Introduction
Ischemic heart disease is a leading cause of
morbidity and mortality worldwide (1). The most effective clinical intervention
for myocardial ischemia is timely revascularization (2). However, myocardial ischemia/reperfusion
(I/R) caused by revascularization can also promote further
myocardial damage (3). The
pathophysiological mechanism of myocardial I/R injury (MIRI) is
complicated; however, numerous studies have revealed that the
inflammatory response serves an important role in I/R-induced
myocardial damage (4-6).
Thus, determining novel approaches to decrease the inflammation may
lead to effective treatments for MIRI.
Deleted in esophageal cancer 1 (DEC1) was first
identified in human embryonic cartilage tissue and it has since
been confirmed to function as a basic helix-loop-helix
transcription factor that can regulate transcription in various
types of cell, including hepatocyte and nerve cells (7). The expression levels of DEC1 have been
identified to significantly increase in response to environmental
stimuli, and DEC1 has been demonstrated to be involved in various
physiological activities, such as chondrocyte differentiation
(8), lipid metabolism (9), cell proliferation (10) and the immune response (11). Increasing evidence has suggested that
DEC1 is also an important transcription factor in inflammation
(12-14).
The continuous activation of DEC1 and the consequent elevation of
the inflammatory cascade events has served as a crucial pathogenic
mechanism in a wide range of human inflammatory diseases, such as
periodontitis (12), autoimmune
encephalomyelitis (13) and
rheumatoid arthritis (15).
It is well established that the activation of the
Toll-like receptor 4 (TLR4)/NF-κB signaling pathway induces the
transcription of numerous cytokines to promote inflammatory
responses (16,17). For example, Zhang et al
(12) reported that DEC1 modulated
Porphyromonas gingivalis-induced periodontitis via the
TLR4/NF-κB signaling pathway. In fact, the excessive activation of
the TLR4/NF-κB inflammatory pathway has been identified to be
closely associated with the development of MIRI (18,19).
However, whether the activation of TLR4/NF-κB could also be
triggered by DEC1 during MIRI remains largely unknown. Thus,
further study into the physiological function and underlying
mechanisms of DEC1 in MIRI is required. The present study
demonstrated that the genetic silencing of DEC1 using RNA
interference technology reduced inflammation in MIRI by repressing
the TLR4/NF-κB signaling pathway. Thus, the present study provided
a novel insight into the potential mechanisms of DEC1 in MIRI and
may provide a novel therapeutic target for preventing MIRI.
Materials and methods
Animal studies
The procedures for the experiments and animal care
were approved by the Animal Care and Use Committee of Hubei
Polytechnic University and conformed to the Guide for the Care and
Use of Laboratory Animals produced by the National Institutes of
Health (20). A total of 48 adult
male Sprague-Dawley rats (age, 8 weeks; weight, about 250 g) were
purchased from Vital River Laboratory Animal Technology Co., Ltd.
and housed at 23±2˚C with 50% relative humidity, with a 12-h
light/dark cycle and ad libitum access to food and
water.
Small interfering RNA (siRNA)
interference
The adenovirus (RNA interference) containing siRNA
against DEC1 was synthesized and generated with a pBHGlox_E1,3Cre
plasmid (Microbix Biosystems) using the AdMax system (Microbix
Biosystems) according to the manufacturer's protocols. A scrambled
sequence (500 µl; Sangon Biotech Co., Ltd.) was used as the
negative control. 293T cells (cat. no. CRL-1573; American Type
Culture Collection; 4x105 cells/ml) were used to package
and amplify the adenoviruses (500 µl) with
Lipofectamine™ 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) in DMEM (Gibco; Thermo Fisher Scientific, Inc.),
supplemented with 10% FBS (Gibco; Thermo Fisher Scientific, Inc.)
at 37˚C in a 5% CO2 incubator for 10-15 days. After the
majority of 293T cells exhibited typical cytopathic effects,
samples were frozen-thawed at -70˚C/37˚C three times and
centrifuged at 7,000 x g at 4˚C for 5 min. Virus supernatants were
subsequently collected. The final virus concentration was
1x1011 plaque-forming units, as using the endpoint
dilution method (21).
