Introduction
Cancer is a complex genetic disease in addition to
being one of the leading cause of mortality worldwide (1,2). Despite
improving diagnostic techniques and therapeutics, cancer affects
the quality of life of those patients affected, and creates serious
social and economic burdens. Therefore, there is an urgent
requirement to elucidate the molecular mechanisms underlying cancer
development, and to identify novel biomarkers to improve diagnosis,
treatment and prognosis. The transient receptor potential (TRP)
gene was first cloned in 1989 and categorized into a nonselective
cation channel superfamily (3). The
human TRP family is divided into six subfamilies: TRPC, TPRV, TRPM,
TRPP, TRPML and TRPA. TRPM for ‘melastatin’ contains 8 members,
namely TRPM1, TRPM2, TRPM3, TRPM4, TRPM5, TRPM6, TRPM7 and
TRPM8(4). TRPM2 has been
demonstrated to promote the growth of prostate cancer cells
(5). TRPM4 was suggested to enhance
cancer cell proliferation via upregulating the β-catenin signaling
pathway (6). TRPM7 has been
considered to regulate the migration and invasion of metastatic
breast cancer cells (7). Taken
together, the TRPM protein family may be attractive targets for
anticancer therapies or prognostic biomarkers in certain types of
human cancer (8,9). However, to the best of our knowledge, a
systematic study on the transcriptional expression and prognostic
value of the TRPM protein family members in human tumors has not
been conducted yet. In the present study, the mRNA expression
patterns of the TRPM protein family between tumor tissues were
investigated and compared with normal tissues through the Oncomine
database. Furthermore, the present study analyzed prognostic values
using The Cancer Genome Atlas (TCGA) database.
Materials and methods
Oncomine analysis
In the present study, Oncomine (https://www.oncomine.org), an online cancer microarray
database, was used to analyze the mRNA expression levels of TRPMs
in different types of cancer. The cut-offs were set as fold change
(FC) =2 and P<0.01, the analysis type was set as cancer vs.
normal analysis, and data type as mRNA. The significant differences
between cancer and normal tissues, genes, datasets, sample sizes,
FC, Student's t-test and P-values were presented.
Kaplan-Meier plotter analysis
Kaplan-Meier plotter (https://www.kmplot.com) (10), which contains gene expression data
and clinical data, was used to evaluate the prognostic value of
TRPMs mRNA levels. The present study focused on overall survival
(OS) patient information with a 10-year follow-up. Patient samples
were separated into two groups based on their median expression
(high and low expression, the median group was included in the high
group) in order to estimate the prognostic value of a certain gene.
Kaplan-Meier plots were created by analyzing the OS of patients
with cancer. P<0.05 was considered to indicate a statistically
significant difference. Both log rank P-value and hazard ratio (HR)
with 95% confidence intervals were calculated and summarized. The
present study used the best specific probes (JetSet probes) that
recognized the TRPM protein family (11).
PrognoScan analysis
The results of the survival analyses were downloaded
from PrognScan database (http://dna00.bio.krytech.ac.jp/PrognoScan/index.html/)
(12), which is a public microarray
database containing clinical annotations of gene expression and the
prognostic value of genes, was used to evaluate the prognostic
effects of TRPMs in certain types of cancer in the present study.
P<0.05 was considered to indicate a statistically significant
difference.
Gene Expression Profiling Interactive
Analysis (GEPIA)
GEPIA (http://gepia.cancer-pku.cn/) (13), an interactive web server for
analyzing RNA sequencing expression data, was mined to predict the
differential expression levels of TRPM8 in liver and prostate
cancer groups compared with the control group. GEPIA was also used
to validate gene expression and evaluate the survival analysis of
the TRPMs in patients with liver cancer. P<0.05 was considered
to indicate a statistically significant difference.
Results
mRNA expression pattern of the TRPM
protein family in different types of human cancer
In order to investigate the transcriptional levels
of TRPMs in cancerous and control tissues among the multiple
different types of cancer, the present study performed an Oncomine
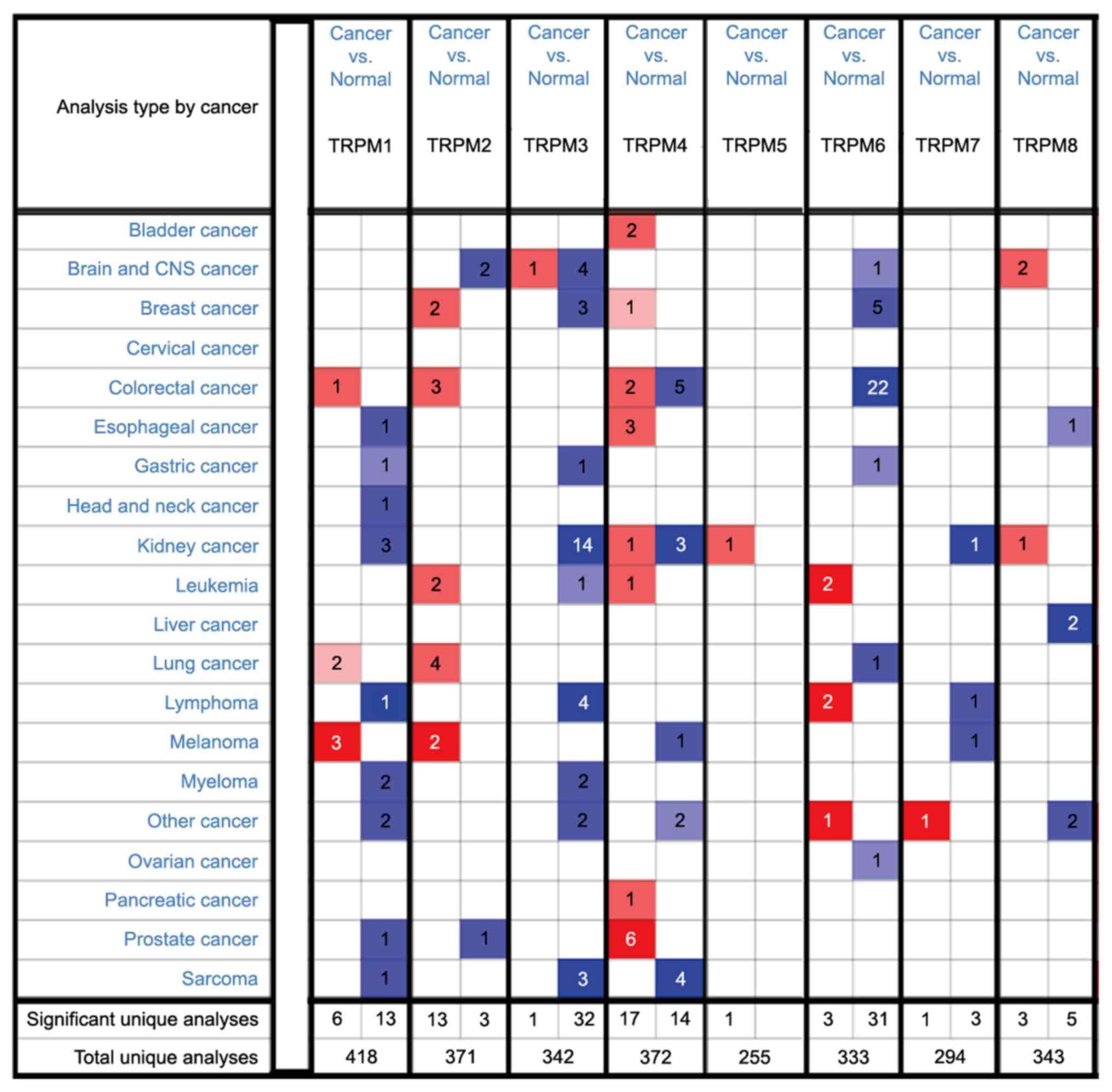
analysis. The database contains a total of 418, 371, 342, 372, 255,
333, 294 and 343 unique analyses for TRPM1, TRPM2, TRPM3, TRPM4,
TRPM5, TRPM6, TRPM7 and TRPM8, respectively (Fig. 1). The transcriptional levels of the
TRPM family members extracted from the Oncomine database were
significantly increased or decreased compared with the normal group
in various types of cancer.
The latest data from GLOBOCAN 2018 has reported that
there were 18.1 million incident cancer cases and 9.6 million
cancer mortalities in 2018(1). The
top 6 types of cancer to be diagnosed in both sexes combined were
lung, breast, prostate, colorectal, stomach and liver cancer
(1). Melanoma is the fifth most
common malignancy in men and the sixth most common in women
(14). Therefore, the present study
underlined the expression level and prognosis of TRPMs family in
these tumors, and certain other common types of solid tumor.
Expression levels and prognostic
values of TRPMs in breast cancer
Firstly, the present study investigated the
expression levels of the TRPMs family in breast cancer using the
Oncomine database. The analysis included 11 datasets in total.
According to the TCGA database, TRPM2 was revealed to be
upregulated in ductal carcinoma and invasive breast cancer.
