Introduction
Developmental dysplasia of the hip (DDH), previously
known as congenital hip dislocation, is a frequently disabling
condition characterized by premature arthritis later in life
(1). The term encompasses a spectrum
of diseases ranging from minor acetabular dysplasia to irreducible
dislocation, which affects 25-50 in 1,000 live births among Lapps
and Native Americans but is very rare among southern Chinese and
African populations (1). A number of
factors, including genetic mutations and intrauterine and postnatal
environmental factors, are thought to contribute to the disease
(2).
Genetic influence on DDH has been long known but is
still poorly understood. Although most clinical cases appear to be
sporadic, 12-33% of patients have a family history of DDH (3,4). One
study reported a 10-fold increase in the incidence of DDH among the
parents of index patients and a seven-fold increase among the
siblings compared with the general population (3), which suggested a complex aetiology
involving multiple genes interacting with environmental factors.
Several genes, such as collagen alpla-1 (I) chain, homeobox protein
Hox-B, basement membrane-specific heparan sulfate proteoglycan core
protein, plasma membrane calcium-transporting ATPase 4,
pregnancy-associated plasma protein-A2 (PAPPA2), teneurin-3
(TENM3), growth/differentiation factor 5 (GDF5) and
transforming growth factor beta-1 proprotein have been investigated
in various populations as candidates (5-11).
However, to the best of our knowledge, few results have been
replicated in other populations, suggesting that geographic and
ethnic factors probably also play a key role in disease aetiology.
Therefore, locating and subsequently identifying the causative
genes for DDH are essential. Two identified genes have been found
based on the genetic analysis of large segregating DDH families
after mapping of the causative regions. A variant in the CX3C
chemokine receptor 1 (CX3CR1) gene was found in all
DDH-affected members of a 72-member, four-generation affected
family (12). Watson et al
(13) identified a mutation in the
UFM1 specific peptidase 2 (UFSP2) gene in a family with
Beukes hip dysplasia. Genetic variations in other diseases were
also found after locating causative regions. Shrimpton et al
(14) found that chromosome 2q31 was
associated with congenital vertical talus, which led to the
detection of a mutation in the homeobox protein Hox-D10 gene
using whole-genome linkage analysis. Additionally, the vitamin D
receptor gene was associated with late-onset Alzheimer's disease
after mapping susceptibility loci to chromosome 12q13(15).
In the present study, a genome-wide linkage scan
with the Affymetrix 10K GeneChip was performed on a four-generation
Chinese family, which included 19 healthy members and 5 patients
affected with DDH. For the first time, at least for this pedigree,
the evidence showed that the disease is mapped to two novel regions
at 8q23-q24 and 12p12.
Materials and methods
Patients.
The proband (III-6), a 28-year-old female, appeared
at the Department of Paediatric Orthopaedics, Shengjing Hospital of
China Medical University for consultation for her two-year-old son
(IV-3), as she had a family history of DDH. The pedigree of the
Chinese family is shown in Fig. 1.
The four-generation Chinese family, including 24 individuals,
resides in a village in northeast China. Family members were
invited to participate in the study, and those aged ≥18 years gave
informed consent. Approval was obtained from the Human Research
Ethics Committee of Shengjing Hospital of China Medical University.
