabs were used to remove non-migrating
cells. Fixation of the cells on the lower membrane that had
migrated or invaded was performed using 4% paraformaldehyde for 30
min at 25˚C followed by crystal violet staining for 30 min at 25˚C.
Invading and migrating cells were counted from 10 randomly selected
fields using a light microscope (magnification, x40; Olympus
Corporation).
Western blotting
RIPA lysis buffer (Beyotime Institute of
Biotechnology) containing protease inhibitor was used to extract
the total proteins from samples for 20 min on ice. After the
measurement of protein concentration using Bradford's reagent
(Beyotime Institute of Biotechnology), Total protein (25 µg) was
separated by 10% SDS-PAGE was run to resolve samples, followed by
transfer onto PVDF membranes. Non-fat milk (5%) in TBST was used
for blocking for 60 min at room temperature, followed by overnight
incubation with primary antibodies at 4˚C. The following antibodies
were used: E-cadherin (1:1,000; cat. no. ab40772; Abcam),
N-cadherin (1:1,000; cat. no. ab18203; Abcam), vimentin (1:1,000;
cat. no. ab92547; Abcam), β-actin (1:5,000; cat. no. 4970T; Cell
Signaling Technologies, Inc.), IL-6 (1:1,000; cat. no. 12912T; Cell
Signaling Technology, Inc.), JAK2 (1:1,000; cat. no. 3230T; Cell
Signaling Technologies, Inc.), phospho-JAK2 (1:1,000; cat. no.
3771T; Cell Signaling Technologies, Inc.), phospho-STAT3 (1:1,000;
cat. no. 9145T; Cell Signaling Technologies, Inc.) and STAT3
(1:1,000; cat. no. MAB1799-SP; Proteintech Group, Inc.). The
secondary antibody used was horseradish peroxidase-conjugated
anti-rabbit (1:10,000; cat. no. ab205718; Abcam) or anti-mouse IgG
(1:10,000; cat. no. HAF007; Proteintech Group, Inc.) for 1 h at
25˚C. Protein expression was normalized to β-actin and were
visualized using an enhanced chemiluminescence system (EMD
Millipore). Relative expression was measured using Bio-Rad Image
Lab software version 5.1 (Bio-Rad Laboratories, Inc.).
Luciferase reporter assay
A pMIR-REPORT luciferase vector (Invitrogen; Thermo
Fisher Scientific, Inc.) was constructed to perform luciferase
reporter assays. PCR amplification of the 3'-UTR fragments of IL-6,
from the RNA samples of 293T cells, containing the potential
miR-449b-3p binding sites was performed, followed by insertion into
the vectors, referred to as the pMIR-IL-6-wild-type (WT)
thereafter. In addition, site-directed mutagenesis of the wild type
binding site was performed using a FAST site-Directed mutagenesis
kit (Tiangen Biotech Co., Ltd.) to generate a vector encoding the
pMIR-IL-6-mutant (MUT). 293T cells (1.5x104) were seeded
into 96-well plates for 24 h at 37˚C followed by co-transfection
with miR-449b-3p (50 nM) or negative control (50 nM) and the
aforementioned constructed vectors (50 nM) using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.). A Dual-Luciferase® reporter assay
(Promega Corporation) was performed to assess the enzyme activity
in accordance with the manufacturer's protocol. Data were
normalized against the activity of the Renilla luciferase
activity.
IL-6 ELISA
A human IL-6 ELISA kit (cat. no. ab46027; Abcam) was
utilized to measure the levels of IL-6 in the normal culture medium
after 48 h of culture of the various cells listed, in accordance
with the manufacturer's protocol.
Bioinformatics analysis
The potential targets of miR-449b-3p were predicted
using the Targetscan database 7.1 (http://www.targetscan.org/vert_72/).
Statistical analysis
SPSS version 25.0 software (IBM Corp.) was employed
for statistical analysis, and data are presented as the mean ± SD.