Establishment of MIRI model rats
The 48 healthy male SD rats were divided into four
groups (n=12/group): The sham group (Sham); I/R group (I/R);
adenovirus expressing green fluorescent protein Control group
(Ad-G-Control); and DEC1-targeting RNA interference group
(Ad-G-DEC1). The rat MIRI model was prepared in the I/R,
Ad-G-Control and Ad-G-DEC1 groups. Briefly, sodium pentobarbital
(30 mg/kg; intraperitoneal) was used to anesthetize the rats and
open the chest. A volume of 80 µl Ad-G-Control or Ad-G-DEC1 diluted
in saline (1x1010 PFU/ml; 10 times diluted) was injected
into the heart wall of 8-week-old rats. A total of 4 days later,
the rats were re-anesthetized, the rats' chests were reopened and
the left anterior descending artery (LAD) was ligated using a 6-0
silk suture. Following 30 min of ligation, the circulation was
restored for ~4 h by removing the silk suture; when the rat's limbs
demonstrated slight movement (~70 min intervals), the rats were
re-anesthetized to keep them under anesthetic during this
procedure. Post-reperfusion, the rats were sacrificed by jugular
vein injection of potassium chloride (75 mg/kg). Death was
confirmed using a standard body part II-lead electrocardiogram.
Both blood (5 ml) and part of the anterior wall of the left
ventricular myocardium near the cardiac apex were subsequently
harvested from the rats. The sham groups served as the control and
no occlusion of the LAD was performed.
Determination of myocardial
enzymes
Harvested blood was centrifuged at 500 x g for 5 min
at 4˚C to obtain serum, which was subsequently used to measure
myocardial enzyme levels, including creatine kinase (CK; cat. no.
QS1107) and lactate dehydrogenase (LDH; cat. no. QS1001) using
commercially available biochemical kits (Beijing Solarbio Science
& Technology Co., Ltd.), according to the manufacturers'
protocols. The data obtained are presented as U/l.
Detection of myocardial infarct area
(IA)/area at risk (AAR)
Evans Blue/triphenyltetrazolium chloride (TTC)
staining was used to determine the IA following MIRI. In brief, the
LADs of the rats were immediately ligated following 4 h of
reperfusion and ~1 ml Evans blue (Sigma-Aldrich; Merck KGaA) was
intravenously injected to distinguish non-ischemic and risk areas.
Next, rats were sacrificed by injecting 75 mg/kg potassium chloride
into the jugular vein, after which hearts were removed, frozen at
-20˚C for 10 min and sliced into 2 mm thick sections. Sections were
subsequently stained with 1.5% TTC (Sigma-Aldrich; Merck KGaA) for
15 min at room temperature. The risk area was stained red, whilst
the IA appeared white. Image-Pro Plus 5.0 software (Media
Cybernetics, Inc.) was used to calculate the AAR (red + white) and
IA (white). The IA/AAR ratio was calculated for quantitative
analysis.
Histological examination
The harvested hearts were fixed in 10% formalin for
48 h at room temperature and embedded in paraffin.
Paraffin-embedded tissues were sliced into 4-µm-thick sections and
stained with hematoxylin and eosin (hematoxylin staining, 5 min at
room temperature; eosin staining, 2 min at room temperature). Light
microscopy (magnification, x200) was used to observe the
morphological changes in the myocardial tissue following
reperfusion in each group.
Reverse transcription-quantitative PCR
(RT-qPCR)
RT-qPCR was used to detect mRNA levels as previously
described (19). In detail, total
RNA was extracted from the harvested hearts using
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc.) and reverse-transcribed into cDNA using the SuperScript IV
Reverse Transcriptase kit (Thermo Fisher Scientific, Inc.) at 37˚C
for 60 min according to the manufacturer's protocol. qPCR was
subsequently performed using the SYBR Green Master Mix kit (Thermo
Fisher Scientific, Inc.) on the 7500 ABI Prism system in accordance
with the manufacturer's protocol. The following thermocycling
conditions were used for the qPCR: Initial denaturation at 95˚C for
10 min; followed by 40 of cycles of denaturation at 95˚C for 30
sec, annealing at 60˚C for 30 sec and elongation at 72˚C for 30
sec. Data were analyzed using the
2-ΔΔCq method (22). The following primer pairs were used
for the qPCR: TLR4 forward, 5'-AGTGTATCGGTGGTCAGTGTGCT-3' and
reverse, 5'-AAACTCCAGCCACACATTCC-3'; NF-κB forward,
5'-AAGCACAGATACCACTAAGACGCA -3' and reverse,
5'-TTCAGCCTCATAGAAGCCATCC-3'; DEC1 forward,
5'-GTCTGTGAGTCACTCTTCAG-3' and reverse,
5'-GAGTCTAGTTCTGTTTGAAGG-3'; and GAPDH forward,
5'-TGGCCTTCCGTGTTCCTAC-3' and reverse, 5'-GAGTTGCTGTTGAAGTCGCA-3'.