The status of the 24 individuals was established on the basis of
clinical and radiological examinations. Five individuals (aged
23-72 years) had DDH, including two males (III-9 and III-10) and
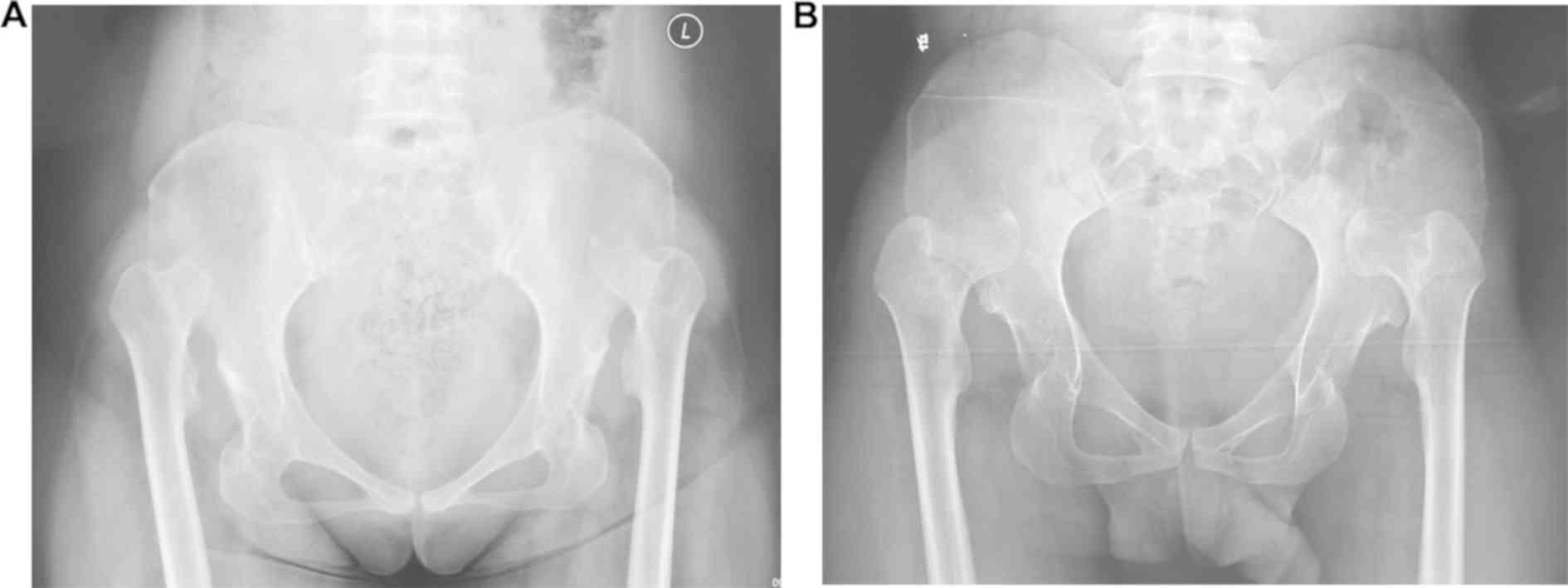
three females (I-1, III-6 and III-7). Anterior-posterior
radiography of the pelvis revealed that all affected members had
similar bilateral hip dislocation, which was classified as grade IV
according to the Tönnis classification system (Fig. 2) (16). These individuals did not have any
other system abnormalities associated with Down syndrome, Marfan
syndrome, Ehlers-Danlos syndrome or dyschondroplasia and were
therefore diagnosed with isolated DDH. All other members of the
pedigree had normal appearance and height (163-175 cm).
Genome-wide scan and linkage
analysis.
Blood samples were obtained from all members of the
family except for two individuals (III-3 and III-5). DNA was
extracted using QIAamp DNA Blood mini kit (Qiagen GmbH) through a
standard procedure. A genome-wide scan was performed on the 22
individuals using Affymetrix 10K single nucleotide polymorphism
(SNP) arrays at GeneTech Biotechnology Co., Ltd. Parametric and
nonparametric multipoint linkage analyses were performed using
Genespring GT v.2.0 software (Agilent Technologies, Inc.), and the
logarithm of odds (LOD) and NPL scores were calculated (Fig. 3). Parametric linkage analysis was
performed, assuming an autosomal recessive trait with full
penetrance and Affymetrix ‘Asian’ allele frequencies.
Results
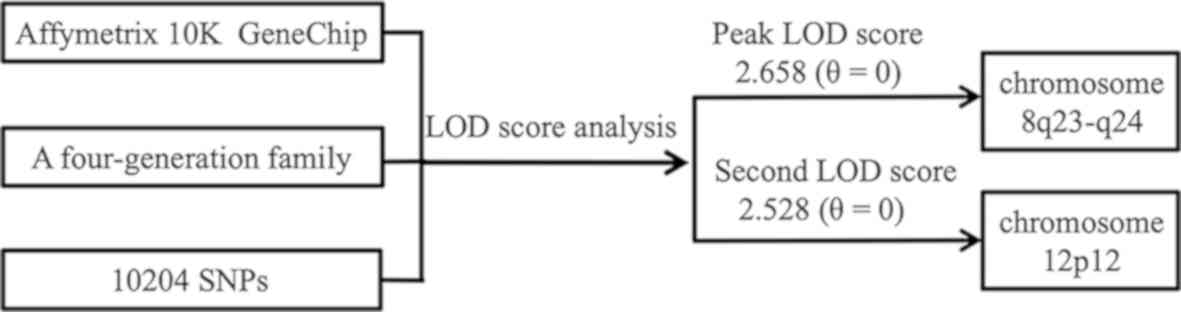
The strongest evidence for linkage was found on
chromosome 8q23-24, with a peak LOD score of 2.658 (θ=0), covering
2.377 Mb from SNP rs724717 to rs720132. This interval included nine
additional successive SNPs: rs1566071, rs1902121, rs756404,
rs702768, rs777813, rs2033995, rs147959, rs2884367 and rs1898287.
