Introduction
Coronary artery disease (CAD) is the first cause of
morbidity and mortality all around the world. During the last 30
years, important progress was made in order to reduce the
morbi-mortality of atherosclerotic disease. Correction of
cardiovascular risk factors and improving treatments for acute
coronary syndromes have represented the first line of study. An
important goal is to diagnose patients in early stages, in order to
reduce acute cardiovascular events. Latest discoveries revealed
that mutations in the angiotensin-converting enzyme (ACE) gene may
influence the onset and the severity of CAD (1). Therefore, studying the association
between genotype and phenotype would be an important cornerstone in
cardiology.
The angiotensin-converting enzyme is common to RAAS
(renin-angiotensin-aldosterone system) and the kinin-kalicrein
system. ACE is a zinc metalloproteinase that cleaves the terminal
dipeptide (His-Leu) of Ang I, turning it into Ang II, a highly
constricting substance. Due to interference on the two systems, ACE
also inactivates bradykinin, a vasodilating substance. Through its
functions, ACE maintains the hydro-salin balance and vascular tone
(2,3).
The ACE gene is found on the long arm of chromosome
17, at heading 23 (17q23), measures 21 kb and includes 26 exons and
25 introns (4).
ACE polymorphism consists of insertions
(I)/deletions (D) at the level of intron 16. Thus, from the
polymorphism of intron 16, there are 3 genotypes: Insertional
homozygote (II), heterozygote (ID) and teletional homozygote (DD).
Serum levels of ACE are determined by genetic polymorphism in the
following order: DD>ID>II.
The central goal of our study was to investigate the
correlation between the presence of the D-allele and severe
stenosis or occlusion of one or several coronary arteries.
Patients and methods
The present study included 154 patients with acute
coronary syndroms (acute myocardial infarction and unstable angina)
admitted to the Institute for Cardiovascular Disease ‘George I.M.
Georgescu’, Iasi. The study was approved by the Ethics Committee of
the ‘Grigore T. Popa’ University of Medicine and Pharmacy (Iasi,
Romania) on 09.06.2015, according to the law of medical research,
no. 206 from 27.05.2004. Informed consent was obtained from all
patients included in the study. The inclusion criteria were the
following, according to the European Guidelines (5): i) diagnosed coronary artery disease;
ii) exposure to minimum two cardiovascular risk factors:
dyslipidemia (total cholesterol >200 mg/dl, LDL-cholesterol
>100 mg/dl, HDL-cholesterol <40 mg/dl for males and <50
mg/dl for females, triglycerides >150 mg/dl or normal lipid
profile under cholesterol lowering medication), high blood pressure
(>140/90 mmHg or normal blood pressure under medication),
diabetes mellitus, family history of cardiovascular disease at
young age (<50 year-old for male relatives or at climax for
female relatives), smoking, overweight (BMI between 25-29.9) or
obesity (BMI >30).
Anamnesis helped us extract the following
information: symptoms, family history (CAD, stroke, lower extremity
artery disease and other consequences of severe atherosclerosis),
personal cardiovascular history, work exposure to toxic substances
and smoking habit.
Through clinical examination (for the BMI
calculation) weight and height, blood pressure, heart rate and
cardiac ascultation were recorded.
Basic haematological and biochemical blood analysis
included: Red blood cell count, haemoglobin, mean haemoglobin
concentration, mean erythrocyte volume, white blood cell count and
leucocitary formula, platelet count, mean platelet volume,
inflamatory markers [erythrocyte sedimentation rate (ESR); C
reactive protein (CRP); fibrinogen (Fg)], glycemia, glicated
haemoglobin, lipid profile (total cholesterol, LDL cholesterol, HDL
cholesterol, triglycerides), renal function (urea, creatinine),
liver function (AST, ALT), cardiac necrosis enzymes (CK-MB, LDH,
TGO).
An electrocardiogram was recorded for each patient
(for rythm, ST segment and T wave analysis) and echocardiography
(for ejection fraction, left ventricle contractility, cardiac
dimension and valve analysis) was performed.
All patients underwent coronary angiography in order
to adequately define the coronary status and those who met the
criteria from the revascularisation guidelines, were implanted
percutaneous stents in order to restore myocardial perfusion.
Apart from the blood for standard analysis, 2 extra
mililiters of blood for genetic analysis was also collected for the
determination of angiotensin-converting enzyme gene polymorphisms,
deletions or insertions of genes.
Polymerase chain reaction (PCR) was used for the
amplification of genetic material. The method is based on the
DNA-polymerase ability to synthesize new strands of complementary
DNA, starting from a base chain. DNA-polymerase can only add
nuleotide to a pre-existing 3'-OH group, so it takes a primer to
add the first nucleotide. In the end, the amplified sequence will
be found in billions of copies. The process has several stages: DNA
distortion, bonding of primers, extension of primers and consists
of several cycles.