The mRNA expression levels of DEC1, TLR4 and NF-κB were normalized
to GAPDH.
Western blotting
Western blotting was performed to detect the protein
expression levels of DEC1, TLR4, NF-κB and GAPDH in the harvested
hearts. Briefly, total protein was extracted using the protein
extraction kit (cat. no. P0028; Beyotime Institute of
Biotechnology). Total protein (40 µg) was quantified using a
bicinchoninic acid assay (Beyotime Institute of Biotechnology) and
separated via 10% SDS-PAGE. The separated proteins were
subsequently transferred onto PVDF membranes electrophoretically
(EMD Millipore). The membranes were blocked with 5% non-fat dry
milk in PBS with 0.05% Tween-20 for 2 h at room temperature.
Samples were then incubated with the following primary antibodies
at 4˚C overnight: Anti-DEC1 (1:600; cat. no. sc-101023; Santa Cruz
Biotechnology, Inc.), anti-TLR4 (1:500; cat. no. sc-10741; Santa
Cruz Biotechnology, Inc.), anti-NF-κB (1:600; cat. no. sc-514451;
Santa Cruz Biotechnology, Inc.) and anti-GADPH (1:1,000; cat. no.
sc-48166; Santa Cruz Biotechnology, Inc.). Following the primary
antibody incubation, the membranes were washed 3 times with TBS
with 0.05% Tween and incubated with horseradish peroxidase
(HRP)-conjugated anti-mouse (1:5,000; cat. no. AS014),
HRP-conjugated anti-goat (1:5,000; cat. no. AS064) or
HRP-conjugated anti-rabbit (1:5,000; cat. no. AS063; all, ABclonal
Biotech Co., Ltd.) IgG secondary antibodies for 2 h at room
temperature. Finally, protein bands were visualized using an
enhanced chemiluminescence system (Thermo Fisher Scientific, Inc.).
Quantity One 1-D software (version 4.6.9; Bio-Rad Laboratories,
Inc.) was used for densitometric analysis.
ELISAs
ELISA kits (R&D Systems, Inc.) were used to
determine the concentrations of interleukin (IL)-1β (cat. no.
SRLB00) and tumor necrosis factor (TNF)-α (cat. no. SRTA00) in the
harvested hearts, according to the manufacturer's protocol.
Statistical analysis
Statistical analysis was performed using SPSS 21.0
software (IBM Corp.) and data are presented as the mean ± standard
deviation (n=5). Statistical differences between two groups were
determined using a Student's t-test, whereas statistical
differences between more than two groups were analyzed using a
one-way ANOVA, followed by a Tukey's post hoc test for multiple
comparisons. P<0.05 was considered to indicate a statistically
significant difference.