The same region also yielded the highest NPL score of 2.883
(P=0.0156) from non-parametric multipoint linkage analysis.
Additionally, the second highest NPL score of 2.727 (P=0.0156) and
an LOD score of 2.528 (θ=0) were obtained on chromosome 12p12 for
three consecutive markers (rs1919980, rs763853 and rs725124;
Table I). This region overlapped a
narrow distance of 0.642 Mb. Notably, in addition to the two
regions, no significant linkage was identified for other
chromosomal regions (with both LOD and NPL scores >2.0).
 | Table ILOD score of two novel chromosome
regions. |
Table I
LOD score of two novel chromosome
regions.
| | LOD at θ |
|---|
| Chromosome
region | Length (Mb) | Variation | 0 | 0.05 | 0.1 | 0.2 | 0.3 | 0.4 |
|---|
| 8q23-q24 | 2.377 | 1-9 | 2.658 | 2.361 | 2.058 | 1.442 | 0.851 | 0.349 |
| 12p12 | 0.642 | 10-12 | 2.528 | 2.240 | 1.946 | 1.349 | 0.777 | 0.297 |
Discussion
The aetiology of DDH may involve complex
interactions between many genes and the environment. Carter and
Wilkinson (17) postulated the
existence of two genetic systems underlying the disease: The
former, polygenic, is related to dysplasia of the acetabulum, and
the latter, possibly dominant, controls the capsule around the hip
joint. In accordance with the above hypothesis, Wynne-Davies
(18) proposed that two aetiologic
subtypes of DDH may be observed: One with acetabular dysplasia and
another with joint laxity. During the past decade, genome-wide
scans and association studies of large families have mapped DDH to
chromosome regions 3p22 (American), 4q35 (Africans), 16p
(Icelandic), 13q22 (Japanese), 20q11 (Chinese) and 15q13 and 19p13
(Saudi). Genetic mutations, such as CX3CR1, UFSP2,
ubiquinol-cytochrome-c reductase complex, PAPPA2, GDF5 and
TENM3, were identified in these regions. The present results
and previously identified chromosome regions may be related to the
growing development of the hip joint according to the function of
genes included in these regions (10,12,13,19-22). DDH was
previously associated with the D17S1820 marker from chromosome
region 17q21 by analysing 303 individuals from 101 Chinese trios
(data not shown). The results were further confirmed by Feldman
et al (23) through analysis
of a multi-generation American family with 18 members by
linkage.
The Chinese family, as reported here, features a
rare example of non-syndromic DDH suggestive of an uncommon
autosomal recessive model, which is different from previously
reported autosomal dominant traits (4,12).
However, two unaffected members of the family (III-3 and III-5)
refused to provide blood samples, which has limited the power of
linkage analysis. Nevertheless, this large pedigree may still be
useful for genetic linkage studies to locate candidate genes. For
the family, genome-wide scanning has provided strong evidence for
regions linked with the disease. Although neither LOD nor NPL
scores exceeded 3.0, this may provide a basis for the accumulation
of more evidence for the linkage.
Maximum LOD and NPL scores (2.658 and 2.883,
respectively) were obtained on chromosome 8q23-24. The region spans
2.377 Mb from SNPs rs724717 to rs720132. Four genes are known to
reside in this region, including transcriptional repressor GATA
binding 1 (TRPS1), eukaryotic translation initiation factor
3 subunit H (EIF3H), RAD21 cohesin complex component
(RAD21) and rRNA-binding ribosome biosynthesis protein UTP23
(UTP23). Among the four candidates, UTP23 encodes a
small subunit processome component, which is required for pre-18S
rRNA maturation (24). The product
of RAD21 is involved in the repair of double-strand DNA
breaks and chromatid cohesion during mitosis (25). Eukaryotic translation initiation
factor 3 subunit H (EIF3H) is a subunit of the eukaryotic
translation initiation factor 3 complex. The intact EIF3H
protein contributes to efficient translation initiation on 5'
leader sequences harbouring multiple upstream open reading frames,
and normal levels of EIF3H are implicated in growth control through
protein synthesis (26,27).