However, TRPM3 and TRPM6 were downregulated in a variety of
different types of breast cancer. Furthermore, TRPM4 was
significantly increased only in male breast cancer. No significant
differences in TRPM1, TRPM5, TPRM7 and TRPM8 levels were observed
between cancerous and control tissues. All the statistically
significant results are summarized in Table I.
 | Table IDatasets of TRPM protein family in
breast cancer. |
Table I
Datasets of TRPM protein family in
breast cancer.
| Gene | Dataset | Normal (n) | Tumor (n) | Fold change | t-test | P-value |
|---|
| TRPM2 | TCGA | Breast (61) | Invasive ductal
breast carcinoma (389) | 2.158 | 14.181 |
1.69x10-28 |
| | | Breast (61) | Invasive breast
carcinoma (76) | 2.083 | 7.516 |
3.63x10-12 |
| TRPM3 | TCGA | Breast (61) | Invasive breast
carcinoma (76) | -2.185 | -12.894 |
1.07x10-22 |
| | | Breast (61) | Male vreast
carcinoma (3) | -2.007 | -7.258 |
8.00x10-4 |
| | | Breast (61) | Invasive ductal
nreast carcinoma (389) | -2.185 | -14.073 |
2.56x10-22 |
| TRPM4 | TCGA | Breast (61) | Male breast
carcinoma (3) | 2.108 | 6.148 |
5.00x10-3 |
| TRPM6 | TCGA | Breast (61) | Invasive ductal
breast carcinoma (389) | -5.432 | -18.443 |
3.50x10-37 |
| | | Breast (61) | Invasive lobular
breast carcinoma (36) | -2.803 | -8.379 |
5.73x10-12 |
| | | Breast (61) | Mixed lobular and
ductal breast carcinoma (7) | -3.793 | -6.679 |
7.29x10-5 |
| | | Breast (61) | Intraductal
cribriform breast adenocarcinoma (3) | -5.308 | -9.154 |
2.00x10-3 |
| | | Breast (61) | Invasive breast
carcinoma (76) | -3.548 | -8.71 |
9.25x10-15 |
Breast cancer is now described in terms of intrinsic
biological subtypes and is defined into the following four main
subtypes: Basal-like
(ER-/PR-/HER2-), luminal A
(ER+/HER2-/grade 1 or 2), Basal-like B
(ER+/HER2-/grade 3) and HER2-enriched (any
HER2+ tumor) (15). The
Kaplan-Meier curves presenting the OS of four breast cancer
subtypes with a 10-year follow-up are presented in Fig. 2. Poor patient outcome was found to be
associated with high expression levels of TRPM2 in the patients
with HER2+ subtype (Fig.
2D) and low expression of TRPM2 in patients with luminal B
subtype (Fig. 2C). High TRPM3
expression was associated with increased OS in luminal B breast
carcinoma subtype (Fig. 2G).
However, in the patients with the basal (Fig. 2I and M) and HER2+ (Fig. 2L and P) subtypes with a 10-year follow-up, high
expression levels of TRPM4 and TRPM6 indicated decreased survival
rates.
Expression levels and prognostic
values of TRPMs in lung cancer
Using the Oncomine database, the present study
analyzed the transcriptional expression of TRPM members in lung
cancer. In a group of datasets including Bhattacharjee et al
(16) and Garbe et al
(17), the transcriptional
expression levels of TRPM1 and TRPM2 in small cell lung carcinoma
were significantly increased compared with that in the control
tissues. According to Bhattacharjee et al (16), TRPM1 also was upregulated in lung
carcinoid tumor. According to the dataset from Garber et al
(17), it was revealed that TPRM2
was elevated in squamous cell lung carcinoma and large cell lung
carcinoma when compared with the control group. According to
Okayama et al (18), TRPM6
was decreased in lung adenocarcinoma; however, this was increased
in the lung adenocarcinoma samples in the study of Garber et
al (17). However, no
statistical differences were observed between lung cancer and
control tissue groups for TRPM3, TRPM4, TRPM5, TRPM7 and TRPM8 in
the present study. The detailed results are presented in Table II.
 | Table IIDatasets of TRPM protein family in
lung cancer. |
Table II
Datasets of TRPM protein family in
lung cancer.
| Gene | Dataset | Normal (n) | Tumor type (n) | Fold change | t-test | P-value |
|---|
| TRPM1 | Bhattacharjee et
al (16) | Lung (17) | Small cell lung
carcinoma (6) | 2.24 | 2.664 | 0.007 |
| | | Lung (17) | Lung carcinoid
tumor (20) | 2.366 | 2.766 | 0.005 |
| TRPM2 | Garber et al
(17) | Lung (5) Fetal lung
(1) | Large cell lung
carcinoma (4) | 4.561 | 5.046 |
5.28x10-4 |
| | | Lung (5) Fetal lung
(1) | Small cell lung
carcinoma (4) | 4.068 | 4.404 | 0.001 |
| | | Lung (5) Fetal lung
(1) | Lung adenocarcinoma
(39) | 3.586 | 5.194 |
5.85x10-4 |
| | | Lung (5) Fetal lung
(1) | Squamous cell lung
carcinoma (13) | 3.53 | 3.927 |
7.23x10-4 |
| TRPM6 | Okayama et al
(18) | Lung (20) | Lung adenocarcinoma
(226) | -2.363 | -9.369 | 6.51x10-12 |
The present study employed the Kaplan-Meier plotter
to identify the role of TRPM protein family in lung adenocarcinoma
and squamous cell lung carcinoma, which are the most common types
of lung cancer (19). For patients
with lung adenocarcinoma, decreased TRPM1 (Fig. 3A) and TRPM2 (Fig. 3C) levels were associated with
improved OS. However, decreased TRPM6 (Fig. 3E) were associated with lower OS rates
in the patients with lung adenocarcinoma with a 10-year follow-up.
No statistical difference was observed for patients with squamous
cell lung carcinoma when regarding OS (Fig. 3B, D
and F).
Expression levels and prognostic
values of TRPMs in colorectal cancer
As for colon and rectal carcinoma, all statistically
significant datasets are summarized in Table III. According to TCGA datasets,
TRPM1 was increased in rectosigmoid cancer compared with the
control tissues. It was revealed that TRPM2 was increased in colon
and cecum adenocarcinoma from TCGA datasets. According to
Skrzypczak et al (20), TRPM4
was elevated in colon adenoma epithelia, but decreased in colon
carcinoma epithelial and colorectal carcinoma compared with colon
tissues. In a group of datasets including TCGA and Hong et
al (21), TRPM4 was decreased in
colon adenocarcinoma, rectal adenocarcinoma and colorectal
carcinoma compared with control tissues. This analysis involved 7
datasets in total (20-25),
TRPM6 was observed to be upregulated in colon and rectal carcinomas
compared with control tissues. There were no differences in the