Results
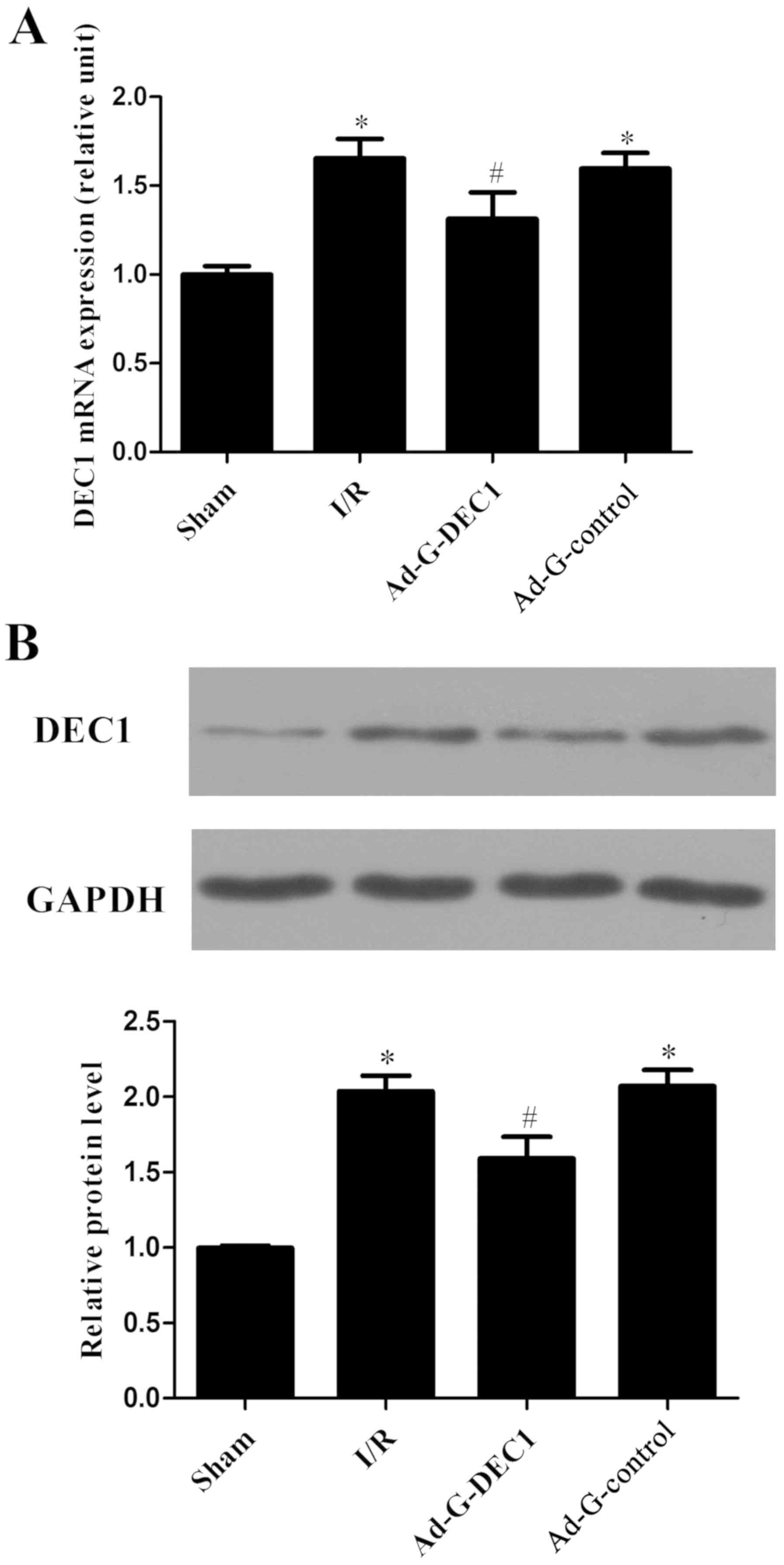
Delivery of DEC1-targeting RNA
interference decreases the increased DEC1 expression levels induced
by I/R
Following reperfusion, compared with the sham group,
the mRNA and protein expression levels of DEC1 were significantly
increased in the I/R group (P<0.05; Fig. 1A and B). Notably, following the delivery of the
DEC1-targeting RNA inference, the expression levels of DEC1 were
significantly decreased compared with the I/R and Ad-G-Control
groups (P<0.05; Fig. 1).
Delivery of DEC1-targeting RNA
interference decreases the concentration of serum marker
enzymes
LDH and CK are commonly used as molecular markers
for myocardial injury (23). The
activities of CK and LDH were significantly increased in the I/R
group compared with the sham group (P<0.05; Fig. 2A and B); however, following the delivery of
DEC1-targeting RNA interference, the concentration of CK and LDH in
the Ad-G-DEC1 group was significantly decreased compared with the
I/R and Ad-G-Control groups (P<0.05; Fig. 2).
