Introduction
Colorectal cancer (CRC) is one of the most common
malignancies worldwide (1,2). The routine therapeutic strategy for CRC
is surgery combined with platinum-based treatment (3,4).
Oxaliplatin, as a platinum-based drug, inhibits the progression of
CRC by binding to and cross-linking the DNA of cancer cells
(3). Although advances have been
made in the treatment of CRC, the overall survival rate of CRC
patients remains unsatisfactory due to chemoresistance and
recurrence (5,6). A previous study indicated that
chemoresistance emerges in over 20% of all CRC patients within 6
months of therapy (7). Therefore,
exploration of novel therapies to enhance the chemosensitivity of
CRC cells to oxaliplatin is imperative.
MicroRNAs (miRNAs/miRs) are a class of non-coding
RNAs. Increasing evidence demonstrated that miRNAs are involved in
various physiological and pathological processes, including cell
proliferation, metastasis, invasion and apoptosis, and
dysregulation of certain miRNAs is associated with multiple types
of cancer by acting as either oncogenic or tumor-suppressive agents
(8-12).
miR-96 plays a vital role in the progression of various cancer
types (13,14). Studies indicated that miR-96 has a
role as an oncogene or tumor suppressor, depending on the type of
cancer (13-16).
In CRC, miR-96 demonstrated to act as an oncogene and increase cell
proliferation (17,18). However, the potential function of
miR-96 in regulating the sensitivity of CRC to oxaliplatin and the
associated mechanisms remain to be elucidated.
In the present study, the possible role of miR-96 in
regulating the resistance of CRC cells to oxaliplatin was
investigated. The proliferation and apoptotic rates of CRC cells
treated with oxaliplatin and miR-96 inhibitor was determined.
Subsequently, the expression of proliferation- and
apoptosis-associated genes was examined after treatment with
oxaliplatin and miR-96 inhibitor. The results indicated that
knockdown of miR-96 enhances the sensitivity of CRC cells to
oxaliplatin.
Materials and methods
Tissue samples
A total of 40 CRC tissues and 40 adjacent tumor
tissues were obtained from 40 patients (25 males and 15 females;
age range 48-74 years) diagnosed with CRC at the First People's
Hospital of Lianyungang (China) between Jan 2016 and Oct 2017. The
tissue samples were confirmed by pathological examination and
immediately stored in liquid nitrogen after surgery. Tumor size was
recorded as the diameter of the tumor; metastasis was confirmed as
positive when tumor cells were found on the nearby lymph nodes and
the degree of differentiation of the tumor was determined by
histological examination. All patients provided written informed
consent. The present study was approved by the Ethics Committee of
the First People's Hospital of Lianyungang (China).
Cell culture and transfection
The CRC cell line SW480 was purchased from the
American Type Culture Collection. Cells were cultured in RPMI-1640
medium (Gibco; Thermo Fisher Scientific, Inc.) containing 10% FBS
(Gibco; Thermo Fisher Scientific, Inc.) and 1%
penicillin/streptomycin (Gibco; Thermo Fisher Scientific, Inc.) at
35˚C in an atmosphere with 5% CO2. miR-96 inhibitor,
miR-negative control (miR-NC), small interfering RNA (siRNA)
targeting tropomyosin 1 (TPM1) and siRNA NC were designed and
provided by Guangzhou RiboBio Co., Ltd. The cells were transfected
with miR-96 inhibitor, miR-NC, miR-96 mimics, siRNA TPM1 and siRNA
NC using Lipofectamine™ 3000 (Invitrogen; Thermo Fisher Scientific,
Inc.) for 48 h. A total of 20 µmol/l oxaliplatin (Sigma-Aldrich;
Merck KGaA) was used to investigate oxaliplatin sensitivity. The
sequences of the miRNAs and siRNAs used were: miR-96 inhibitor,
5'-GCAAAAAUGUGCUAGUGCCAAA-3'; miR-NC, 5'-CAGUACUUUUGUGUAGUACAA-3';
miR-96 mimic, 5'-UUUGGCACUAGCACAUUUUUGC-3', siRNA TPM1
5'-CCCGTAAGCTGGTCATCAT-3' and siRNA NC,
5'-CCCAACGGTTGACTGTCAT-3'.