expression levels of TRPM3, TRPM5, TRPM7 and TRPM8 between
colorectal cancer and control tissues.
 | Table IIIDatasets of TRPM protein family in
colorectal cancer. |
Table III
Datasets of TRPM protein family in
colorectal cancer.
| Gene | Dataset | Normal (n) | Tumor type (n) | Fold change | t-test | P-value |
|---|
| TRPM1 | TCGA | Colon (19) Rectum
(3) | Rectosigmoid
adenocarcinoma (3) | 3.482 | 9.492 |
9.11x10-5 |
| TRPM2 | TCGA | Colon (19) Rectum
(3) | Colon mucinous
adenocarcinoma (22) | 3.352 | 7.898 |
1.71x10-9 |
| | | Colon (19) Rectum
(3) | Colon
adenocarcinoma (101) | 3.045 | 9.359 |
6.79x10-15 |
| | | Colon (19) Rectum
(3) | Cecum
adenocarcinoma (22) | 3.063 | 7.595 |
3.31x10-9 |
| TRPM4 | Skrzypczak et al
(20) | Colon (10) | Colon adenoma
(5) | 2.923 | 10.06 |
1.18x10-7 |
| TRPM4 | Skrzypczak et al
(20) | Colon (10) | Colon carcinoma
epithelia (5) | -2.232 | -10.598 |
7.31x10-7 |
| | TCGA | Colon (19) Rectum
(3) | Colon
adenocarcinoma (101) | -2.297 | -10.673 |
1.32x10-16 |
| | | Colon (19) Rectum
(3) | Rectal
adenocarcinoma (60) | -2.264 | -9.519 |
1.17x10-14 |
| | Hong et al
(21) | Colon (12) | Colorectal
carcinoma (70) | -2.67 | -6.172 |
4.22x10-8 |
| | Skrzypczak et al
(20) | Colorectal Tissue
(24) | Colorectal
carcinoma (36) | -2.133 | -4.864 |
5.00x10-6 |
| TRPM6 | Skrzypczak et al
(20) | Colorectal Tissue
(24) | Colorectal
carcinoma (36) | -15.311 | -12.377 |
7.31x10-18 |
| | | Colorectal Tissue
(24) | Colorectal
adenocarcinoma (45) | -23.416 | -17.382 |
3.14x10-20 |
| | Sabates-Bellver
et al (22) | Ascending Colon (4)
Sigmoid colon (15) Descending colon (5) Transverse colon (1) Rectum
(7) | Rectal adenoma
(7) | -10.094 | -13.134 |
2.76x10-9 |
| | | Ascending colon (4)
Sigmoid colon (15) Descending colon (15) Transverse colon (1)
Rectum (7) | Colon adenoma
(25) | -29.071 | -14.421 |
2.09x10-17 |
| | Hong et al
(21) | Colon (12) | Colorectal
carcinoma (70) | -17.076 | -18.137 |
2.29x10-26 |
| | TCGA | Colon (19) Rectum
(3) | Cecum
adenocarcinoma (22) | -7.558 | -15.345 |
6.67x10-19 |
| | TCGA | Colon (19) Rectum
(3) | Colon mucinous
adenocarcinoma (22) | -7.851 | -13.889 |
6.58x10-17 |
| | TCGA | Colon (19) Rectum
(3) | Colon
adenocarcinoma (101) | -15.955 | -17.402 |
4.79x10-27 |
| | TCGA | Colon (19) Rectum
(3) | Rectal
adenocarcinoma (60) | -16.23 | -16.009 |
1.81x10-25 |
| | TCGA | Colon (19) Rectum
(3) | Rectosigmoid
adenocarcinoma (3) | -6.986 | -13.172 |
5.47x10-9 |
| | TCGA | Colon (19) Rectum
(3) | Rectal mucinous
adenocarcinoma (6) | -2.486 | -6.936 |
3.47x10-6 |
| | Skrzypczak et al
(23) | Colon (10) | Colon adenoma
(5) | -14.37 | -18.47 |
8.42x10-11 |
| | Skrzypczak et al
(23) | Colon (10) | Colon adenoma
epithelia (5) | -23.56 | -17.083 |
6.31x10-10 |
| | Skrzypczak et al
(23) | Colon (10) | Colon carcinoma
(5) | -41.197 | -25.401 |
1.64x10-10 |
| | Skrzypczak et al
(23) | Colon (10) | Colon carcinoma
epithelia (5) | -15.226 | -15.723 |
7.73x10-10 |
| | Gaedcke et al
(24) | Rectum (65) | Rectal
adenocarcinoma (65) | -2.345 | -17.187 |
2.83x10-10 |
| | Kaiser et al
(25) | Colon (5) | Rectal mucinous
adenocarcinoma (4) | -3.254 | -10.995 |
6.20x10-5 |
| | Kaiser et al
(25) | Colon (5) | Rectal
adenocarcinoma (8) | -2.893 | -8.959 |
2.39x10-5 |
| | Kaiser et al
(25) | Colon (5) | Rectosigmoid
adenocarcinoma (10) | -2.579 | -6.526 |
1.92x10-5 |
| | Kaiser et al
(25) | Colon (5) | Colon mucinous
adenocarcinoma (13) | -13.448 | -9.454 |
1.72x10-5 |
| | Kaiser et al
(25) | Colon (5) | Cecum
adenocarcinoma (17) | -2.538 | -7.776 |
3.94x10-5 |
| | Kaiser et al
(25) | Colon (5) | Colon
adenocarcinoma (41) | -2.785 | -9.252 |
6.54x10-5 |
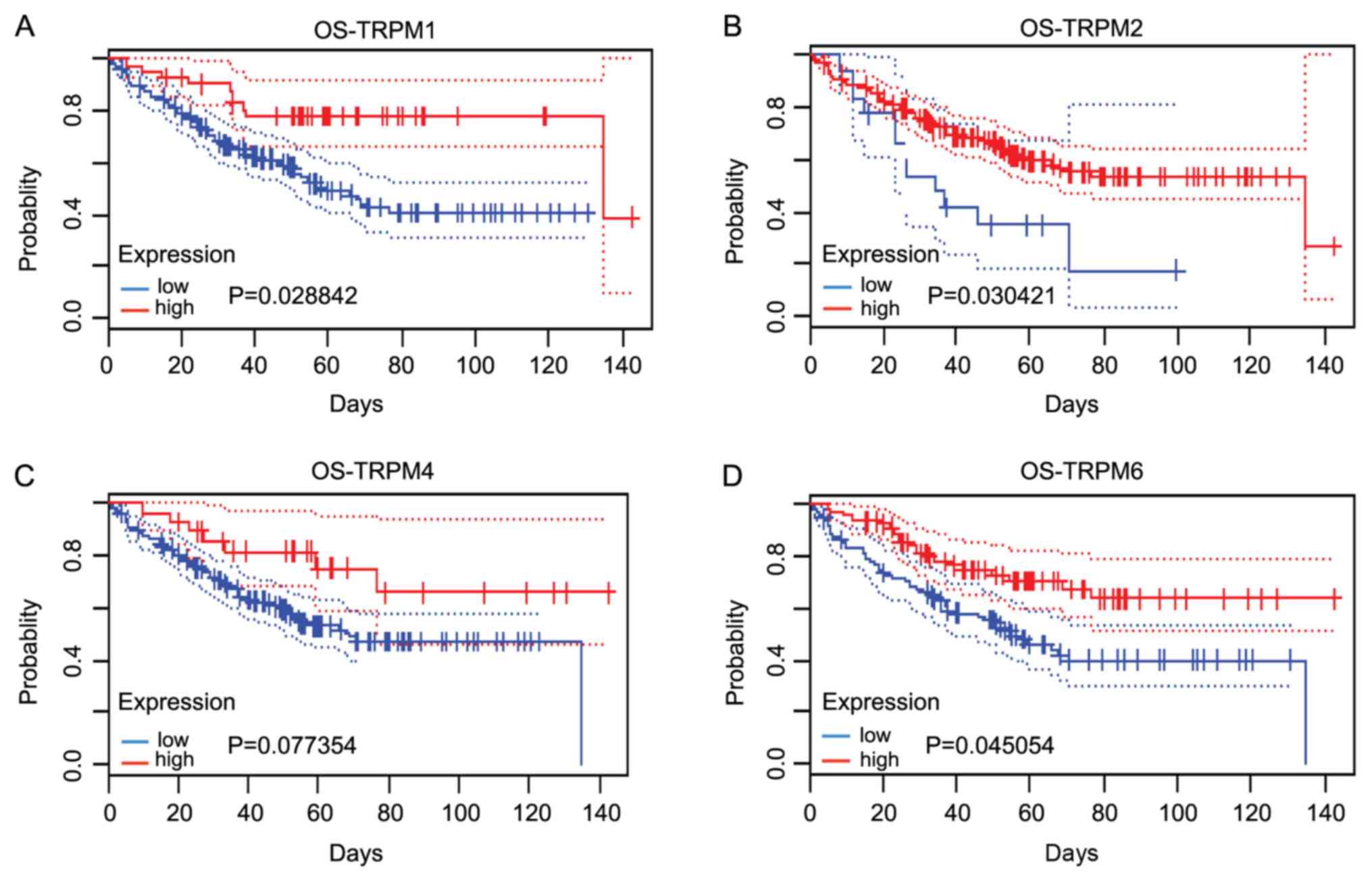
The associations between TRPM protein family (TRPM1,
TRPM2, TRPM4 and TRPM6) and the survival outcomes of patients with
colorectal cancer involving OS were determined using the PrognScan
database (12). It was revealed that
lower expression levels of TRPM1, TRPM2 and TRPM6 were associated
with poor prognoses in patients with colorectal cancer. The
aberrant regulation of TRPM1, TRPM2 and TRPM6 may contribute to the
tumorigenesis and development of colorectal cancer (Fig. 4A, B
and D). However, the expression of
TRPM4 was not statistically significant in terms of patient
prognoses (Fig. 4C).
Expression levels and prognostic
values of TRPMs in gastric cancer
In the dataset from D'Errico et al (26), TRPM1 and TRPM6 were decreased in
gastric mixed adenocarcinoma compared with in gastric mucosa.
According to Wang et al (27), it was revealed that TRPM3 levels were
decreased in gastric cancer compared with gastric mucosa and
gastric tissue. However, there was no difference observed in the
expression levels between gastric cancer and control tissue groups
in the other members of the TRPM protein family. The detailed
results are presented in Table
IV.
 | Table IVDatasets of TRPM protein family in
gastric cancer. |
Table IV
Datasets of TRPM protein family in
gastric cancer.
| Gene | Dataset | Normal (n) | Tumor type (n) | Fold change | t-test | P-value |
|---|
| TPRM1 | D' Errico et al
(26) | Gastric mucosa
(31) | Gastric mixed
adenocarcinoma (4) | -3.576 | -3.638 | 0.008 |
| TPRM3 | Wang et al
(27) | Gastric mucosa (12)
Gastric tissue (3) | Gastric cancer
(12) | -3.251 | -3.398 | 0.001 |
| TRPM6 | D' Errico et al
(26) | Gastric mucosa
(31) | Gastric mixed
adenocarcinoma (4) | -2.025 | -4.013 | 0.004 |
The present study then assessed the prognostic
effects of TRPM1, TRPM3 and TRPM6 in gastric cancer. The prognostic
effects of these genes are presented in Fig. 5. For intestinal-type patients, high
mRNA expression levels of TRPM1, TRPM3 and TPRM6 were significantly
associated with improved OS [TRPM1: Hazard ratio (HR)=1.4
(1.02-1.92); P=0.035; TRPM3: HR, 1.68 (1.23-2.31); P=0.0011; TRPM6:
HR=1.64 (1.11-2.42); P=0.012]. However, it was revealed that the
mRNA expression levels of TRPM1, TRPM3 and TRPM6 were not
associated with longer OS in patients with gastric mixed types and
diffuse types of cancer.