All experiments were repeated three times. Differences between two
groups were evaluated using Student's t-test. Differences between
multiple groups were evaluated using one-way ANOVA followed by
Tukey's post-hoc comparisons. The association between miR-449b-3p
and IL-6 was assessed using Spearman's rank correlation while
patient survival curves (Kaplan-Meier) were assessed using the
log-rank test. The χ2 test was used to analyse the
relationships between miR-449b-3p expression and the
clinicopathological features of patients with NSCLC. P<0.05 was
considered to indicate a statistically significant difference.
Results
Decreased miR-449b-3p in NSCLC samples
and the inverse association with prognosis
RT-qPCR was performed to analyze the miR-449b-3p
levels in NSCLC samples compared with adjacent normal tissues. It
was shown that there were decreased miRNA levels in tumor samples
(73.8% or 45/61 samples) compared to the adjacent normal tissues
(Fig. 1A), which in turn exhibited
approximately double the mRNA expression levels of the tumor
tissues in terms of the overall expression data (Fig. 1B). The level of miR-449b-3p was less
in patients with advanced tumor-node-metastasis (TNM) stage or
those with metastasis to the lymph nodes (Fig. 1C and D). Clinicopathological data were analyzed
after dividing the patients into two groups according to the median
miR-449b-3p expression levels. The expression levels of miR-449b-3p
in the NSCLC tissue were inversely associated with the presence of
lymphatic metastasis (P=0.014) and an advanced TNM stage (P=0.021)
(Table I). Furthermore, the overall
survival and disease-free survival rates were significantly reduced
in patients with low levels of miR-449b-3p compared with the
patients with increased levels (Fig.
1E and F). Overall, a noteworthy
decline in miR-449b-3p was observed in NSCLC clinical samples,
suggesting miR-449b-3p has a potential function as an independent
marker of the prognosis of the disease.
 | Table IAssociation between miR-449b-3p levels
and non-small cell lung cancer patient clinicopathological
findings. |
Table I
Association between miR-449b-3p levels
and non-small cell lung cancer patient clinicopathological
findings.
| | | miR-449b-3p
expression | |
|---|
| Variable | n | High level, n=29 | Low level, n=32 | P-value |
|---|
| Sex | | | | 0.075 |
|
Male | 51 | 22 | 29 | |
|
Female | 11 | 8 | 3 | |
| Age, years | | | | 0.553 |
|
<60 | 39 | 20 | 19 | |
|
>60 | 23 | 10 | 13 | |
| Smoking | | | | 0.851 |
|
Yes | 40 | 19 | 21 | |
|
No | 22 | 11 | 11 | |
| Pathological
type | | | | 0.779 |
|
Adenocarcinoma | 28 | 13 | 15 | |
|
Squamous
carcinoma | 34 | 17 | 17 | |
| Tumor
differentiation | | | | 0.286 |
|
Well +
Moderate | 49 | 22 | 27 | |
|
Poor | 13 | 8 | 5 | |
| Tumor size, cm | | | | 0.830 |
|
<3 | 51 | 25 | 26 | |
|
>3 | 11 | 5 | 6 | |
| Lymphatic
metastasis | | | | 0.014 |
|
Yes | 22 | 6 | 16 | |
|
No | 40 | 24 | 16 | |
| TNM
classification | | | | 0.021 |
|
I-II | 22 | 15 | 7 | |
|
III-IV | 40 | 15 | 25 | |
Inhibition of migration, metastasis
and EMT by microRNA-331-3p
The functions of miR-449b-3p in NSCLC metastasis was
investigated on account of the association between the miRNA levels
and clinicopathological data, such as metastasis. miRNA levels in
the aforementioned tumor cell lines were compared against the
control Beas-2b epithelial line. The tumor cell lines all displayed
decreased miR-449b-3p expression compared with Beas-2b cells. Of
this data, the two cell lines with the highest and lowest levels of
miR-449b-3p were used for subsequent analysis: A549 and H1299 with
the minimum and maximum levels, respectively (Fig. 2A). Lentivirus-based assays were
performed to elevate or reduce miR-449b-3p levels in these two
lines, respectively. The effective change in expression levels were
confirmed by RT-qPCR (Fig. 2B and
C). Transwell assays examining the
cell invasion and migration revealed a decreased number of A549
cells that could migrate and invade with raised miR-449b-3p
expression levels (Fig. 2D). The
opposite response was observed in H1299 cells following knockdown
of miR-449b-3p. Transwell assays revealed increased cell numbers
that could migrate and invade with H1299 transfected with
anti-miR-449b-3p (Fig. 2E). The
transfected A549 and H1299 cells were assessed for markers of EMT.