CCK-8 assay
Cells were seeded into a 96-well plate at a density
of 2x103 cells/well. At 0, 8, 16 and 44 h, the cells
were stained with 20 µl CCK-8 solution (Beyotime Institute of
Biotechnology) for 4 h according to the manufacturer's
instructions. Subsequently, the absorbance rate of the cells at a
wavelength of 450 nm was examined with a spectrophotometer (Thermo
Fisher Scientific, Inc.).
Flow cytometry
Cells were trypsinized and collected by
centrifugation at room temperature at 1,000 x g for 5 min. The
cells were then washed with pre-cooled PBS and resuspended in
binding buffer (Beyotime Institute of Biotechnology) in the
centrifuge tubes. Subsequently, the cells were stained with
Annexin-V FITC and propidium iodide (BD Biosciences). Finally, the
apoptotic rate of SW480 cells was determined with a flow cytometer
(EPICS XL; Beckman Coulter) and analyzed using FlowJo software
(version 10.5.3; FlowJo LLC).
Reverse transcription-quantitative PCR
(RT-qPCR)
Cells were lysed with TRIzol® reagent
(Invitrogen; Thermo Fisher Scientific, Inc.) and total RNA was
extracted according to the manufacturer's instructions. RNA was
reverse transcribed to generate cDNA with the PrimeScript RT
Reagent kit (Takara Bio, Inc.). Subsequently, qPCR was performed on
an ABI Prism 7500 Real-Time PCR system (Thermo Fisher Scientific,
Inc.) using the SYBR™ Green PCR Master Mix (cat. no. 431270;
Applied Biosystems;, Thermo Fisher Scientific, Inc.). The following
thermocycling conditions were used: Initial denaturation at 95˚C
for 10 min; followed by 40 cycles of 95˚C for 15 sec and 60˚C for
30 sec. The relative RNA expression was determined using the
2-∆∆Cq method (19). U6 and GAPDH served as the internal
reference gene for miRNA and mRNA, respectively. Each experiment
was performed in triplicate. The following primer pairs were used
for the qPCR: miR-96 forward, 5'-TTTGGCACTAGCACAT-3' and reverse,
5'-GAGCAGGCTGGAGAA-3'; U6 forward, 5'-ATTGGAACGATACAGAGAAGAT-3' and
reverse, 5'-GGAACGCTTCACGAATTT-3'; TPM1 forward,
5'-CTCTCAACGATATGACTTCCA-3' and reverse,
5'-TTTTTTTAGCTTACACAGTGTT-3'; Bcl-2 forward,
5'-TTCTTTGAGTTCGGTGGGGTC-3' and reverse,
5'-TGCATATTTGTTTGGGGCAGG-3'; BAX forward,
5'-TCCACCAAGAAGCTGAGCGAG-3' and reverse,
5'-GTCCAGCCCATGATGGTTCT-3'; GAPDH forward,
5'-CGAGCCACATCGCTCAGACA-3' and reverse,
5'-GTGGTGAAGACGCCAGTGGA-3'.
Western blotting
Cells or tissues were lysed and total protein was
extracted using RIPA buffer (Sigma-Aldrich; Merck KGaA). Protein
concentration was measured with a bicinchoninic acid kit (Pierce;
Thermo Fisher Scientific, Inc.). Subsequently, 30 µg of
protein/lane was separated by 10% SDS-PAGE and transferred onto
PVDF membranes. The membranes were blocked with 5% non-fat milk for
1 h at 37˚C under exclusion of direct light. Subsequently, the
membranes were incubated with the following primary antibodies (all
purchased from Abcam) overnight at 4˚C: Anti-Bcl-2 (cat. no.
ab59348; 1:1,000), anti-BAX (cat. no. ab32503; 1:1,000), anti-TPM1
(cat. no. ab55915; 1:1,000) and anti-GAPDH (cat. no. ab9485;
1:2,000). The membranes were then incubated with horseradish
peroxidase-labeled secondary antibodies (cat. no. ab6721; 1:5,000;
Abcam) at 37˚C for 1 h. Protein signal was visualized with an ECL
detection system (Thermo Fisher Scientific, Inc.) and the density
of the protein bands was evaluated with a sensitive
chemiluminescent substrate (Sigma-Aldrich; Merck KGaA). GAPDH was
used as an internal control. Densitometric analysis was performed
using Image J (version 2.1.4; National Institutes of Health).