Expression levels and prognostic
values of TRPMs liver cancer
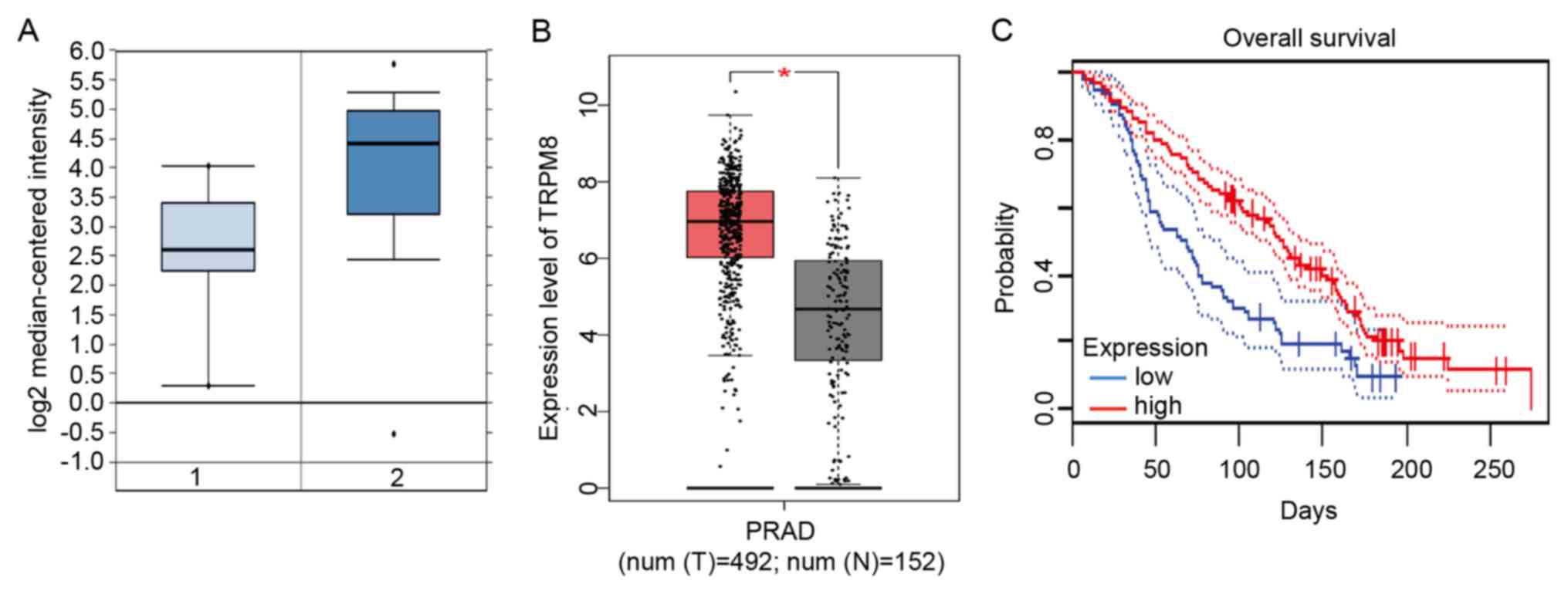
By analyzing the Oncomine database, only TRPM8 was
differentially expressed; its mRNA expression level was
significantly increased compared with that in the control liver
tissues (FC=-6.512; P=7.31E-09; Fig.
6A). In addition, the present study further determined that the
TRPM8 levels were significantly elevated in liver cancer tissues by
analyzing the GEPIA database (P<0.05; Fig. 6B). In addition, the present study
mapped the survival curves of patients with high (red) and low
(black) expression of liver cancer from the GEPIA database,
demonstrating that the OS time of patients with high TRPM8 gene
expression was significantly shorter (HR=0.69; P=0.041; Fig. 6C).
Expression levels and prognostic
values of TRPMs in prostate cancer
In the Oncomine database, 7 datasets possessed
significant differences between prostate cancer and control tissue
in total. According to Tomlins et al (28), TRPM1 and TRPM2 levels were decreased
in prostate cancer (TRPM1: FC=-2.195; t=-3.77;
P=3.12x10-4; TRPM2: FC=-2.455; t=-4.001;
P=1.75x10-4). However, the opposite conclusion was drawn
from a series of databases that demonstrated that TRPM4 and TRPM8
were increased in prostate cancer when compared with the normal
prostate (29-34).
The detailed results are presented in Table V.
 | Table VDatasets of TRPM protein family in
prostate cancer. |
Table V
Datasets of TRPM protein family in
prostate cancer.
| Gene | Dataset | Normal (n) | Tumor type (n) | Fold change | t-test | P-value |
|---|
| TRPM1 | Tomlins et al
(28) | Prostate gland
(23) | Prostatic
intraepithelial neoplasia (13) | -2.195 | -3.77 |
3.12x10-4 |
| TRPM2 | Tomlins et al
(28) | Prostate gland
(21) | Prostatic
intraepithelial neoplasia (13) | -2.455 | -4.001 |
1.75x10-4 |
| TRPM4 | Varambally et al
(29) | Prostate gland
(6) | Prostate carcinoma
(7) | 3.622 | 8.767 |
3.94x10-6 |
| | Liu et al
(30) | Prostate gland
(13) | Prostate carcinoma
(44) | 2.753 | 5.931 |
2.57x10-6 |
| | Vanaja et al
(31) | Prostate gland
(8) | Prostate
adenocarcinoma (27) | 3.937 | 6.464 |
4.24x10-7 |
| | Grasso et al
(32) | Prostate gland
(28) | Prostate carcinoma
(59) | 3.059 | 8.109 |
7.08x10-11 |
| | Arredouani et al
(33) | Prostate gland
(8) | Prostate carcinoma
(13) | 2.761 | 4.796 |
1.13x10-11 |
| | Wallace et al
(34) | Prostate gland
(20) | Prostate
adenocarcinoma (69) | 4.542 | 4.226 |
1.59x10-4 |
| TRPM8 | Vanaja et al
(31) | Prostate gland
(8) | Prostate
adenocarcinoma (27) | 2.883 | 3.082 | 0.005 |
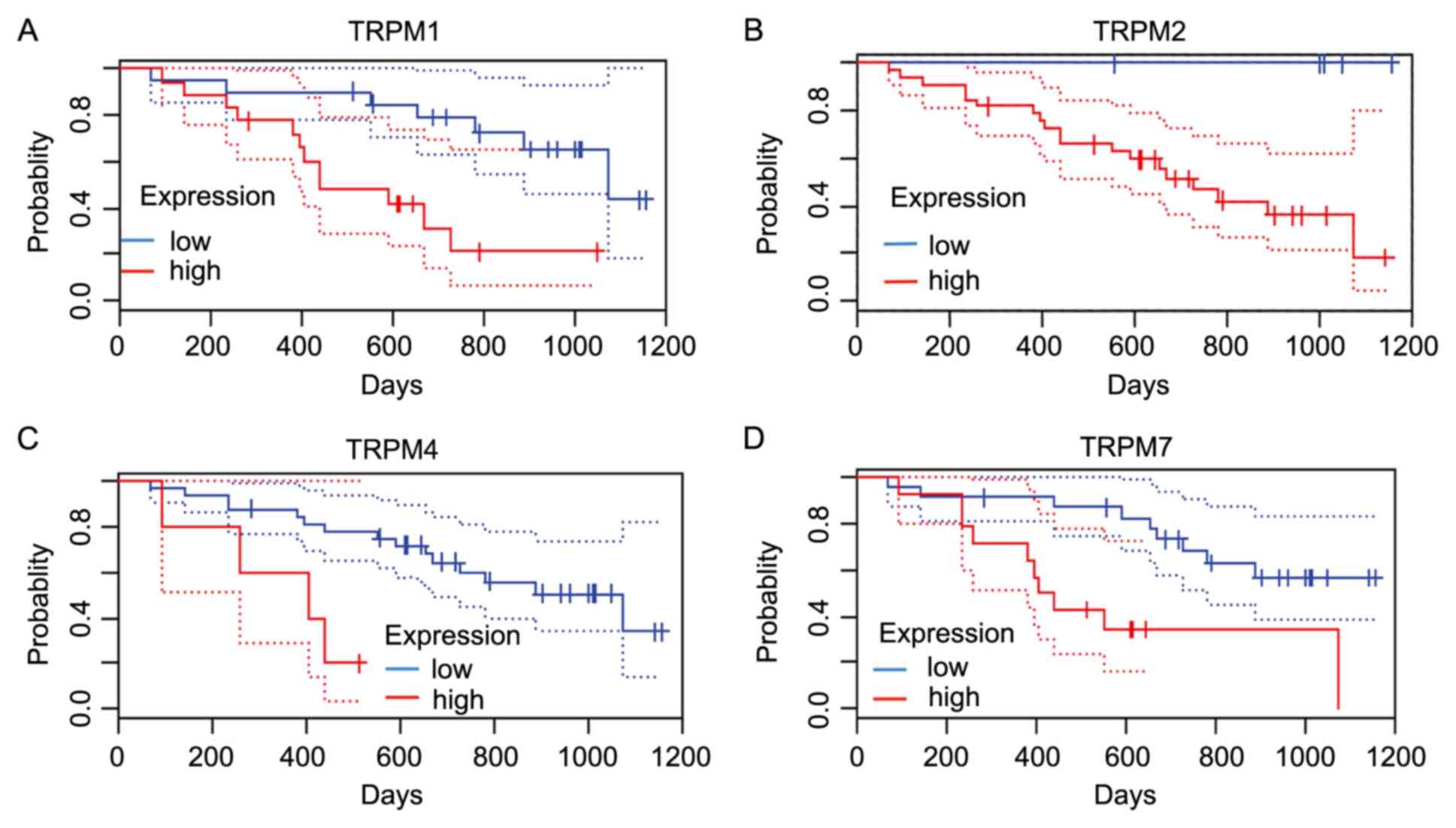
Subsequently, the present study evaluated the
prognostic effect of the TRPM protein family members (TRPM1, TRPM2,
TRPM4 and TRPM8) on the prognosis of prostate cancer through the
PrognoScan database. Only TRPM8 expression exhibited a
statistically significant association with the prognosis of the
patient [P=0.006968; HR=0.87 (0.78-0.96); Fig. 7].