Interestingly, miR-449b-3p overexpression in A549 cells
significantly increased E-cadherin (a marker for epithelial tissue)
expression whilst reducing those of N-cadherin and vimentin
(markers of mesenchymal lineage). By contrast, the opposite trend
was observed in the H1299 cells following knockdown of miR-449b-3p
(Fig. 2F). In conclusion,
miR-449b-3p treatment appeared to suppress EMT of these two NSCLC
lines to reduce their migratory and invasive capabilities.
 | Figure 2.miR-449b-3p decreases the ability of
NSCLC lines to migrate, invade and undergo epithelial-mesenchymal
transition. (A) Detection of miR-449b-3p expression levels in the
five NSCLC and control epithelial cell lines using reverse
transcription-quantitative PCR, as analyzed using one-way ANOVA
with Tukey's post-hoc comparisons. miR-449b-3p expression levels in
(B) A549 and (C) H1299 cells transfected with miR-449b-3p or
anti-miR-449b-3p, respectively. (D) Illustrative images and
quantification of transwell assays of A549 transfected with
miR-449b-3p. Scale bars, 50 µm. (E) Illustrative images and
quantification of transwell assays of H1299 transfected with
anti-miR-449b-3p. Scale bars represent 50 µm. (F) Western blot of
E-cadherin, N-cadherin and vimentin in the aforementioned
transfected cell lines. Right panel: Relative protein levels of
E-cadherin, N-cadherin and vimentin. *P<0.05,
**P<0.01 and ***P<0.001 vs. respective
control. miR, microRNA; NC, negative control; NSCLC, non-small cell
lung cancer. |
miR-449b-3p directly targets IL-6 in
NSCLC
The potential miRNA targets were predicted using the
Targetscan database (http://www.targetscan.org/vert_72/), which revealed
that IL-6 was a target (Fig. 3A).
This was followed by the construction and establishment of
pMIR-IL-6-WT and pMIR-IL-6-MUT (Fig.
3A). 293T cells were co-transfected with miR-449b-3p and these
constructs. Luciferase activity was decreased in the
miR-449b-3p-transfected cells compared with those that received
miR-NC. This was not observed in the pMIR-IL-6-MUT-transfected
cells (Fig. 3B). Thus, the binding
of miR-449b-3p to the 3'-UTR of IL-6 was confirmed. Additionally, a
significant decrease in IL-6 mRNA and protein expression levels was
shown by RT-qPCR and western blotting, respectively, in A549 cells
(high levels of miR-449b-3p) compared with the NC. Similarly, H1299
cells with miR-449b-3p-knockdown displayed augmented levels of IL-6
mRNA and protein expression levels compared with the NC virus group
(Fig. 3C and D). The ELISA also showed similar results,
in that the concentration of IL-6 was significantly inhibited in
the A549 cells, but significantly increased in the H1299 cells
(Fig. 3E). Subsequently, RT-qPCR was
used to examine the IL-6 mRNA expression levels in NSCLC tumor
samples. An inverse correlation between IL-6 and miR-449b-3p was
revealed (r=-0.417, P<0.001; Fig.