Dual-luciferase reporter assay
The online database TargetScan (http://www.targetscan.org/vert_71/) predicted
TPM1 as a target gene of miR-96. The wild-type or mutant sequence
from the 3'-untranslated region (3'-UTR) of TPM1 containing the
binding site for miR-96 was cloned into the luciferase gene
reporter vector pMIR (Thermo Fisher Scientific, Inc.). The reporter
plasmids were then co-transfected with miR-96 mimics or miR-NC into
SW480 cells with Lipofectamine® 2000 transfection
reagent for 48 h (Invitrogen; Thermo Fisher Scientific, Inc.). A
Dual-Lumi™ Dual Luciferase Reporter Assay kit (Beyotime Institute
of Biotechnology) was used to detect luciferase activity on a
GloMax Explorer Multimode microplate reader (Promega Corp.).
Luciferase activity was normalized to Renilla luciferase
activity.
Statistical analysis
SPSS 18.0 (SPSS, Inc.) was used to analyze the data.
Values are expressed as the mean ± SD. Paired Student's t-test was
performed to compare the expression of miR-96 and TPM1 in CRC
tissues and adjacent normal tissues. One-way ANOVA with Tukey's
post hoc test or two-way ANOVA with Sidak's multiple comparisons
test was applied to analyze differences among multiple groups. The
χ2 test was used for the analysis of clinical
information presented in Table I.
Pearson's correlation coefficient was used for correlation
analysis. P<0.05 was considered to indicate a statistically
significant difference.
 | Table IClinical information of patients. |
Table I
Clinical information of patients.
| | miR-96
expression | |
|---|
| Parameters | High (relative
expression of miR-96 in CRC tissue >1; n=25) | Low (relative
expression of miR-96 in CRC tissue <1; n=15) | P-value |
|---|
| Sex | | | 0.4638 |
|
Male | 17 | 8 | |
|
Female | 8 | 7 | |
| Age | | | 0.8003 |
|
<60 | 16 | 9 | |
|
≥60 | 9 | 6 |
| Tumor size (cm) | | | 0.0258a |
|
≥5 | 14 | 3 | |
|
<5 | 11 | 12 | |
| Differentiation | | | 0.4119 |
|
Well-moderate | 10 | 8 | |
|
Poor | 15 | 7 | |
| Metastasis | | | 0.0222a |
|
Negative | 9 | 11 | |
|
Positive | 16 | 4 | |
Results
miR-96 is upregulated in CRC
tissues
RT-qPCR was performed to determine the expression of
miR-96 in CRC tissues and normal tissues. The results indicated
that the expression level of miR-96 in CRC tissues was
significantly higher compared with adjacent tissues (Fig. 1). Moreover, increased expression of
miR-96 was positively associated with the tumor size and metastasis
(Table I).
TPM1 is a direct target of miR-96
The online database TargetScan predicted that miR-96
directly binds to the 3'-UTR of TPM1 (Fig. 2A). The expression level of TPM1 in
CRC tissues was significantly lower compared with adjacent tissues
(Fig. 2B), and the expression of
miR-96 and TPM1 was negatively correlated (r=-0.4459; P=0.0039;
Fig. 2C). To further confirm the
targeting interaction, a dual-luciferase reporter assay was
performed. The results demonstrated that the relative luciferase
activity of CRC cells transfected with miR-96 mimics and a
luciferase reporter vector containing the wild-type sequence of the
3'-UTR of TPM1 was significantly decreased compared with the NC
group, while there were no significant differences between miR-96
mimic- and NC-transfected cells in the mutant group (Fig. 2D).