Expression levels and prognostic
values of TRPMs in melanoma
A total of 3 datasets revealed significant
differences between melanoma and control tissues in the Oncomine
database. In the studies by Talantov et al (35) and Haqq et al (36), it was revealed that the level of
TRPM1 expression in control skin tissues was low, while it
increased markedly in melanoma samples. According to Haqq et
al (36), TRPM2 was also
demonstrated to be upregulated in melanoma tissues when compared
with control skin tissues. However, in the study conducted by Riker
et al (37), the expression
levels of TRPM4 and TRPM7 were markedly elevated in cutaneous
melanoma samples when compared with control tissues. The results
are presented in Table VI.
 | Table VIDatasets of TRPM protein family in
melanoma. |
Table VI
Datasets of TRPM protein family in
melanoma.
| Gene | Dataset | Normal (n) | Tumor type (n) | Fold change | t-test | P-value |
|---|
| TRPM1 | Talantov et al
(35) | Skin (7) | Benign melanocytic
skin nevus (18) | 34.333 | 7.586 |
4.13x10-7 |
| | Talantov et al
(35) | Skin (7) | Cutaneous melanoma
(45) | 19.17 | 8.278 |
2.83x10-5 |
| | Haqq et al
(36) | Skin (3) | Non-neoplastic
nevus (9) | 2.634 | 6.291 |
5.33x10-5 |
| TRPM2 | Haqq et al
(36) | Skin (3) | Melanoma (6) | 3.106 | 10.783 |
6.70x10-6 |
| | Haqq et al
(36) | Skin (3) | Non-neoplastic
nevus (9) | 2.316 | 7.136 |
1.58x10-5 |
| TRPM4 | Riker et al
(37) | Skin (4) | Cutaneous melanoma
(14) | -7.112 | -5.004 |
6.51x10-5 |
| TRPM7 | Riker et al
(37) | Skin (4) | Cutaneous melanoma
(14) | -2.601 | -3.968 |
5.59x10-4 |
To further assess the role of TRPM protein family in
cancer progression of patients with melanoma, the present study
used the PrognoScan database to calculate prognostic values based
on cox P<0.05(12). As presented
in Fig. 8, TRPM1 and TRPM4 were
significantly associated with OS.
Expression levels and prognostic
values of TRPMs in other types of cancer
The present study also analyzed the transcriptional
level of TRPMs in certain other types of solid tumor. It was
suggested that the most marked differences were observed in kidney
cancer, esophageal cancer, brain and central nervous system (CNS)
cancer, and head and neck cancer (Fig.
1). All the detailed analyses of the aforementioned cancer
types are summarized in Table VII.
For clear cell renal cell carcinoma, which is the most common type
of kidney cancer, all genes were downregulated (38-42).
TRPM4, TPRM5 and TRPM8 in the study by Yusenko et al
(43) were increased in renal
oncocytoma samples when compared with the control group. However,
there were no statistically significant differences observed in
TRPM2 and TRPM6 levels between kidney cancer and control tissues.
TRPM4 expression was upregulated in esophageal cancer, while TRPM1
and TRPM8 expression levels were decreased (44-46).
In the cases of brain and CNS cancer, TRPM2, TRPM3 and TRPM6 were
expressed at low levels in different types of brain and CNS cancer
(47-51).
However, TRPM8 was elevated in glioblastoma when compared with
control brain tissues in the data by Murat et al (51) and Lee et al (49). Notably, only TRPM1 was expressed at
an increased level in head and neck squamous cell carcinoma when
compared with buccal mucosa (FC=-5.324; t, -8.031; P=1.52E-10)
according to the study by Ginos et al (52).
 | Table VIIDatasets of TRPM family in other
cancers. |
Table VII
Datasets of TRPM family in other
cancers.
| Cancer type | Gene | Dataset | Normal (n) | Tumor type (n) | Fold change | t-test | P-value |
|---|
| Kidney | TRPM1 | Cutcliffe et al
(38) | Fetal kidney
(3) | Renal wilms tumor
(18) | -2.541 | -3.605 |
9.45x10-4 |
| | | Jones et al
(39) | Kidney (23) | Clear cell renal
cell carcinoma (23) | -4.321 | -16.61 |
3.60x10-16 |
| | | Jones et al
(39) | Kidney (23) | Renal pelvis
urothelial carcinoma (8) | -3.927 | -15.642 |
4.53x10-8 |
| | TRPM3 | Cutcliffe et al
(38) | Fetal kidney
(3) | Renal wilms tumor
(18) | -2.484 | -9.971 |
3.28x10-9 |
| | | Cutcliffe et al
(38) | Fetal kidney
(3) | Clear cell sarcoma
of the kidney (14) | -2.3 | -8.486 |
2.07x10-7 |
| | | Yusenko et al
(43) | Fetal kidney (2)
Kidney (3) | Renal wilms tumor
(4) | -15.445 | -8.899 |
3.90x10-5 |
| | | Yusenko et al
(43) | Fetal kidney (2)
Kidney (3) | Chromophobe renal
cell carcinoma (4) | -24.806 | -7.897 |
3.06x10-4 |
| | | Yusenko et al
(43) | Fetal kidney (2)
Kidney (3) | Renal oncocytoma
(4) | -12.037 | -6.976 |
3.16x10-4 |
| | | Yusenko et al
(43) | Fetal kidney (2)
Kidney (3) | Papillary renal
cell carcinoma (19) | -2.491 | -3.007 | 0.004 |
| | | Yusenko et al
(43) | Fetal kidney (2)
Kidney (3) | Clear cell renal
cell carcinoma (26) | -2.155 | -2.816 | 0.006 |
| | | Jones et al
(39) | Kidney (23) | Renal pelvis
urothelial carcinoma (8) | -2.588 | -10.534 |
1.22x10-11 |
| | | Jones et al
(39) | Kidney (23) | Chromophobe renal
cell carcinoma (6) | -2.097 | -13.197 |
2.10x10-9 |
| | | Jones et al
(39) | Kidney (23) | Clear cell renal
cell carcinoma (23) | -2.48 | -6.97 |
8.02x10-9 |
| | | Gumz et al
(40) | Kidney (10) | Clear cell renal
cell carcinoma (10) | -3.346 | -7.187 |
6.29x10-7 |
| | | Beroukhim et al
(41) | Renal cortex (10)
Renal tissue (1) | Non-hereditary
clear cell renal cell carcinoma (27) | -5.56 | -6.922 |
5.03x10-8 |
| | | Beroukhim et al
(41) | Renal cortex (10)
Renal tissue (1) | Hereditary clear
cell Renal cell carcinoma (32) | -4.687 | -7.061 |
1.26x10-7 |
| | | Lenburg et al
(42) | Kidney (9) | Clear cell renal
cell carcinoma (9) | -2.934 | -5.54 |
6.54x10-5 |
| | TRPM4 | Yusenko et al
(43) | Fetal kidney (2)
Kidney (3) | Renal oncocytoma
(4) | 3.218 | 5.827 |
3.24x10-4 |
| | | Yusenko et al
(43) | Fetal kidney (2)
Kidney (3) | Renal oncocytoma
(4) | -8.912 | -11.645 |
5.16x10-6 |
| | | Beroukhim et al
(41) | Renal cortex (10)
Renal tissue (1) | Hereditary clear
cell Renal cell carcinoma (32) | -2.236 | -8.049 |
9.29x10-8 |
| | | Gumz et al
(40) | Kidney (10) | Clear cell renal
cell carcinoma (10) | -3.562 | -4.352 |
2.26x10-4 |
| | TRPM5 | Yusenko et al
(43) | Fetal kidney (2)
Kidney (3) | Renal wilms tumor
(4) | 9.955 | 3.917 | 0.003 |
| | TRPM7 | Yusenko et al
(43) | Fetal kidney (2)
Kidney (3) | Renal wilms tumor
(4) | -2.16 | -6.705 |
6.05x10-4 |
| | TRPM8 | Yusenko et al
(43) | Fetal kidney (2)
Kidney (3) | Papillary renal
cell carcinoma (19) | 22.217 | 4.332 |
5.07x10-4 |
| Esophageal | TRPM1 | Hao et al
(44) | Duodenum (13)
Esophagus (14) | Esophageal
Adenocarcinoma (5) | -3.222 | -4.282 |
3.87x10-4 |
| | TRPM4 | Kimchi et al
(45) | Esophagus (8) | Barrett's esophagus
(8) | 4.661 | 3.301 | 0.003 |
| | | Kimchi et al
(45) | Esophagus (8) | Esophageal
adenocarcinoma (8) | 4.448 | 3.535 | 0.002 |
| | | Kim et al
(46) | Esophagus (28) | Barrett's esophagus
(15) | 3.233 | 8.597 |
3.60x10-8 |
| | TRPM8 | Kimchi et al
(45) | Esophagus (8) | Esophageal
adenocarcinoma (8) | -3.315 | -3.413 | 0.003 |
| Brain and CNS | TRPM2 | Liang et al
(47) | Brain (2)
Cerebellum (1) | Oligoastrocytoma
(3) | -2.694 | -4.518 | 0.007 |
| | | Bredel et al
(48) | Brain (4) | Anaplastic
oligodendroglioma (3) | -4.071 | -5.62 | 0.005 |
| | TRPM3 | Lee et al
(49) | Neural stem cell
(3) | Glioblastoma
(22) | 4.661 | 3.301 |
5.67x10-5 |
| | | Sun et al
(50) | Brain (23) | Oligodendroglioma
(50) | -2.286 | -8.417 |
3.95x10-12 |
| | | Sun et al
(50) | Brain (23) | Glioblastoma
(81) | -2.447 | -10.147 |
2.32x10-17 |
| | | TCGA | Brain (10) | Brain glioblastoma
(542) | -16.791 | -16.791 |
1.92x10-9 |
| | | Murat et al
(51) | Brain (4) | Glioblastoma
(80) | -2.46 | -5.563 | 0.001 |
| | TRPM6 | Sun et al
(50) | Brain (23) | Diffuse astrocytoma
(7) | -2.5 | -3.865 | 0.001 |
| | TRPM8 | Murat et al
(51) | Brain (4) | Glioblastoma
(80) | 2.49 | 8.432 |
4.39x10-11 |
| | | Lee et al
(49) | Neural stem cell
(3) | Glioblastoma
(22) | 5.257 | 6.712 |
4.98x10-6 |
| Head and neck | TRPM1 | Ginos et al
(52) | Buccal Mucosa
(13) | Head and neck
squamous cell carcinoma (41) | -5.324 | -8.031 | 1.52
x10-10 |
Subsequently, the present study further analyzed the
association between the TRPMs and the survival rate of patients in
all the aforementioned types of cancer using the PrognoScan
database (12). In conclusion, the
effect of the TRPM protein family on the prognosis of renal cancer
was not significant. In particular, the high expression of TRPM8
was associated with poor OS in patients with esophageal cancer
[P=0.001214; HR=225.46 (8.47-6004.05)]. As for brain cancer,
increasing TRPM6 levels were associated with poor prognosis
[P=0.010649; HR=3.70 (1.36-10.09)]. However, there may be no
association between this protein family with the survival outcomes
of patients with head and neck cancer.