3F). Taken together, this data indicated the direct binding of
miR-449b-3p with IL-6.
 | Figure 3.miR-449b-3p directly targets IL-6. (A)
IL-6 WT or MUT vector construction scheme. Red letters represent
bases mutated. (B) miR-449b-3p binding to IL-6, as shown by the
luciferase reporter assay. Data were analyzed using Student's
t-tests. (C) IL-6 mRNA of A549 and H1299 cells transfected with
miR-449b-3p and anti-miR-449b-3p, respectively, assessed using
reverse transcription-quantitative PCR. (D) IL-6 protein expression
levels of A549 and H1299 transfected with miR-449b-3p and
anti-miR-449b-3p, respectively, were assessed using western blot
assays. Right panel: Relative protein expression levels of IL-6.
(E) Quantification of IL-6 in the culture medium of A549 and H1299
cells transfected with miR-449b-3p and anti-miR-449b-3p,
respectively. Data were analyzed using Student's t-tests. (F)
Clinical specimens showing an inverse correlation between
miR-449b-3p and IL-6 in NSCLC samples. Data were assessed using
Spearman's rank correlation analysis. ***P<0.001. IL,
interleukin; miR, microRNA; MUT, mutant; NC, negative control; NS,
not significant; UTR, untranslated region; WT, wild-type. |
miR-449b-3p suppresses EMT through the
IL-6/JAK2/STAT3 signaling pathway
As IL-6 has been identified to be a target of
miR-499b-3p in NSCLC, the mechanism of action behind the
IL-6/JAK2/STAT3 network was further investigated. A549 cells were
transfected with pcDNA-IL-6, which was shown to increase the mRNA
expression levels of IL-6 compared to the negative control
(pcDNA-NC; Fig. 4A). As shown in
Fig. 4B and C, the aforementioned EMT markers and
JAK2/STAT3 circuit activation were inhibited by miR-449b-3p, while
this inhibitory effect on EMT and JAK2/STAT3 circuit activation was
impeded by the restoration of IL-6 in the NSCLC cell lines. To
summarize, the anti-cancer effects of miR-449b-3p (in terms of
invasion and EMT) were shown to be mediated through JAK2/STAT3
signaling, which regulates IL-6 in NSCLC cells.
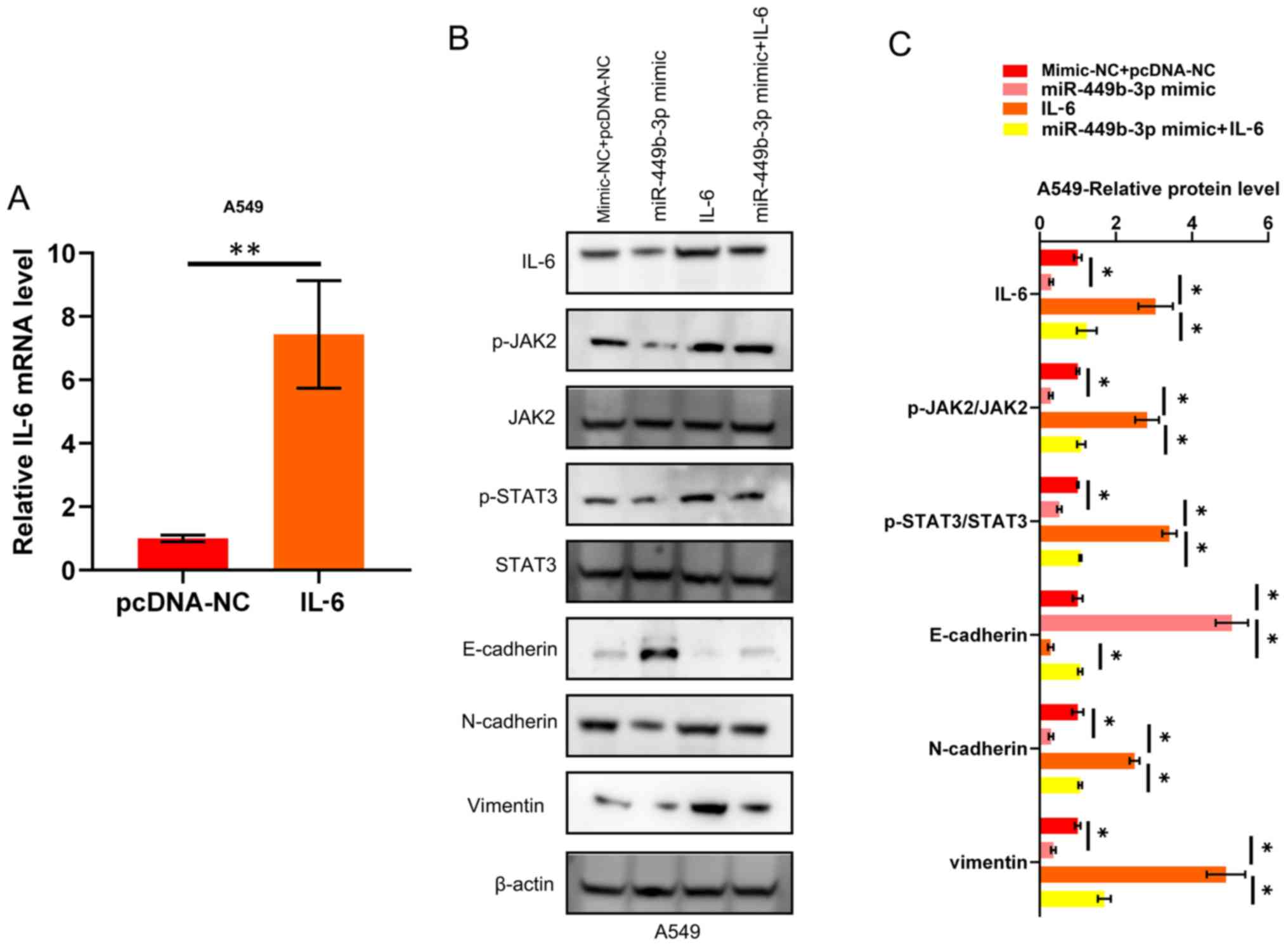 | Figure 4.miR-449b-3p suppresses EMT through the
IL-6/JAK2/STAT3 pathway. (A) IL-6 mRNA expression in A549
transfected with IL-6. Data were analyzed using Student's t-tests.
(B) Western blotting and (C) quantification showing the activation
of JAK2, and STAT3, through the phosphorylation of these proteins,
as well as the suppression of EMT markers, by miR-449b-3p. This