Downregulation of miR-96 increases the
sensitivity of CRC cells to oxaliplatin via targeting TPM1
Cells were transfected with miR-96 inhibitor and a
CCK-8 assay and flow cytometry were performed to determine the cell
viability and apoptosis of CRC cells following different
treatments. As presented in Fig. 3,
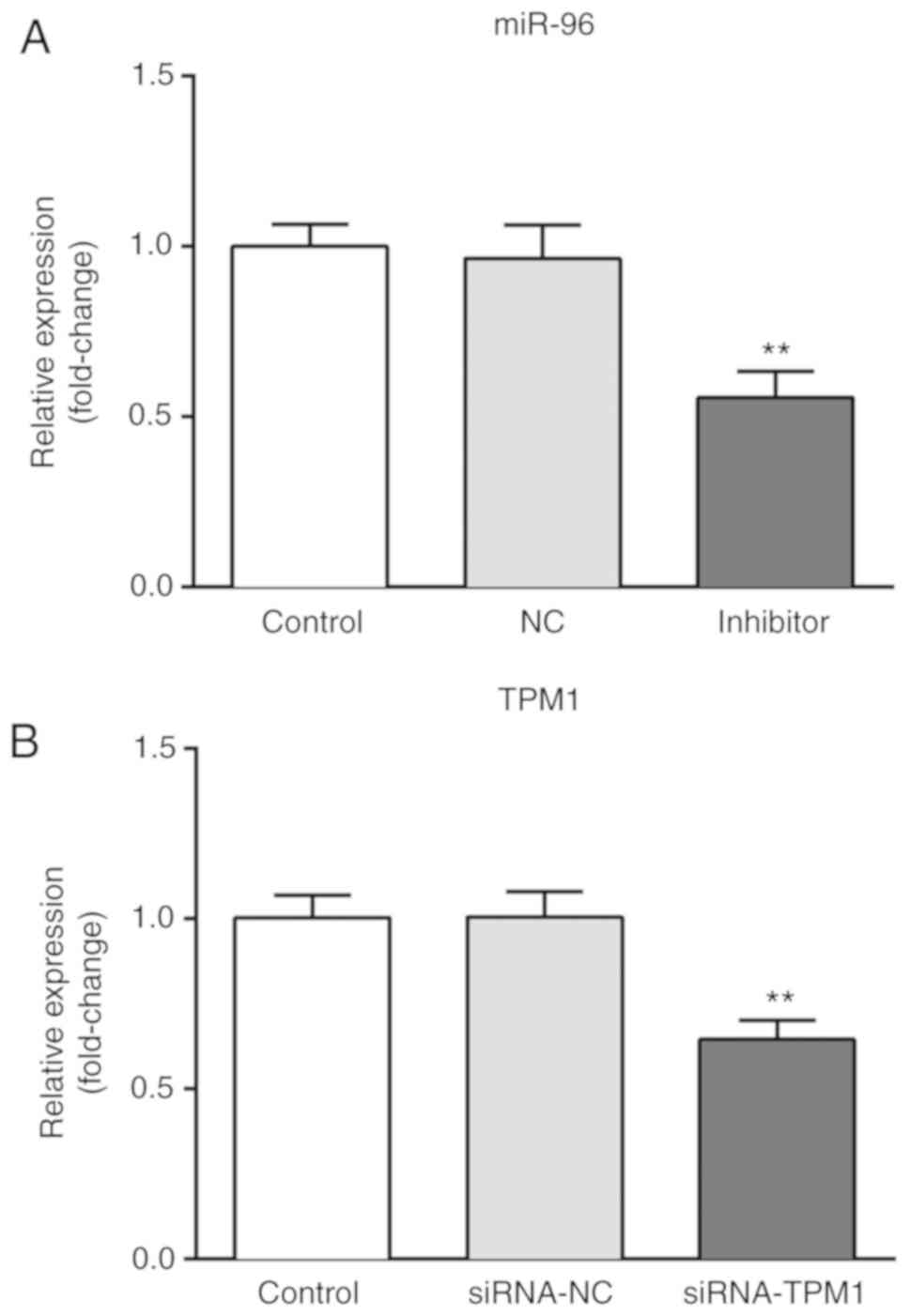
the expression of miR-96 was significantly downregulated following
transfection with miR-96 inhibitor compared with the miR-NC group
(Fig. 3A), and the expression of
TPM1 was significantly downregulated following transfection with
siRNA-TPM1 compared with the siRNA-NC group (Fig. 3B), suggesting that TPM1 knockdown was
successfully performed. Furthermore, the results of the CCK-8 assay
indicated that oxaliplatin significantly inhibited the viability of
SW480 cells compared with the blank control group, and the cell
viability was markedly decreased in the oxaliplatin + miR-96
inhibitor group compared with the oxaliplatin group (Fig. 4A). In addition, the apoptotic rate of
the SW480 cells in the oxaliplatin group was significantly higher
compared with the control group, and the apoptotic rate in the
oxaliplatin + miR-96 inhibitor group significantly increased
compared with the oxaliplatin group (Fig. 4B). Transfection of siRNA TPM1 in
oxaliplatin + miR-96 inhibitor-treated cells significantly
decreased cell viability and increased cell apoptosis compared with
the oxaliplatin + miR-96 inhibitor group (Fig. 4).