Decreased expression levels of DEC1
decreases the IA
To quantitatively determine the extent of myocardial
injury, the IA/AAR was calculated. The IA/AAR was identified to be
56.4±6.2% in the I/R group (Fig. 3).
Notably, the downregulation of DEC1 expression in Ad-G-DEC1 group
exerted a cardioprotective effect by reducing IA/AAR during MIRI
compared with the I/R group (P<0.05; Fig. 3). However, no significant effect on
IA/AAR was observed in the Ad-G-Control group compared with the I/R
group (P>0.05; Fig. 3).
Histopathological evaluation of the
heart tissue
In the I/R group, the myocardial fibers were
observed to be disorganized and ruptured, exhibiting signs of edema
(Fig. 4). Notably, the transfection
of the DEC1-targeting RNA interference vector partially decreased
the myocardial damage. Furthermore, no significant effect was
observed in the histological morphology of the heart tissue in the
Ad-G-Control group compared with the I/R group (Fig. 4).
Determination of the levels of IL-1β
and TNF-α
ELISAs were used to detect the concentrations of
IL-1β and TNF-α in the harvested specimens. It was revealed that
the levels of IL-1β and TNF-α were significantly increased in the
I/R group compared with the sham group, whereas the downregulation
of DEC1 expression significantly decreased their levels compared
with the I/R and Ad-G-Control groups (P<0.05; Fig. 5).
Downregulation of DEC1 expression
inhibits the TLR4/NF-κB signaling pathway
To investigate the underlying mechanisms of
DEC1-induced I/R injury, the expression levels of TLR4 and NF-κB
were determined. I/R significantly increased the mRNA and protein
expression levels of TLR4 and NF-κB compared with the sham group
(P<0.05; Fig. 6A-C). However, the
delivery of the DEC1-targeting RNA interference at the onset of I/R
significantly decreased the expression levels of TLR4 and NF-κB
compared with those in the I/R and Ad-G-Control groups. Therefore,
these results suggested that I/R injury may be suppressed via the
downregulation of DEC1, which may occur through its ability to
inhibit the TLR4/NF-κB signaling pathway.
Discussion
MIRI is an important cause of myocardial damage and
it is an unsolved problem in clinical practice (24). Inflammation has been identified to
serve a crucial role in MIRI and a persistent proinflammatory
reaction promotes a series of complications following reperfusion
therapy (25,26). Therefore, effectively decreasing
inflammation may be an important therapeutic target to improve the
outcomes for MIRI. A previous study has found that DEC1 served an
important role in various physiological functions, including cell
differentiation, circadian rhythm regulation, hypoxia and
inflammatory responses (27). Thus,
in the past decade, DEC1-mediated inflammatory activation has
become an important topic of research (12,14);
however, it is unclear whether there is an association between
DEC1-mediated inflammatory activation and MIRI. In the present
study, it was observed that the downregulation of DEC1 by RNA
interference decreased MIRI, which is characterized by the
decreased release of LDH/CK and infarct area. In addition, DEC1
silencing inhibited the protein expression levels of
inflammation-related cytokines, such as IL-1β and TNF-α. In the
mechanistic studies, it was observed that the downregulation of
DEC1s significantly inhibited the TLR4/NF-κB signaling pathway. The
aforementioned results therefore indicated that DEC1 silencing may
possess a marked ability to ameliorate I/R-induced damage and
inflammation by inhibiting the TLR4/NF-κB signaling pathway.
TLR4 is a major cell surface initiator of
inflammatory responses (28).
Accumulating evidence has suggested that TLR4-mediated signal
transduction is activated during the process of MIRI (29,30).
Notably, Zhang et al (12)
identified that the inflammation-associated transcription factor
DEC1 regulated the cellular responses in P. gingivalis
infection by promoting the production of IL-1β and TNF-α through
the activation of TLR4. Meanwhile, the downregulation of DEC1
expression also decreased their expression levels and decrease the
severity of periodontitis (12).
With regards to the possible involvement of DEC1 in TLR4-associated
inflammation, it was thus hypothesized that DEC1 may also serve a
proinflammatory role in response to an I/R insult. Consistent with
the aforementioned data, the results of the present study indicated
that the DEC1-associated inflammatory signaling pathway was
activated in the process of MIRI and inhibiting DEC1 expression via
RNA interference decreased the expression levels of IL-1β, TNF-α
and TLR4, which weakened the myocardial damage observed.