Discussion
Despite increasing advances in early diagnosis and
treatment options, cancer remains a significant cause of morbidity
and mortality worldwide (1,53,54).
Tumor formation and metastasis is a complex process, and is the
result of various gene dysregulation events and cellular processes,
including tumorigenesis, basement membrane degradation, matrix
permeability, cell adhesion and angiogenesis. Ca2+
signaling pathways are necessary for regulation of the cell cycle,
cell proliferation and apoptosis, and are involved in the process
of tumorigenesis (55). TRPM, one
superfamily of the TRP cation channel, contributes to the
regulation of intracellular Ca2+ concentration (56,57). The
TRPM protein family consists of 8 structural and functional
channels that are widely expressed in a number of different types
of tissue and have diverse physiological functions (8). The TRPM protein family may serve as
triggers for enhanced proliferation and aberrant differentiation,
which leads to the pathogenic proliferative and invasive
characteristics of cancer (58).
Differences in expression of the TRPM channels may provide a new
basis for tumor diagnosis, and may be a novel target for cancer
therapy. The present study used the Oncomine database to
systematically analyze the mRNA expression levels of the TRPM
protein family in different types of tumor, and assessed the
prognostic values using the Kaplan-Meier plotter, and PrognoScan
and GEPIA databases.
Breast cancer remains the most common type of
malignant tumor and the leading cause of cancer-associated
mortality among women worldwide (1,59). Due
to its high heterogeneity, it is necessary to constantly
investigate new biomarkers in order to distinguish different
subtypes and predict their clinical behavior and therapeutic
response (60-62).
The expression of TRPM2 and TRPM4 were upregulated in ductal
carcinoma and invasive breast cancer when compared with control
breast tissue. By contrast, TCGA database demonstrated that TRPM3
and TRPM6 were downregulated in invasive breast tumors. The results
from the present study suggested that TRPM2, TRPM3, TRPM4 and TRPM6
may be used as molecular biomarkers to identify breast cancer
invasion (63). The Kaplan-Meier
analysis demonstrated that decreased TRPM2 may be used to predict
prognosis in patients with Luminal B breast cancer and
HER2+ breast cancer subtypes. High expression of TRPM4
and TRPM6 indicated lower survival rates in patients with basal and
HER2+ subtypes. In addition, according to the analysis
of the present study, TPRM3 may be used as a biomarker for the
Luminal B breast cancer subtype. The results suggested that certain
members of the TRPM family may be potential biomarkers and targets
for new breast cancer therapies. Associations between TRPM proteins
and breast cancer continue to be identified as a result of rapid
advances in molecular biology and genetics research (9). In addition, TRPM6 somatic mutations
have also been observed in an independent cohort of breast cancer
samples (64).
Lung cancer has the highest rates of incidence and
mortality in China (65) and around
the world (66). The present study
systemically analyzed the expression and prognostic value of TRPMs
in lung cancer. The results indicated that the decreased expression
levels of TRPM1 and TRPM2, and increased expression levels of TRPM6
in lung adenocarcinoma may serve an important role in lung cancer
tumorigenesis. The present study revealed that transcriptional
TRPM2 is a novel prognostic biomarker for lung adenocarcinoma;
consistent with the results of Huang et al (67), which demonstrate that the knockdown
of TRPM2-antisense also significantly inhibited cell
proliferation.
Colorectal cancer is the third most common diagnosed
type of cancer in humans which poses a significant public health
issue worldwide, with >1.8 million cases diagnosed each year
(1,68). As a result of the numerous studies
investigating TRPM channels and colorectal cancer, tumor treatment
options are becoming more diverse and accurate for colorectal
cancer. The present study revealed that TRPM1, TRPM2 and TRPM6 may
serve as diagnostic markers for the prognosis of colorectal cancer
development and are useful targets for pharmaceutical
interventions. These data provide evidence to support the
hypothesis that TRPM1, TRPM2 and TRPM6 serve a crucial role in
tumor growth and metastasis formation (8).
The results of the present study may contribute to a
more complete understanding of the expression levels and prognostic
values of TRPM family members in certain solid tumors, including
gastric cancer, which causes nearly 1 million mortalities worldwide
each year (1,69,70).
Certain cell channels, including TRP, are more active or are
upregulated in gastric cancer cells (70). The present study suggested that the
abnormal regulation of TRPM1, TRPM2 and TRPM3 may be vital in the
development of intestinal type gastric cancer. They may participate
in different stages of tumorigenesis. Not all TRPM channels have
been investigated thoroughly and the current literature base
remains inadequate. The results from the present study regarding
TRPM2 expression are in concordance with the data from the study by
Almasi et al (71), which
suggested that TRPM2 knockdown inhibits cell proliferation, and
promotes apoptosis in gastric cancer cells. However, research is
currently focused on TRPM7, and there are few studies on TRPM1 and
TRPM3. Therefore, future studies investigating these specific
proteins are required.
The present study aimed to assess the importance of
TRPMs in liver cancer, the fourth most common cause of mortality
associated with cancer (1). The most
significant finding from the present study was that the OS time of
patients with liver cancer exhibiting increased TRPM8 expression
levels was significantly shorter compared with those patients with
decreased TRPM8 expression. This suggested that TRPM8 may be a
novel marker for liver cancer survival and prognostic accuracy.
However, these results must be interpreted with caution, as further
work is required in order to establish the viability of this new
marker.
Prostate cancer is a common form of cancer in adult
males which is responsible for one-fourth of all incident cancer
cases in western countries, with its incidence continuing to
increase (66,72). The present study suggested that TRPM8
served a key role in mediating the biological behavior of prostate
tumors, consistent with the results of numerous independent studies
demonstrating that TRPM8 was important for the survival, migration
and invasion of prostate cancer cells (73,74).
Deeds et al (75) demonstrated that TRPM1 was expressed
at high levels in poorly metastatic variants of the melanoma cell
line. The Oncomine database and the Kaplan-Meier plotter survival
analyses performed in the present study also demonstrated that
TRPM1 was considered to be a tumor activator of melanoma. The data
implied that low TRPM1 expression levels were associated with
decreased OS rates in comparison with high TRPM1 levels. When
examining TRPM4, the results of the present study also suggested
that it may serve as a factor in regulating melanoma proliferation,
apoptosis and necrosis.
In the present study, it was also revealed that the
downregulation of TRPM4 expression was associated with improved OS
in patients with glioma. In addition, the potential association
between TRPM8 and esophageal cancer OS was measured, and the
results implied that TRPM8 may be a prognostic marker and potential
therapeutic target for esophageal cancer. However, this family
appears to not be associated with OS in kidney cancer and head and
neck cancer.
In summary, the results of the present study
indicated that certain members of the TRPM protein family exhibit
significant differences in mRNA expression levels between cancer
and control tissues. A number of these proteins may be useful
biomarkers for cancer prognosis, and may represent novel anticancer
targets.
Acknowledgements
Not applicable.
Funding
The present study was supported by Projects from the
Scientific Research Fund of Zhejiang Provincial Education
Department (grant no. Y201941836), the Natural Science Foundation
of Zhejiang Province (grant no. LGF19H090007) and the Traditional
Chinese Medicine Science and Technology Plan of Zhejiang Province
(grant no. 2019ZB030).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
FHQ and XLM were responsible for the Oncomine and
Kaplan-Meier plotter analysis and writing the original draft. LDL
was involved in the PrognoScan analysis and interpretation of the
data, and revising the manuscript critically for important
intellectual content. MHH was involved in revising the manuscript
and participated in the interpretation of data. HT made
contributions to the acquistion of data. XHJ was involved in the
GEPIA analysis and interpretation of the data. JPZ was responsible
for the conception, design of the study and revising the manuscript
critically for important intellectual content. All authors reviewed
and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Fidler MM, Soerjomataram I and Bray F: A
global view on cancer incidence and national levels of the human
development index. Int J Cancer. 139:2436–2446. 2016.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Montell C and Rubin GM: Molecular
characterization of the Drosophila trp locus: A putative integral
membrane protein required for phototransduction. Neuron.
2:1313–1323. 1989.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Schmitz C and Perraud AL: The TRPM cation
channels in the immune context. Curr Pharm Design. 11:2765–2778.
2005.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Zeng X, Sikka SC, Huang L, Sun C, Xu C,
Jia D, Abdel-Mageed AB, Pottle JE, Taylor JT and Li M: Novel role
for the transient receptor potential channel TRPM2 in prostate
cancer cell proliferation. Prostate Cancer Prostatic Dis.