observation was reversed by IL-6 restoration. Analyzed using
one-way ANOVA with Tukey's post-hoc comparisons.
*P<0.05, **P<0.01. EMT,
epithelial-mesenchymal transition; IL, interleukin; JAK, Janus
kinase; miR, microRNA; NC, negative control. |
Discussion
Research is yet to cast a complete light on the
mechanisms behind NSCLC. As research has highlighted the
involvement of miRNAs in the advancement and tumorigenesis of NSCLC
(19), this present study explored
the mechanisms of action of miR-449b-3p in NSCLC. While previous
studies have shown altered levels of miR-449b-3p in several tumors
including NSCLC, the exact working in terms of targets and
signaling remained to be studied (14-17).
This current study corroborated the reduced miR-449b-3p levels in
NSCLC samples, with its levels associated with the clinical TNM
stage/metastasis as well as the overall and disease-free survival.
The ability of the NSCLC cell lines studied in this present study
to migrate, invade and undergo EMT was suppressed by raising the
expression levels of miR-449b-3p. Furthermore, lowering the miRNA
levels boosted the ability of the NSCLC cell lines to migrate and
invade via EMT. This work demonstrates the cancer-inhibitory role
of miR-449b-3p, as well as its role as a potential marker for the
prognosis of NSCLC.
mRNAs may be targeted by miRNAs to decrease their
expression. This holds true for miR-449b-3p, with research
outlining several targets such as hepatocyte growth factor receptor
in hepatocellular carcinoma (15),
proto-oncogene RET in papillary thyroid carcinoma (16) and regulation of nuclear pre-mRNA
domain containing 1B in breast cancer cells (17). In this present study, the results
showed and confirmed that the cytokine IL-6 was directly targeted
and regulated by the miR-449b-3p. The effects of IL-6 are
pleiotropic, controlling immune and tumor functions (20). The JAK-signal transducer and STAT
circuit is activated by IL-6 via the broad spectrum receptor gp130
and the specific IL-6Rα co-receptor (21). This is followed by dimerization and
nuclear translocation of the phosphorylated STATs, leading to the
expression of downstream genes of IL-6(22). While the complete methods of action
of miR-449b-3p in NSCLC are yet to be uncovered, the current
results suggest that the molecule may inhibit the ability of the
studied NSCLC cell lines to migrate and invade through IL-6.