TRPS1 seems to be the most attractive
candidate from the 8q23-24 region. TRPS1 binds DNA through a
single GATA-type zinc-finger domain that recognizes a consensus DNA
sequence. Mutations in the TRPS1 gene may cause
tricho-rhino-phalangeal syndrome (TRPS) (28), an autosomal dominant type
craniofacial and skeletal dysplasia. In addition to craniofacial
deformities, patients with TRPS generally have skeletal anomalies,
including short stature, hip abnormalities (dysplasia, dislocation
of the hip or joint laxity), cone-shaped epiphyses and premature
closure of growth plates, which have reflected defects in
endochondral ossification. Long bones of the vertebrate
appendicular skeleton are formed through the process of
endochondral ossification when initial cartilaginous anlagen are
replaced by bone (29). Various
transcription factors and signalling pathways may be involved in
the regulation of endochondral bone formation. The Runx2
transcription factor is the master regulator of osteoblast
differentiation and is required for chondrocyte hypertrophy.
TRPS1 physically interacts with Runx2 and represses
Runx2-mediated trans-activation. Transgenic mice experiments
demonstrated that loss of repression of the Runx2-Ihh-positive
regulatory loop induced by TRPS1 mutation can result in
altered endochondral bone formation, which is characterized by the
dysregulation of chondrocyte differentiation and uncoupling of
processes of perichondrial mineralization and chondrocyte
maturation (30). Thus, TRPS1
is the most promising gene and should be investigated in future
studies.
SOX5 is the only gene known to reside in the
12p12 region. The gene encodes a member of the SOX (SRY-related
HMG-box) family of transcription factors involved in the regulation
of embryonic development and cell fate determination. Sox5,
together with Sox6, is essential for the establishment of
cartilage growth plates and thus the proper and timely development
of endochondral bones through regulation of chondrocyte
differentiation and proliferation (31). Foetuses of Sox5-Sox6 double
null mice may die with severe, generalized chondrodysplasia and
fail to form the epiphyseal plates and endochondral bones. Notably,
chondrodysplasia can also be observed at the dysplastic acetabulum
in DDH, albeit mild and partial. SOX5 gene knockdown
inhibited MMP-9 gene expression, contributing to fibroblast
survival, proliferation, migration and invasion in a rheumatoid
arthritis study (32). Therefore,
the potential effects of the SOX5 gene on the development of
non-syndrome DDH cannot be excluded.
According to the analysis above, the two novel
regions may affect articular cartilage formation, not only through
regulating chondrocyte differentiation and proliferation, but also
by uncoupling processes of perichondrial mineralization and
chondrocyte maturation. They may also be related to joint
development by promoting fibroblast growth.
As a complex disease, DDH is genetically
heterogeneous due to population differences and the complexity of
hip development. It is likely that different candidate genes have
contributed to the susceptibility to this disorder in different
populations. The present study showed a genome-wide linkage
analysis for a Chinese DDH family and the identification of two
novel loci at 8q23-24 and 12p12. The present results indicated the
presence of a disease-associated mutation or polymorphism in the
mapped regions of the affected members. With a number of candidate
genes identified from the two regions (TRPS1 and SOX5
in particular), mutations in these genes can be screened through
sequence analysis. These studies may lead to the eventual
identification of novel mutations or polymorphisms predisposing
individuals to the disease, which may enable early diagnosis and
optimal treatment.
To conclude, the evidence presented showed that
developmental dysplasia of the hip is mapped to two novel regions
at 8q23-q24 and 12p12. The present results indicate the presence of
a disease-associated mutation or polymorphism in the mapped regions
for the affected members.
Acknowledgements
The authors would like to thank Professor Lili Wang
(Key Laboratory of the Health Ministry for Congenital Malformation,
Shengjing Hospital of China Medical University) for her help in the
preparation of the manuscript.
Funding
The study was supported by the National Natural
Science Foundation of China (grant no, 81772296).
Availability of data and materials
The datasets used and/or analysed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
LL contributed to the conception of the study. YC
and QL contributed significantly to data analysis and manuscript
preparation. XX and LXZ analysed the data and wrote the manuscript.
LJZ helped perform data analysis with constructive discussions. All
authors read and approved the final manuscript.
Ethics approval and consent to
participate
Ethics approval was obtained from the Human Research
Ethics Committee of Shengjing Hospital of China Medical University.
Family members agreed to be included in this research study.