13:195–201. 2010.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Armisen R, Marcelain K, Simon F, Tapia JC,
Toro J, Quest AF and Stutzin A: TRPM4 enhances cell proliferation
through up-regulation of the β-catenin signaling pathway. J Cell
Physiol. 226:103–109. 2011.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Meng X, Cai C, Wu J, Cai S, Ye C, Chen H,
Yang Z, Zeng H, Shen Q and Zou F: TRPM7 mediates breast cancer cell
migration and invasion through the MAPK pathway. Cancer Lett.
333:96–102. 2013.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Hantute-Ghesquier A, Haustrate A,
Prevarskaya N and Lehen'kyi V: TRPM family channels in cancer.
Pharmaceuticals (Basel). 11(2)2018.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Wong KK, Banham AH, Yaacob NS and Nur
Husna SM: The oncogenic roles of TRPM ion channels in cancer. J
Cell Physiol, 2019.
|
|
10
|
Nagy Á, Lánczky A, Menyhárt O and Győrffy
B: Validation of miRNA prognostic power in hepatocellular carcinoma
using expression data of independent datasets. Sci Rep.
8(9227)2018.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Li Q, Birkbak NJ, Gyorffy B, Szallasi Z
and Eklund AC: Jetset: Selecting an optimal microarray probe set to
represent a gene. BMC Bioinformatics. 12(474)2011.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Mizuno H, Kitada K, Nakai K and Sarai A:
PrognoScan: A new database for meta-analysis of the prognostic
value of genes. BMC Med Genomics. 2(18)2009.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Tang Z, Li C, Kang B, Gao G, Li C and
Zhang Z: GEPIA: A web server for cancer and normal gene expression
profiling and interactive analyses. Nucleic Acids Res. 45:W98–W102.
2017.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2012. CA Cancer J Clin. 62:10–29. 2012.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Gyorffy B, Lanczky A, Eklund AC, Denkert
C, Budczies J, Li Q and Szallasi Z: An online survival analysis
tool to rapidly assess the effect of 22,277 genes on breast cancer
prognosis using microarray data of 1,809 patients. Breast Cancer
Res Treat. 123:725–731. 2010.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Bhattacharjee A, Richards WG, Staunton J,
Li C, Monti S, Vasa P, Ladd C, Beheshti J, Bueno R, Gillette M, et
al: Classification of human lung carcinomas by mRNA expression
profiling reveals distinct adenocarcinoma subclasses. Proc Natl
Acad Sci USA. 98:13790–13795. 2001.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Garber ME, Troyanskaya OG, Schluens K,
Petersen S, Thaesler Z, Pacyna-Gengelbach M, van de Rijn M, Rosen
GD, Perou CM, Whyte RI, et al: Diversity of gene expression in
adenocarcinoma of the lung. Proc Natl Acad Sci USA. 98:13784–13789.
2001.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Okayama H, Kohno T, Ishii Y, Shimada Y,
Shiraishi K, Iwakawa R, Furuta K, Tsuta K, Shibata T, Yamamoto S,
et al: Identification of genes upregulated in ALK-positive and
EGFR/KRAS/ALK-negative lung adenocarcinomas. Cancer Res.
72:100–111. 2012.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Gyorffy B, Surowiak P, Budczies J and
Lanczky A: Online survival analysis software to assess the
prognostic value of biomarkers using transcriptomic data in
non-small-cell lung cancer. PLoS One. 8(e82241)2013.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Skrzypczak M, Goryca K, Rubel T, Paziewska
A, Mikula M, Jarosz D, Pachlewski J, Oledzki J and Ostrowski J:
Modeling oncogenic signaling in colon tumors by multidirectional
analyses of microarray data directed for maximization of analytical
reliability. PLoS One. 5(pii: e13091)2010.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Hong Y, Downey T, Eu KW, Koh PK and Cheah
PY: A ‘metastasis-prone’ signature for early-stage mismatch-repair
proficient sporadic colorectal cancer patients and its implications
for possible therapeutics. Clin Exp Metastasis. 27:83–90.
2010.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Sabates-Bellver J, Van der Flier LG, de
Palo M, Cattaneo E, Maake C, Rehrauer H, Laczko E, Kurowski MA,
Bujnicki JM, Menigatti M, et al: Transcriptome profile of human
colorectal adenomas. Mol Cancer Res. 5:1263–1275. 2007.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Skrzypczak M, Goryca K, Rubel T, Paziewska
A, Mikula M, Jarosz D, Pachlewski J, Oledzki J and Ostrowski J:
Modeling oncogenic signaling in colon tumors by multidirectional
analyses of microarray data directed for maximization of analytical
reliability. PLoS One. 5(pii: e13091)2010.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Gaedcke J, Grade M, Jung K, Camps J, Jo P,
Emons G, Gehoff A, Sax U, Schirmer M, Becker H, et al: Mutated KRAS
results in overexpression of DUSP4, a MAP-kinase phosphatase and
SMYD3, a histone methyltransferase, in rectal carcinomas. Genes
Chromosomes Cancer. 49:1024–1034. 2010.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Kaiser S, Park YK, Franklin JL, Halberg
RB, Yu M, Jessen WJ, Freudenberg J, Chen X, Haigis K, Jegga AG, et
al: Transcriptional recapitulation and subversion of embryonic
colon development by mouse colon tumor models and human colon
cancer. Genome Biol. 8(R131)2007.PubMed/NCBI View Article : Google Scholar
|
|
26
|
D'Errico M, de Rinaldis E, Blasi MF, Viti
V, Falchetti M, Calcagnile A, Sera F, Saieva C, Ottini L, Palli D,
et al: Genome-wide expression profile of sporadic gastric cancers
with microsatellite instability. Eur J Cancer. 45:461–469.
2009.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Wang Q, Wen YG, Li DP, Xia J, Zhou CZ, Yan
DW, Tang HM and Peng ZH: Upregulated INHBA expression is associated
with poor survival in gastric cancer. Med Oncol. 29:77–83.
2012.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Tomlins SA, Mehra R, Rhodes DR, Cao X,
Wang L, Dhanasekaran SM, Kalyana-Sundaram S, Wei JT, Rubin MA,
Pienta KJ, et al: Integrative molecular concept modeling of
prostate cancer progression. Nat Genet. 39:41–51. 2007.PubMed/NCBI View Article : Google Scholar
|
|
29
|
VVarambally S, Yu J, Laxman B, Rhodes DR,
Mehra R, Tomlins SA, Shah RB, Chandran U, Monzon FA, Becich MJ, et
al: Integrative genomic and proteomic analysis of prostate cancer
reveals signatures of metastatic progression. Cancer Cell.
8:393–406. 2005.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Liu P, Ramachandran S, Ali Seyed M,
Scharer CD, Laycock N, Dalton WB, Williams H, Karanam S, Datta MW,
Jaye DL, et al: Sex-determining region Y box 4 is a transforming
oncogene in human prostate cancer cells. Cancer Res. 66:4011–4019.
2006.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Vanaja DK, Cheville JC, Iturria SJ and
Young CYF: Transcriptional silencing of zinc finger protein 185
identified by expression profiling is associated with prostate
cancer progression. Cancer Res. 63:3877–3882. 2003.PubMed/NCBI
|
|
32
|
Grasso CS, Wu YM, Robinson DR, Cao X,
Dhanasekaran SM, Khan AP, Quist MJ, Jing X, Lonigro RJ, Brenner JC,
Asangani IA, et al: The mutational landscape of lethal
castration-resistant prostate cancer. Nature. 487:239–243.
2012.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Arredouani MS, Lu B, Bhasin M, Eljanne M,
Yue W, Mosquera JM, Bubley GJ, Li V, Rubin MA, Libermann TA, et al:
Identification of the transcription factor single-minded homologue
2 as a potential biomarker and immunotherapy target in prostate
cancer. Clin Cancer Res. 15:5794–5802. 2009.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Wallace TA, Prueitt RL, Yi M, Howe TM,
Gillespie JW, Yfantis HG, Stephens RM, Caporaso NE, Loffredo CA and
Ambs S: Tumor immunobiological differences in prostate cancer
between African-American and European-American men. Cancer Res.
68:927–936. 2008.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Talantov D, Mazumder A, Yu JX, Briggs T,
Jiang Y, Backus J, Atkins D and Wang Y: Novel genes associated with
malignant melanoma but not benign melanocytic lesions. Clin Cancer
Res. 11:7234–7242. 2005.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Haqq C, Nosrati M, Sudilovsky D, Crothers
J, Khodabakhsh D, Pulliam BL, Federman S, Miller JR III, Allen RE,
Singer MI, et al: The gene expression signatures of melanoma
progression. Proc Natl Acad Sci USA. 102:6092–6097. 2005.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Riker AI, Enkemann SA, Fodstad O, Liu S,
Ren S, Morris C, Xi Y, Howell P, Metge B, Samant RS, et al: The
gene expression profiles of primary and metastatic melanoma yields
a transition point of tumor progression and metastasis. BMC Med
Genomics. 1(13)2008.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Cutcliffe C, Kersey D, Huang CC, Zeng Y,
Walterhouse D and Perlman EJ: Clear cell sarcoma of the kidney:
up-regulation of neural markers with activation of the sonic
hedgehog and Akt pathways. Clin Cancer Res. 11:7986–7994.
2005.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Jones J1, Otu H, Spentzos D, Kolia S, Inan
M, Beecken WD, Fellbaum C, Gu X, Joseph M, Pantuck AJ, et al: Gene
signatures of progression and metastasis in renal cell cancer. Clin
Cancer Res. 11:5730–5739. 2005.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Gumz ML1, Zou H, Kreinest PA, Childs AC,
Belmonte LS, LeGrand SN, Wu KJ, Luxon BA, Sinha M, Parker AS, et
al: Secreted frizzled-related protein 1 loss contributes to tumor
phenotype of clear cell renal cell carcinoma. Clin Cancer Res.