Consequently, the present study provided evidence to suggest that
DEC1-mediated TLR4 activation may participate in myocardial I/R;
however, the potential mechanisms remain unknown.
NF-κB has a broad role in inflammation and other
natural and pathological processes; its important role in the
cardiovascular system following TLR4 activation been well
established (31,32). Notably, numerous studies have
provided evidence that the TLR4/NF-κB signaling pathway served an
important role in the pathological process of MIRI (18,19). For
example, Olkkonen et al (15)
indicated that the DEC gene enhanced NF-κB binding to DNA to
increase IL-1β expression levels in human fibroblasts. Therefore,
it was necessary to investigate whether DEC1-mediated TLR4 signal
transduction in MIRI was activated synergistically with NF-κB; the
data from the present study demonstrated that DEC1 silencing also
significantly decreased the expression levels of NF-κB.
The cytokine cascade in the inflammatory processes
is a highly complex system (33).
IL-1β and TNF-α are both proinflammatory cytokines, which have been
previously implicated in the pathophysiology of MIRI (34). In addition, DEC1 has been confirmed
to be an important transcription factor in inflammation by
activating immune cells (CD4+ T cells) and controlling
the expression of multiple immunomodulatory genes, such as IL-1β
and TNF-α (12,14). Martínez-Llordella et al
(13) also reported that
DEC1-deficient mice exhibited an increased production of IL-10, but
no effect on IFN-γ levels. Based on these results, DEC1 was
silenced and the concentrations of IL-1β and TNF-α were detected in
the myocardium following I/R in the present study. The results
revealed that DEC1 silencing exerted protective effects against I/R
injury by inhibiting the production of IL-1β and TNF-α. IL-1β and
NF-κB have been identified to independently contribute to the
increased expression of DEC1, an effect that is dependent upon the
PI3K/AKT signaling pathways, amongst others (14,15).
Taken together, these results suggested that the downregulation of
DEC1 may not only decrease the levels of IL-1β and TNF-α directly,
but it may also disturb the positive feedback loop between these
inflammatory factors and DEC1.
In conclusion, the results of the present study
demonstrated that the downregulation of DEC1 expression alleviated
MIRI by attenuating inflammation in a TLR4/NF-κB-dependent manner.
However, multiple other signaling pathways are considered to be
involved in the pathophysiological processes of MIRI. Thus, further
studies are required to investigate whether there are any other
signaling pathways involved in the regulation over DEC1 in MIRI.
Taken together, the present results suggested that DEC1 may be a
prospective therapeutic option for MIRI.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Hubei
Provincial Natural Science Foundation of China (grant no.
2017CFB211) and the Health Commission of Hubei Province Scientific
Research Project (grant no. WJ2019Q011).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
WPX and KZ wrote the manuscript, interpreted the
data and performed the experiments. YZ and SXM acquired and
analyzed the data. WPX and DQJ performed the literature search,
designed the study and revised the manuscript. All authors read and
approval the final manuscript.
Ethics approval and consent to
participate
The procedures for the experiments and animal care
were approved by the Animal Care and Use Committee of Hubei
Polytechnic University and conformed to the Guide for the Care and
Use of Laboratory Animals produced by the National Institutes of
Health.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Nelson CP, Goel A, Butterworth AS, Kanoni
S, Webb TR, Marouli E, Zeng L, Ntalla I, Lai FY, Hopewell JC, et
al: Association analyses based on false discovery rate implicate
new loci for coronary artery disease. Nat Genet. 49:1385–1391.
2017.PubMed/NCBI View
Article : Google Scholar
|
|
2
|
Godoy LC, Lawler PR, Farkouh ME, Hersen B,
Nicolau JC and Rao V: Urgent revascularization strategies in
patients with diabetes mellitus and acute coronary syndrome. Can J
Cardiol. 35:993–1001. 2019.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Shen Y, Liu X, Shi J and Wu X: Involvement
of Nrf2 in myocardial ischemia and reperfusion injury. Int J Biol
Macromol. 125:496–502. 2019.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Wu MY, Yiang GT, Liao WT, Tsai AP, Cheng
YL, Cheng PW, Li CY and Li CJ: Current mechanistic concepts in
ischemia and reperfusion injury. Cell Physiol Biochem.