The JAK/STAT pathway is an evolutionarily conserved
signaling pathway that is involved in a number of basic cellular
functions, such as cell growth and metastasis, which lead to the
development and progression of cancer (23,24).
IL-6 stimulation leads to the autophosphorylation and activation of
JAK, which in turn phosphorylates STAT3. STAT3 then forms a
homodimer and translocates into the cell nucleus to promote the
transcription of responsive oncogenes, including c-Myc, Bcl-1,
cyclin D1/D2 and Bcl-xl (25). In
this present study, overexpressing miR-449b-3p was shown to inhibit
the activation of the JAK2/STAT3 signaling pathway, which was
rescued by overexpression of IL-6. Therefore, it was speculated
that miR-449b-3p may exert a tumor suppressor role in NSCLC through
the IL-6/JAK2/STAT3 pathway, which necessitates future
analysis.
To summarize, downregulation of miR-449b-3p in NSCLC
samples was shown in this present study, which correlated with the
survival of patients and the ability of the cancer to metastasize,
as well as to display EMT characteristics, through the
IL-6/JAK2/STAT3 pathway.
Acknowledgements
Not applicable.
Funding
This study was supported by grants from the Chinese
Medicine Science and Technology Special of Guangxi Autonomous
Region (grant no. G2K210-096).
Availability of data and materials
The datasets used and/or analyzed in the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
YY conceived the study and designed the experiments.
KC and HXL provided the experimental materials. KC, HXL, and PPL
performed the experiments. ZJG performed the data analysis. KC
wrote the manuscript. All authors contributed to the interpretation
and discussion of the results and reviewed the manuscript. All
authors read and approved the final manuscript.
Ethics approval and consent to
participate
The ethics committee of the First Affiliated
Hospital of Guangxi University of Traditional Chinese Medicine
(Nanning, China) issued approval of the patient samples. All
patients provided written informed consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2018. CA Cancer J Clin. 68:7–30. 2018.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Heist RS and Engelman JA: SnapShot:
Non-small cell lung cancer. Cancer Cell. 21(448.e2)2012.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297.
2004.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Croce CM: Causes and consequences of
microRNA dysregulation in cancer. Nat Rev Genet. 10:704–714.
2009.PubMed/NCBI View
Article : Google Scholar
|
|
6
|
Wen J, Hu Y, Liu Q, Ling Y, Zhang S, Luo
K, Xie X, Fu J and Yang H: miR-424 coordinates multilayered
regulation of cell cycle progression to promote esophageal squamous
cell carcinoma cell proliferation. EBioMedicine. 37:110–124.
2018.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Zhou YW, Zhang H, Duan CJ, Gao Y, Cheng
YD, He D, Li R and Zhang CF: miR-675-5p enhances tumorigenesis and
metastasis of esophageal squamous cell carcinoma by targeting
REPS2. Oncotarget. 7:30730–30747. 2016.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Chang RM, Xiao S, Lei X, Yang H, Fang F
and Yang LY: miRNA-487a promotes proliferation and metastasis in
hepatocellular carcinoma. Clin Cancer Res. 23:2593–2604.