To determine the genetic polymorphism of the ACE
gene, the MutaGel ACE kit was used. It determines mutations such as
insertions (I)/deletions (D) based on analysis of the ALU sequence
at intron 16. A kit allows 24 determinations. The MutaGel ACE kit
contains specific primers for identifying both mutations: I-allele
(fragment 490 bp) and D-allele (190 bp fragment), master mixture
(Master Mix) for chain polymerization (Taq enzyme,
MgCl2, dNTP, buffer solution), positive control for
genotypes containing I-allele, positive control for genotypes
containing D-allele, negative controls and high purity solution for
PCR. In addition to this kit KBR3005 DNA extraction kit and gel
electrophoresis reagents were also used. In the first stage, the
purity and concentration of the DNA was checked. Then the following
amplification program was used: initial hold step (initial
distortion) at 95 degrees for 5 min, followed by 30 exponential
amplification cycles, each cycle having 3 temperature variations
(distortion at 94 degrees for 60 sec, hybridization at 57 degrees
for 60 sec and elongation at 72 degrees for 90 sec) and final hold
step 72 degrees 5 min and then 4 degrees. Next the amplified
material was separated by electrophoresis in the 1.5% agarose gel,
coloring SYBR Green fluorochrome fragments and analyzing them using
UV spectrophotometry with 312 nm wavelengths. Finally, the
identification of the fragments was made by comparing them with the
DNA ladder.
Statistical analysis was performed using SPSS 18.0.
Primary processing, i.e. data systematization through
centralization and grouping, led to the obtaining of primary
indicators, which are presented in the form of absolute sizes. On
the basis of the primary indicators, different statistical
processes of comparison, abstraction and generalization were
obtained of the derivative indicators. Derivative indicators are
intended to highlight the qualitative aspects of an ensemble,
targeting the relationship between different parts of a group of
patients or different characteristics, interdependence links
between variables. The following derivative indicators were used,
described by the ANOVA test: Indicators of the mean value (simple
arithmetic mean, median, module, minimum and maximum values) and
indicators of dispersion (standard deviation, coefficient of
variation). The following statistical tests and correlation methods
were also used: i) Chi-square test, qualitative non-parametric
test, compares frequency distributions. ii) Student's t-test,
parametric test that compares the average values recorded in 2
groups with normal distributions. iii) The F test (ANOVA) used when
comparing 3 or more groups with normal distributions. iv) The
Kruskall-Wallis correlation compares ordinal variables in 3 or more
groups. v) Correlation coefficient ‘Pearson’ (r) is the correlation
of 2 variables in the same group, the direct/indirect correlation
given by the coefficient sign.
Results
The patients were divided into four groups,
according to the severity of the CAD, as revealed by the coronary
angiography: 0C (non-significant stenosis, considered control
group); 1C (one-vessel disese); 2C (two-vessel disease); and 3C
(three-vessel disease).
The sex distribution revealed that patients were
predominantly males (74%), sex ratio M/F=2.85/1, regardless of the
coronary status (Chi-square=1.688; df=3; P=0.640) (Fig. 1).
Age ranged between 35 and 85, the group mean was
64.5±10,85 years, slightly elevated for the one-vessel disease
patients (65.44 years; P=0.931) (Table
I).
 | Table IAge descriptive indicator (years)
comparison between groups. |
Table I
Age descriptive indicator (years)
comparison between groups.