Inhibition of miR-96 decreases the
expression of Bcl-2 and BAX, and increases expression of TPM1
As presented in Fig.
5, oxaliplatin significantly decreased the mRNA and protein
expression of Bcl-2, and increased the expression of TPM1 and BAX
compared with the control group. Changes in protein expression were
more apparent following treatment with oxaliplatin + miR-96
inhibitor and were attenuated in cells co-transfected with siRNA
TPM1.
Discussion
Although advances have been made in the treatment of
CRC in recent years (20), multidrug
resistance remains the major obstacle of chemotherapy (3). Aberrant expression of certain miRNAs
has crucial roles in the development of chemoresistance (21). The tumor suppressor gene
miR-410 is downregulated in lung cancer tissues and induced
apoptosis in lung cancer cells (22). miR-25 is downregulated in melanoma
cells and promoted the progression of melanoma (23). In the present study, the potential
roles of miR-96 in CRC were investigated. The results indicated
that miR-96 was upregulated in CRC tissues. Previous studies also
showed that the expression of miR-96 is increased in CRC tissues
and that miR-96 enhanced the viability of CRC cells (17,18).
Therefore, it was hypothesized that knockdown of miR-96 may reverse
the tumorigenic behavior of CRC cells. Considering the potential of
miRNA in the chemotherapy of cancers, the present study focused on
the effects of downregulation of miR-96 on the sensitivity of CRC
cells to oxaliplatin.
Oxaliplatin, as a first-line anticancer agent, is a
preferential therapeutic for CRC (3). In the present study, oxaliplatin
suppressed the viability and promoted the apoptosis of CRC cells.
Of note, miR-96 was reported to be associated with the sensitivity
of cancer cells to oxaliplatin (24); however, whether miR-96 affects the
chemosensitivity of CRC cells remains to be elucidated. The present
study indicated that miR-96 inhibitor + oxaliplatin was more potent
in inhibiting cell viability and increasing cell apoptosis of CRC
cells compared with oxaliplatin alone, which demonstrated that
knockdown of miR-96 enhanced the sensitivity of CRC cells to
oxaliplatin. However, the underlying molecular mechanisms remained
elusive.
Previous findings demonstrated that miRNAs regulate
the expression of genes by targeting the 3'-UTR of their target
genes (9). TPM1, predicted as a
target gene of miR-96 with the online database TargetScan 7.1, has
a crucial role in mediating cell proliferation and differentiation
(25). Furthermore, TPM1 is
downregulated in CRC, suggesting that TPM1 may act as a tumor
suppressor in CRC (25). Of note,
knockdown of TPM1 in miR-96 inhibitor-treated CRC cells regulated
the behavior of CRC cells, including proliferation and apoptosis.
Treatment with oxaliplatin + miR-96 inhibitor reduced the
proliferation and promoted apoptosis of CRC cells, while this
effect was inhibited by silencing of TPM1. Therefore, it may be
inferred that TPM1 has positive effects on the chemosensitivity of
CRC cells.