13:4740–4749. 2007.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Beroukhim R1, Brunet JP, Di Napoli A,
Mertz KD, Seeley A, Pires MM, Linhart D, Worrell RA, Moch H, Rubin
MA, et al: Patterns of gene expression and copy-number alterations
in von-hippel lindau disease-associated and sporadic clear cell
carcinoma of the kidney. Cancer Res. 69:4674–4681. 2009.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Lenburg ME, Liou LS, Gerry NP, Frampton
GM, Cohen HT and Christman MF: Previously unidentified changes in
renal cell carcinoma gene expression identified by parametric
analysis of microarray data. BMC Cancer. 3(31)2003.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Yusenko MV, Kuiper RP, Boethe T, Ljungberg
B, van Kessel AG and Kovacs G: High-resolution DNA copy number and
gene expression analyses distinguish chromophobe renal cell
carcinomas and renal oncocytomas. BMC Cancer. 9(152)2009.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Hao Y, Triadafilopoulos G, Sahbaie P,
Young HS, Omary MB and Lowe AW: Gene expression profiling reveals
stromal genes expressed in common between Barrett's esophagus and
adenocarcinoma. Gastroenterology. 131:925–933. 2006.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Kimchi ET, Posner MC, Park JO, Darga TE,
Kocherginsky M, Karrison T, Hart J, Smith KD, Mezhir JJ,
Weichselbaum RR, et al: Progression of Barrett's metaplasia to
adenocarcinoma is associated with the suppression of the
transcriptional programs of epidermal differentiation. Cancer Res.
65:3146–3154. 2005.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Kim SM, Park YY, Park ES, Cho JY, Izzo JG,
Zhang D, Kim SB, Lee JH, Bhutani MS, Swisher SG, Wu X, Coombes KR,
et al: Prognostic biomarkers for esophageal adenocarcinoma
identified by analysis of tumor transcriptome. PLoS One.
5(e15074)2010.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Liang Y, Diehn M, Watson N, et al: Gene
expression profiling reveals molecularly and clinically distinct
subtypes of glioblastoma multiforme. Proc Natl Acad Sci U S A.
102:5814–5819. 2005.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Bredel M1, Bredel C, Juric D, Harsh GR,
Vogel H, Recht LD and Sikic BI: Functional network analysis reveals
extended gliomagenesis pathway maps and three novel MYC-interacting
genes in human gliomas. Cancer Res. 65:8679–8689. 2005.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Lee J, Kotliarova S, Kotliarov Y, Li A, Su
Q, Donin NM, Pastorino S, Purow BW, Christopher N, Zhang W, et al:
Tumor stem cells derived from glioblastomas cultured in bFGF and
EGF more closely mirror the phenotype and genotype of primary
tumors than do serum-cultured cell lines. Cancer Cell. 9:391–403.
2006.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Sun L1, Hui AM, Su Q, Vortmeyer A,
Kotliarov Y, Pastorino S, Passaniti A, Menon J, Walling J, Bailey
R, et al: Neuronal and glioma-derived stem cell factor induces
angiogenesis within the brain. Cancer Cell. 9:287–300.
2006.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Murat A, Migliavacca E, Gorlia T, Lambiv
WL, Shay T, Hamou MF, de Tribolet N, Regli L, Wick W, Kouwenhoven
MC, et al: Stem cell-related ‘self-renewal’ signature and high
epidermal growth factor receptor expression associated with
resistance to concomitant chemoradiotherapy in glioblastoma. J Clin
Oncol. 26:3015–3024. 2008.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Ginos MA, Page GP, Michalowicz BS, Patel
KJ, Volker SE, Pambuccian SE, Ondrey FG, Adams GL and Gaffney PM:
Identification of a gene expression signature associated with
recurrent disease in squamous cell carcinoma of the head and neck.
Cancer Res. 64:55–63. 2004.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Maule M and Merletti F: Cancer transition
and priorities for cancer control. Lancet Oncol. 13:745–746.
2012.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Bray F, Soerjomataram I, Mery L and Ferlay
J: Improving the quality and coverage of cancer registries
globally. Lancet. 386:1035–1036. 2015.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Chen J, Luan Y, Yu R, Zhang Z, Zhang J and
Wang W: Transient receptor potential (TRP) channels, promising
potential diagnostic and therapeutic tools for cancer. Biosci
Trends. 8:1–10. 2014.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Clapham DE: TRP channels as cellular
sensors. Nature. 426:517–524. 2003.PubMed/NCBI View Article : Google Scholar
|
|
57
|
Pedersen SF, Owsianik G and Nilius B: TRP
channels: An overview. Cell Calcium. 38:233–252. 2005.PubMed/NCBI View Article : Google Scholar
|
|
58
|
Gkika D and Prevarskaya N: Molecular
mechanisms of TRP regulation in tumor growth and metastasis.
Biochim Biophys Acta. 1793:953–958. 2009.PubMed/NCBI View Article : Google Scholar
|
|
59
|
Winters S, Martin C, Murphy D and Shokar
NK: Breast cancer epidemiology, prevention and screening. Prog Mol
Biol Transl. 151:1–32. 2017.PubMed/NCBI View Article : Google Scholar
|
|
60
|
Dai X, Xiang L, Li T and Bai Z: Cancer
hallmarks, biomarkers and breast cancer molecular subtypes. J
Cancer. 7:1281–1294. 2016.PubMed/NCBI View Article : Google Scholar
|
|
61
|
Dai X, Li T, Bai Z, Yang Y, Liu X, Zhan J
and Shi B: Breast cancer intrinsic subtype classification, clinical
use and future trends. Am J Cancer Res. 5:2929–2943.
2015.PubMed/NCBI
|
|
62
|
Spitale A, Mazzola P, Soldini D,
Mazzucchelli L and Bordoni A: Breast cancer classification
according to immunohistochemical markers: Clinicopathologic
features and short-term survival analysis in a population-based
study from the South of Switzerland. Ann Oncol. 20:628–635.
2009.PubMed/NCBI View Article : Google Scholar
|
|
63
|
Sumoza-Toledo A, Espinoza-Gabriel MI and
Montiel-Condado D: Evaluation of the TRPM2 channel as a biomarker
in breast cancer using public databases analysis. Bol Med Hosp
Infant Mex. 73:397–404. 2016.PubMed/NCBI View Article : Google Scholar
|
|
64
|
Stephens PJ, Tarpey PS, Davies H, Van Loo
P, Greenman C, Wedge DC, Nik-Zainal S, Martin S, Varela I, Bignell
GR, et al: The landscape of cancer genes and mutational processes
in breast cancer. Nature. 486:400–404. 2012.PubMed/NCBI View Article : Google Scholar
|
|
65
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016.PubMed/NCBI View Article : Google Scholar
|
|
66
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2017. CA Cancer J Clin. 67:7–30. 2017.PubMed/NCBI View Article : Google Scholar
|
|
67
|
Huang C, Qin Y, Liu H, Liang N, Chen Y, Ma
D, Han Z, Xu X, Zhou X, He J and Li S: Downregulation of a novel
long noncoding RNA TRPM2-AS promotes apoptosis in non-small cell
lung cancer. Tumour Biol. 39(1010428317691191)2017.PubMed/NCBI View Article : Google Scholar
|
|
68
|
Arnold M, Sierra MS, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global patterns and trends in
colorectal cancer incidence and mortality. Gut. 66:683–691.
2017.PubMed/NCBI View Article : Google Scholar
|
|
69
|
Bray F, Ren JS, Masuyer E and Ferlay J:
Global estimates of cancer prevalence for 27 sites in the adult
population in 2008. Int J Cancer. 132:1133–1145. 2013.PubMed/NCBI View Article : Google Scholar
|
|
70
|
Kim BJ, Kim SY, Lee S, Jeon JH, Matsui H,
Kwon YK, Kim SJ and So I: The role of transient receptor potential
channel blockers in human gastric cancer cell viability. Can J
Physiol Pharmacol. 90:175–186. 2012.PubMed/NCBI View Article : Google Scholar
|
|
71
|
Almasi S, Kennedy BE, El-Aghil M, Sterea
AM, Gujar S, Partida-Sánchez S and El Hiani Y: TRPM2
channel-mediated regulation of autophagy maintains mitochondrial
function and promotes gastric cancer cell survival via the
JNK-signaling pathway. J Biol Chem. 293:3637–3650. 2018.PubMed/NCBI View Article : Google Scholar
|
|
72
|
Jemal A, Siegel R, Ward E, Hao Y, Xu J and
Thun MJ: Cancer statistics, 2009. CA Cancer J Clin. 59:225–249.
2009.PubMed/NCBI View Article : Google Scholar
|
|
73
|
Bidaux G, Borowiec AS, Dubois C, Delcourt
P, Schulz C, Vanden Abeele F, Lepage G, Desruelles E, Bokhobza A,
Dewailly E, et al: Targeting of short TRPM8 isoforms induces
4TM-TRPM8-dependent apoptosis in prostate cancer cells. Oncotarget.
7:29063–29080. 2016.PubMed/NCBI View Article : Google Scholar
|
|
74
|
Peng M, Wang Z, Yang Z, Tao L, Liu Q, Yi
LU and Wang X: Overexpression of short TRPM8 variant a promotes
cell migration and invasion and decreases starvation-induced
apoptosis in prostate cancer LNCaP cells. Oncol Lett. 10:1378–1384.
2015.PubMed/NCBI View Article : Google Scholar
|
|
75
|
Deeds J, Cronin F and Duncan LM: Patterns
of melastatin mRNA expression in melanocytic tumors. Hum Pathol.
31:1346–1356. 2000.PubMed/NCBI
|