2017.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Song H, Li D, Wu T, Xie D, Hua K, Hu J,
Deng X, Ji C, Deng Y and Fang L: MicroRNA-301b promotes cell
proliferation and apoptosis resistance in triple-negative breast
cancer by targeting CYLD. BMB Rep. 51:602–607. 2018.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Jiang LH, Zhang HD and Tang JH: MiR-30a: A
novel biomarker and potential therapeutic target for cancer. J
Oncol. 2018(5167829)2018.PubMed/NCBI View Article : Google Scholar
|
|
11
|
He D, Wang J, Zhang C, Shan B, Deng X, Li
B, Zhou Y, Chen W, Hong J, Gao Y, et al: Down-regulation of
miR-675-5p contributes to tumor progression and development by
targeting pro-tumorigenic GPR55 in non-small cell lung cancer. Mol
Cancer. 14(73)2015.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Chen W, Wang J, Liu S, Wang S, Cheng Y,
Zhou W, Duan C and Zhang C: MicroRNA-361-3p suppresses tumor cell
proliferation and metastasis by directly targeting SH2B1 in NSCLC.
J Exp Clin Cancer Res. 35(76)2016.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Iqbal MA, Arora S, Prakasam G, Calin GA
and Syed MA: MicroRNA in lung cancer: Role, mechanisms, pathways
and therapeutic relevance. Mol Aspects Med. 70:3–20.
2019.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Jang SG, Yoo CW, Park SY, Kang S and Kim
HK: Low expression of miR-449 in gynecologic clear cell carcinoma.
Int J Gynecol Cancer. 24:1558–1563. 2014.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Buurman R, Gürlevik E, Schäffer V, Eilers
M, Sandbothe M, Kreipe H, Wilkens L, Schlegelberger B, Kühnel F and
Skawran B: Histone deacetylases activate hepatocyte growth factor
signaling by repressing microRNA-449 in hepatocellular carcinoma
cells. Gastroenterology. 143:811–820.e15. 2012.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Li Z, Huang X, Xu J, Su Q, Zhao J and Ma
J: miR-449 overexpression inhibits papillary thyroid carcinoma cell
growth by targeting RET kinase-β-catenin signaling pathway. Int J
Oncol. 49:1629–1637. 2016.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Jiang J, Yang X, He X, Ma W, Wang J, Zhou
Q, Li M and Yu S: MicroRNA-449b-5p suppresses the growth and
invasion of breast cancer cells via inhibiting CREPT-mediated
Wnt/β-catenin signaling. Chem Biol Interact. 302:74–82.
2019.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expres- sion data using real-time quantitative PCR
and the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Legras A, Pécuchet N, Imbeaud S, Pallier
K, Didelot A, Roussel H, Gibault L, Fabre E, Le Pimpec-Barthes F,
Laurent-Puig P and Blons H: Epithelial-to-mesenchymal transition
and MicroRNAs in lung cancer. Cancers (Basel). 9(pii:
E101)2017.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Mantovani A, Allavena P, Sica A and
Balkwill F: Cancer-related inflammation. Nature. 454:436–44.
2008.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Heinrich PC, Behrmann I, Haan S, Hermanns
HM, Müller-Newen G and Schaper F: Principles of interleukin
(IL)-6-type cytokine signalling and its regulation. Biochem J.
374:1–20. 2003.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Naugler WE and Karin M: The wolf in
sheep's clothing: The role of interleukin-6 in immunity,
inflammation and cancer. Trends Mol Med. 14:109–119.
2008.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Bowman T, Garcia R, Turkson J and Jove R:
STATs in oncogenesis. Oncogene. 19:2474–2488. 2000.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Marotta LL, Almendro V, Marusyk A,
Shipitsin M, Schemme J, Walker SR, Bloushtain-Qimron N, Kim JJ,
Choudhury SA, Maruyama R, et al: The JAK2/STAT3 signaling pathway
is required for growth of CD44(+) CD24(-) stem cell-like breast
cancer cells in human tumors. J Clin Invest. 121:2723–2735.
2011.PubMed/NCBI View
Article : Google Scholar
|
|
25
|
Bromberg J and Darnell JE Jr: The role of
STATs in transcriptional control and their impact on cellular
function. Oncogene. 19:2468–2473. 2000.PubMed/NCBI View Article : Google Scholar
|


