To further investigate the underlying molecular
mechanisms, the expression of proliferation- and
apoptosis-associated factors, BAX and Bcl-2 was then examined. The
results indicated that compared with oxaliplatin alone, treatment
with oxaliplatin + miR-96 inhibitor was more potent in
downregulating the expression of Bcl-2 and upregulating TPM1 and
Bax in CRC cells, while this effect was inhibited by
co-transfection of siRNA TPM1. These results suggested that
suppression of miR-96 enhanced the chemosensitivity of CRC cells,
which was negatively affected by siRNA TPM1.
As a limitation, the present study was limited to
in vitro experimentation using cell lines. miRNAs may have
various targets, or a target may be regulated by various upstream
genes, which is to be further investigated in the future. Moreover,
the current study only included one cell line. The roles of miR-96
should also be investigated in other CRC cells in future work.
In conclusion, the present study indicated that
miR-96 was upregulated in CRC. Knockdown of miR-96 enhanced the
chemosensitivity of CRC cells to oxaliplatin via targeting TPM1.
These results may lay a foundation for further studies for the
potential clinical application of miR-96 as a therapeutic for the
treatment of CRC.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
TG performed most experiments and statistical
analysis, and revised the manuscript. PX and HM performed cell
experiments. ST performed statistical analysis. JZ performed
clinical experiments, literature research and wrote part of the
manuscript. YZ designed the study and drafted the manuscript. All
authors read and approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of the First People's Hospital of Lianyungang. All
patients provided written informed consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Liebs S, Keilholz U, Kehler I, Schweiger
C, Hayback J and Nonnenmacher A: Detection of mutations in
circulating cell-free DNA in relation to disease stage in
colorectal cancer. Cancer Med. 8:3761–3769. 2019.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Lei L, Zhao X, Liu S, Cao Q, Yan B and
Yang J: MicroRNA-3607 inhibits the tumorigenesis of colorectal
cancer by targeting DDI2 and regulating the DNA damage repair
pathway. Apoptosis. 24:662–672. 2019.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Wang Q, Wei J, Wang C, Zhang T, Huang D,
Wei F, He F, Cai W, Yang P, Zeng S, et al: Gambogic acid reverses
oxaliplatin resistance in colorectal cancer by increasing
intracellular platinum levels. Oncol Lett. 16:2366–2372.
2018.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Chibaudel B, Tournigand C, Bonnetain F,
Maindrault-Goebel F, Lledo G, André T, Larsen AK, Bengrine-Lefevre
L, Louvet C and de Gramont A: Platinum-sensitivity in metastatic
colorectal cancer: Towards a definition. Eur J Cancer.
49:3813–3820. 2013.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Huang CY, Chiang SF, Chen WT, Ke TW, Chen
TW, You YS, Lin CY, Chao KSC and Huang CY: HMGB1 promotes
ERK-mediated mitochondrial Drp1 phosphorylation for chemoresistance
through RAGE in colorectal cancer. Cell Death Dis.
9(1004)2018.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Ren J, Ding L, Zhang D, Shi G, Xu Q, Shen
S, Wang Y, Wang T and Hou Y: Carcinoma-associated fibroblasts
promote the stemness and chemoresistance of colorectal cancer by
transferring exosomal lncRNA H19. Theranostics. 8:3932–3948.
2018.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Zhang Z, Feng L, Liu P and Duan W: ANRIL
promotes chemoresistance via disturbing expression of ABCC1 by
regulating the expression of Let-7a in colorectal cancer. Biosci
Rep. 38(pii: BSR20180620)2018.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Gupta P, Sata TN, Yadav AK, Mishra A, Vats
N, Hossain MM, Sanal MG and Venugopal SK: TGF-β induces liver
fibrosis via miRNA-181a-mediated down regulation of augmenter of
liver regeneration in hepatic stellate cells. PLoS One.
14(e0214534)2019.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Huerta-Zavala ML, Lopez-Castillejos ES,
Requenez-Contreras JL, Granados-Riveron JT and Aquino-Jarquin G: A
single miRNA and miRNA sponge expression system for efficient
modulation of miR-223 availability in mammalian cells. J Gene Med.