Patient consent for publication
Consent for publication was obtained from
participants.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Kotlarsky P, Haber R, Bialik V and
Eidelman M: Developmental dysplasia of the hip: What has changed in
the last 20 years? World J Orthop. 6:886–901. 2015.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Stevenson DA, Mineau G, Kerber RA,
Viskochil DH, Schaefer C and Roach JW: Familial predisposition to
developmental dysplasia of the hip. J Pediatr Orthop. 29:463–466.
2009.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Benson MKD: Developmental dysplasia of the
hip: Early diagnosis and management. Curr Paediatr. 6:2–8.
1996.PubMed/NCBI
|
|
4
|
Ceylaner G, Ceylaner S, Ustünkan F and
Inan M: Autosomal dominant inheritance of congenital dislocation of
the hip in 16 members of a family. Acta Orthop Traumatol Turc = 42.
289–291. 2008.(In Turkish). PubMed/NCBI View Article : Google Scholar
|
|
5
|
Zhao L, Tian W, Pan H, Zhu X, Wang J,
Cheng Z, Cheng L, Ma X and Wang B: Variations of the COL1A1 gene
promoter and the relation to developmental dysplasia of the hip.
Genet Test Mol Biomarkers. 17:840–843. 2013.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Wang K, Shi D, Zhu P, Dai J, Zhu L, Zhu H,
Lv Y, Zhao B and Jiang Q: Association of a single nucleotide
polymorphism in Tbx4 with developmental dysplasia of the hip: A
case-control study. Osteoarthritis Cartilage. 18:1592–1595.
2010.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Jia J, Li L, Zhao Q, Zhang L, Ru J, Liu X,
Li Q and Shi L: Association of a single nucleotide polymorphism in
pregnancy-associated plasma protein-A2 with developmental dysplasia
of the hip: A case-control study. Osteoarthritis Cartilage.
20:60–63. 2012.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Zhao L, Pan H, Wang J, Cheng Z, Cheng L,
Wang B and Ma X: Two single nucleotide polymorphisms in the GDF5
gene are associated with development dysplasia of the hip in
Chinese female population. Sci China Life Sci. 56:1063–1065.
2013.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Kolundžić R, Trkulja V, Mikolaučić M,
Kolundžić MJ, Pavelić SK and Pavelić K: Association of
interleukin-6 and transforming growth factor-β1 gene polymorphisms
with developmental hip dysplasia and severe adult hip
osteoarthritis: A preliminary study. Cytokine. 54:125–128.
2011.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Basit S, Albalawi AM, Alharby E and
Khoshhal KI: Exome sequencing identified rare variants in genes
HSPG2 and ATP2B4 in a family segregating developmental dysplasia of
the hip. BMC Med Genet. 18(34)2017.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Feldman G, Kappes D, Mookerjee-Basu J,
Freeman T, Fertala A and Parvizi J: Novel mutation in Teneurin 3
found to co-segregate in all affecteds in a multi-generation family
with developmental dysplasia of the hip. J Orthop Res. 37:171–180.
2019.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Feldman GJ, Parvizi J, Levenstien M, Scott
K, Erickson JA, Fortina P, Devoto M and Peters CL: Developmental
dysplasia of the hip: Linkage mapping and whole exome sequencing
identify a shared variant in CX3CR1 in all affected members of a
large multigeneration family. J Bone Miner Res. 28:2540–2549.
2013.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Watson CM, Crinnion LA, Gleghorn L, Newman
WG, Ramesar R, Beighton P and Wallis GA: Identification of a
mutation in the ubiquitin-fold modifier 1-specific peptidase 2
gene, UFSP2, in an extended South African family with Beukes hip
dysplasia. S Afr Med J. 105:558–563. 2015.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Shrimpton AE, Levinsohn EM, Yozawitz JM,
Packard DS Jr, Cady RB, Middleton FA, Persico AM and Hootnick DR: A
HOX gene mutation in a family with isolated congenital vertical
talus and Charcot-Marie-Tooth disease. Am J Hum Genet. 75:92–96.
2004.PubMed/NCBI View
Article : Google Scholar
|
|
15
|
Wang L, Hara K, Van Baaren JM, Price JC,
Beecham GW, Gallins PJ, Whitehead PL, Wang G, Lu C, Slifer MA, et
al: Vitamin D receptor and Alzheimer's disease: A genetic and
functional study. Neurobiol Aging. 33(1844.e1-9)2012.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Bolland BJ, Wahed A, Al-Hallao S,
Culliford DJ and Clarke NM: Late reduction incongenital dislocation
of the hip and the need for secondary surgery: Radiologic
predictors and confounding variables. J Pediatr Orthop. 30:676–682.