46:1650–1667. 2018.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Ruan Z, Wang S, Yu W and Deng F: LncRNA
MALAT1 aggravates inflammation response through regulating PTGS2 by
targeting miR-26b in myocardial ischemia-reperfusion injury. Int J
Cardiol. 288(122)2019.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Yao L, Chen H, Wu Q and Xie K:
Hydrogen-rich saline alleviates inflammation and apoptosis in
myocardial I/R injury via PINK-mediated autophagy. Int J Mol Med.
44:1048–1062. 2019.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Sato F, Kohsaka A, Bhawal UK and Muragaki
Y: Potential roles of Dec and Bmal1 genes in interconnecting
circadian clock and energy metabolism. Int J Mol Sci. 19(pii:
E781)2018.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Liu Q, Wu Y, Seino H, Haga T, Yoshizawa T,
Morohashi S and Kijima H: Correlation between DEC1/DEC2 and
epithelial-mesenchymal transition in human prostate cancer PC-3
cells. Mol Med Rep. 18:3859–3865. 2018.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Noshiro M, Kawamoto T, Nakashima A, Ozaki
N, Ueno T, Saeki M, Honda K, Fujimoto K and Kato Y: Deficiency of
the basic helix-loop-helix transcription factor DEC1 prevents
obesity induced by a high-fat diet in mice. Genes Cells, Jul 3,
2018 (Epub ahead of print).
|
|
10
|
Jia Y, Hu R, Li P, Zheng Y, Wang Y and Ma
X: DEC1 is required for anti-apoptotic activity of gastric cancer
cells under hypoxia by promoting Survivin expression. Gastric
Cancer. 21:632–642. 2018.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Camponeschi A, Todi L, Cristofoletti C,
Lazzeri C, Carbonari M, Mitrevski M, Marrapodi R, Del Padre M,
Fiorilli M, Casato M and Visentini M: DEC1/STRA13 is a key negative
regulator of activation-induced proliferation of human B cells
highly expressed in anergic cells. Immunol Lett. 198:7–11.
2018.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Zhang F, Suzuki M, Kim IS, Kobayashi R,
Hamada N, Sato F and Bhawal UK: Transcription factor DEC1 is
required for maximal experimentally induced periodontal
inflammation. J Periodontal Res. 53:883–893. 2018.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Martínez-Llordella M, Esensten JH, Lipsky
RH, Marini A, Chen J, Mughal M, Mattson MP, Taub DD and Bluestone
JA: CD28-inducible transcription factor DEC1 is required for
efficient autoreactive CD4+ T cell response. J Exp Med.
210:1603–1619. 2013.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Bhawal UK, Ito Y, Tanimoto K, Sato F,
Fujimoto K, Kawamoto T, Sasahira T, Hamada N, Kuniyasu H, Arakawa
H, et al: IL-1β-mediated up-regulation of DEC1 in human gingiva
cells via the Akt pathway. J Cell Biochem. 113:3246–3253.
2012.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Olkkonen J, Kouri VP, Hynninen J,
Konttinen YT and Mandelin J: Differentially expressed in
chondrocytes 2 (DEC2) increases the expression of IL-1β and Is
abundantly present in synovial membrane in rheumatoid arthritis.
PLoS One. 10(e0145279)2015.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Zusso M, Lunardi V, Franceschini D,
Pagetta A, Lo R, Stifani S, Frigo AC, Giusti P and Moro S:
Ciprofloxacin and levofloxacin attenuate microglia inflammatory
response via TLR4/NF-kB pathway. J Neuroinflammation.
16(148)2019.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Rahimifard M, Maqbool F, Moeini-Nodeh S,
Niaz K, Abdollahi M, Braidy N, Nabavi SM and Nabavi SF: Targeting
the TLR4 signaling pathway by polyphenols: A novel therapeutic
strategy for neuroinflammation. Ageing Res Rev. 36:11–19.