21(e3100)2019.PubMed/NCBI View
Article : Google Scholar
|
|
10
|
Wu T, Cao Y, Yang Y and Zhang X, Wang S,
Xu LP and Zhang X: A three-dimensional DNA walking machine for the
ultrasensitive dual-modal detection of miRNA using a fluorometer
and personal glucose meter. Nanoscale. 11:11279–11284.
2019.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Powrozek T, Brzozowska A, Mazurek M, Mlak
R, Sobieszek G and Malecka-Massalska T: Combined analysis of
miRNA-181a with phase angle derived from bioelectrical impedance
predicts radiotherapy-induced changes in body composition and
survival of male patients with head and neck cancer. Head Neck.
41:3247–3257. 2019.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Vos PD, Leedman PJ, Filipovska A and
Rackham O: Modulation of miRNA function by natural and synthetic
RNA-binding proteins in cancer. Cell Mol Life Sci. 76:3745–3752.
2019.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Xie W, Sun F, Chen L and Cao X: miR-96
promotes breast cancer metastasis by suppressing MTSS1. Oncol Lett.
15:3464–3471. 2018.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Bao YH, Wang Y, Liu Y, Wang S and Wu B:
MiR-96 expression in prostate cancer and its effect on the target
gene regulation. Eur Rev Med Pharmacol Sci. 21:4548–4556.
2017.PubMed/NCBI
|
|
15
|
Hong Y, Liang H, Uzair Ur Rehman, Wang Y,
Zhang W, Zhou Y, Chen S, Yu M, Cui S, Liu M, et al: miR-96 promotes
cell proliferation, migration and invasion by targeting PTPN9 in
breast cancer. Sci Rep. 6(37421)2016.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Wu L, Pu X, Wang Q, Cao J, Xu F, Xu LI and
Li K: miR-96 induces cisplatin chemoresistance in non-small cell
lung cancer cells by downregulating SAMD9. Oncol Lett. 11:945–952.
2016.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Rapti SM, Kontos CK, Papadopoulos IN and
Scorilas A: High miR-96 levels in colorectal adenocarcinoma predict
poor prognosis, particularly in patients without distant metastasis
at the time of initial diagnosis. Tumour Biol. 37:11815–11824.
2016.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Ress AL, Stiegelbauer V, Winter E,
Schwarzenbacher D, Kiesslich T, Lax S, Jahn S, Deutsch A,
Bauernhofer T, Ling H, et al: MiR-96-5p influences cellular growth
and is associated with poor survival in colorectal cancer patients.
Mol Carcinog. 54:1442–1450. 2015.PubMed/NCBI View
Article : Google Scholar
|
|
19
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Wu J, Wang F, Liu X, Zhang T, Liu F, Ge X,
Mao Y and Hua D: Correlation of IDH1 and B7H3 expression with
prognosis of CRC patients. Eur J Surg Oncol. 44:1254–1260.
2018.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Fang L, Li H, Wang L, Hu J, Jin T, Wang J
and Yang BB: MicroRNA-17-5p promotes chemotherapeutic drug
resistance and tumour metastasis of colorectal cancer by repressing
PTEN expression. Oncotarget. 5:2974–2987. 2014.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Zhang JR, Zhu RH and Han XP: MiR-410
affects the proliferation and apoptosis of lung cancer A549 cells
through regulation of SOCS3/JAK-STAT signaling pathway. Eur Rev Med
Pharmacol Sci. 22:5994–6001. 2018.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Jiang QQ and Liu WB: miR-25 promotes
melanoma progression by regulating RNA binding motif protein 47.
Med Sci (Paris). 34 Focus issue F1:59–65. 2018.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Guo Y, Pang Y, Gao X, Zhao M, Zhang X,
Zhang H, Xuan B and Wang Y: MicroRNA-137 chemosensitizes colon
cancer cells to the chemotherapeutic drug oxaliplatin (OXA) by
targeting YBX1. Cancer Biomark. 18:1–9. 2017.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Mlakar V, Berginc G, Volavsek M, Stor Z,
Rems M and Glavac D: Presence of activating KRAS mutations
correlates significantly with expression of tumour suppressor genes
DCN and TPM1 in colorectal cancer. BMC Cancer.
9(282)2009.PubMed/NCBI View Article : Google Scholar
|