2010.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Carter C and Wilkinson J: Persistent joint
laxity and congenital dislocation of the hip. J Bone Joint Surg Br.
46:40–45. 1964.PubMed/NCBI
|
|
18
|
Wynne-Davies R: A family study of neonatal
and late-diagnosis congenital dislocation of the hip. J Med Genet.
7:315–333. 1970.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Sun Y, Wang C, Hao Z, Dai J, Chen D, Xu Z,
Shi D, Mao P, Teng H, Gao X, et al: A common variant of
ubiquinol-cytochrome c reductase complex is associated with DDH.
PLoS One. 10(e0120212)2015.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Ingvarsson T, Stefánsson SE, Gulcher JR,
Jónsson HH, Jónsson H, Frigge ML, Pálsdóttir E, Olafsdóttir G,
Jónsdóttir T, Walters GB, Lohmander LS and Stefánsson K: A large
Icelandic family with early osteoarthritis of the hip associated
with a susceptibility locus on chromosome 16p. Arthritis Rheum.
44:2548–2555. 2001.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Zamborsky R, Kokavec M, Harsanyi S, Attia
D and Danisovic L: Developmental dysplasia of hip: Perspectives in
genetic screening. Med Sci (Basel). 7(pii: E59)2019.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Basit S, Alharby E, Albalawi AM and
Khoshhal KI: Whole genome SNP genotyping in a family segregating
developmental dysplasia of the hip detected runs of homozygosity on
chromosomes 15q13.3 and 19p13.2. Congenit Anom (Kyoto). 58:56–61.
2018.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Feldman G, Dalsey C, Fertala K, Azimi D,
Fortina P, Devoto M, Pacifici M and Parvizi J: The Otto Aufranc
Award: Identification of a 4 Mb region on chromosome 17q21 linked
to developmental dysplasia of the hip in one 18-member,
multigeneration family. Clin Orthop Relat Res. 468:337–344.
2010.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Lu J, Sun M and Ye K: Structural and
functional analysis of Utp23, a yeast ribosome synthesis factor
with degenerate PIN domain. RNA. 19:1815–1824. 2013.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Beauchene NA, Díaz-Martínez LA, Furniss K,
Hsu WS, Tsai HJ, Chamberlain C, Esponda P, Giménez-Abián JF and
Clarke DJ: Rad21 is required for centrosome integrity in human
cells independently of its role in chromosome cohesion. Cell Cycle.
9:1774–1780. 2010.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Choudhuri A, Maitra U and Evans T:
Translation initiation factor eIF3h targets specific transcripts to
polysomes during embryogenesis. Proc Natl Acad Sci USA.
110:9818–9823. 2013.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Hronová V, Mohammad MP, Wagner S, Pánek J,
Gunis̆ová S, Zeman J, Poncová K and Valás̆ek LS: Does eIF3 promote
reinitiation after translation of short upstream ORFs also in
mammalian cells? RNA Biol. 14:1660–1667. 2017.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Smaili W, Elalaoui SC, Meier S, Zerkaoui
M, Sefiani A and Heinimann K: A novel TRPS1 mutation in a Moroccan
family with Tricho-rhino-phalangeal syndrome type III: Case report.
BMC Med Genet. 18(50)2017.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Lui JC, Nilsson O and Baron J: Recent
research on the growth plate: Recent insights into the regulation
of the growth plate. J Mol Endocrinol. 53:T1–T9. 2014.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Zhang Y, Xie RL, Gordon J, LeBlanc K,
Stein JL, Lian JB, van Wijnen AJ and Stein GS: Control of
mesenchymal lineage progression by microRNAs targeting skeletal
gene regulators Trps1 and Runx2. J Biol Chem. 287:21926–21935.
2012.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Liu CF and Lefebvre V: The transcription
factors SOX9 and SOX5/SOX6 cooperate genome-wide through
super-enhancers to drive chondrogenesis. Nucleic Acids Res.
43:8183–8203. 2015.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Shi Y, Wu Q, Xuan W, Feng X, Wang F, Tsao
BP, Zhang M and Tan W: Transcription factor SOX5 promotes the
migration and invasion of fibroblast-like synoviocytes in part by
regulating MMP-9 expression in collagen-induced arthritis. Front
Immunol. 9(749)2018.PubMed/NCBI View Article : Google Scholar
|