2017.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Yuan L, Dai X, Fu H, Sui D, Lin L, Yang L,
Zha P, Wang X and Gong G: Vaspin protects rats against myocardial
ischemia/reperfusion injury (MIRI) through the TLR4/NF-κB signaling
pathway. Eur J Pharmacol. 835:132–139. 2018.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Guo X, Jiang H, Yang J, Chen J, Yang J,
Ding JW, Li S, Wu H and Ding HS: Radioprotective 105 kDa protein
attenuates ischemia/reperfusion-induced myocardial apoptosis and
autophagy by inhibiting the activation of the TLR4/NF-κB signaling
pathway in rats. Int J Mol Med. 38:885–893. 2016.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Bayne K: Revised guide for the care and
use of laboratory animals available. American physiological
society. Physiologist. 39: 199:208–211. 1996.PubMed/NCBI
|
|
21
|
Mittereder N, March KL and Trapnell BC:
Evaluation of the concentration and bioactivity of adenovirus
vectors for gene therapy. J Virol. 70:7498–7509. 1996.PubMed/NCBI
|
|
22
|
Livak JK and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Yang J, Fan ZX, Yang J, Ding J, Yang C and
Chen L: microRNA-22 attenuates myocardial ischemia-reperfusion
injury via an anti-inflammatory mechanism in rats. Exp Ther Med.
12:3249–3255. 2016.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Neri M, Riezzo I, Pascale N, Pomara C and
Turillazzi E: Ischemia/reperfusion injury following acute
myocardial infarction: A critical issue for clinicians and forensic
pathologists. Mediators Inflamm. 2017(7018393)2017.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Chorawala MR, Prakash P, Doddapattar P,
Jain M, Dhanesha N and Chauhan AK: Deletion of extra domain A of
fibronectin reduces acute myocardial ischaemia/reperfusion injury
in hyperlipidaemic mice by limiting thrombo-inflammation. Thromb
Haemost. 118:1450–1460. 2018.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Hernandez-Resendiz S, Chinda K, Ong SB,
Cabrera-Fuentes H, Zazueta C and Hausenloy DJ: The role of redox
dysregulation in the inflammatory response to acute myocardial
ischaemia-reperfusion injury-adding fuel to the fire. Curr Med
Chem. 25:1275–1293. 2018.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Ivanov SV, Salnikow K, Ivanova AV, Bai L
and Lerman MI: Hypoxic repression of STAT1 and its downstream genes
by a pVHL/HIF-1 target DEC1/STRA13. Oncogene. 26:802–812.
2007.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Knowlton AA: Paying for the tolls: The
high cost of the innate immune system for the cardiac myocyte. Adv
Exp Med Biol. 1003:17–34. 2017.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Fang Y and Hu J: Toll-like receptor and
its roles in myocardial ischemic/reperfusion injury. Med Sci Monit.
17:RA100–RA109. 2011.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Xue J, Ge H, Lin Z, Wang H, Lin W, Liu Y,
Wu G, Xia J and Zhao Q: The role of dendritic cells regulated by
HMGB1/TLR4 signalling pathway in myocardial ischaemia reperfusion
injury. J Cell Mol Med. 23:2849–2862. 2019.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Su Q, Li L, Sun Y, Yang H, Ye Z and Zhao
J: Effects of the TLR4/Myd88/NF-κB signaling pathway on NLRP3
inflammasome in coronary microembolization-induced myocardial
injury. Cell Physiol Biochem. 47:1497–1508. 2018.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Tang ZH, Peng J, Ren Z, Yang J, Li TT, Li
TH, Wang Z, Wei DH, Liu LS, Zheng XL and Jiang ZS: New role of
PCSK9 in atherosclerotic inflammation promotion involving the
TLR4/NF-κB pathway. Atherosclerosis. 262:113–122. 2017.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Chousterman BG, Swirski FK and Weber GF:
Cytokine storm and sepsis disease pathogenesis. Semin Immunopathol.
39:517–528. 2017.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Ma C, Xu Z and Lv H: Low n-6/n-3 PUFA
ratio improves inflammation and myocardial ischemic reperfusion
injury. Biochem Cell Biol. 97:621–629. 2019.PubMed/NCBI View Article : Google Scholar
|