| | Confidence interval
95% | |
|---|
| Group | N | Mean | Standard
deviation | Standard error | -95% CI | +95% CI | Min | Max | Test F (ANOVA)
P-value |
|---|
| 0C | 24 | 64.13 | 14.21 | 2.90 | 58.12 | 70.13 | 36 | 78 | 0.931 |
| 1C | 48 | 65.44 | 9.53 | 1.38 | 62.67 | 68.20 | 35 | 85 | |
| 2C | 48 | 64.21 | 12.29 | 1.77 | 60.64 | 67.78 | 34 | 80 | |
| 3C | 34 | 64.15 | 7.70 | 1.32 | 61.46 | 66.83 | 44 | 76 | |
| Total | 154 | 64.56 | 10.85 | 0.87 | 62.84 | 66.29 | 34 | 85 | |
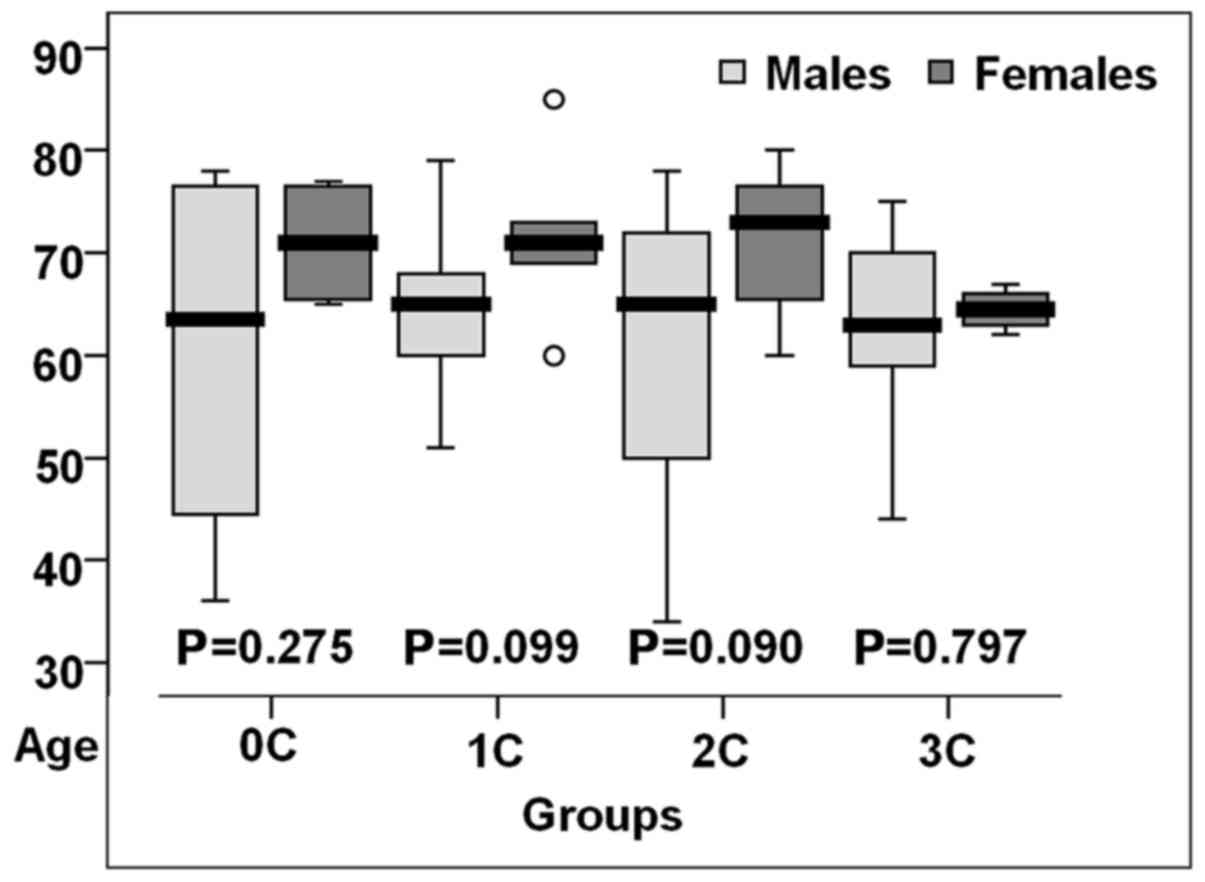
Women with altered coronary status were older than
those in the control group (P>0.05) (Fig. 2).
In conjunction with the average age of the study
group (~65 years), the age group distribution shows a 62.5% share
of patients in group 1C over 65 years of age, while in group 3C the
frequency of patients under 65 years of age was 47.1%. However,
these frequency distributions showed no statistically significant
differences, suggesting the homogeneity of the study group by age
group (Chi-square=1.007; df=3; P=0.800) (Table II).
 | Table IIGroup structure according to age. |
Table II
Group structure according to age.
| | 0C | 1C | 2C | 3C |
|---|
| Age group | n | % | n | % | n | % | n | % |
|---|
| <65 years | 9 | 37.5 | 18 | 37.5 | 21 | 43.8 | 16 | 47.1 |
| ≥65 years | 15 | 62.5 | 30 | 62.5 | 27 | 56.2 | 18 | 52.9 |
The identification of ACE genotypes was carried out
according to the length of the amplicons after migration into the
gel. The 190 bp amplicon signals the presence of allele D and the
480 bp amplicon signals the presence of allele I. Positive control
is heterozygous. The three possible genotype variants: II, ID and
DD were identified (Fig. 3). In the
OC group 21 patients with genotype II, 3 patients with genotype ID
and no patients with genotype DD were identified. In the 1C group 8
patients with genotype II, 40 patients with genotype ID and no
patients with genotype DD were identified. In the 2C group 4
patients with genotype II, 36 patients with genotype ID and 8
patients with genotype DD were identified. In the 3C group no
patients with genotype II, 6 patients with genotype ID and 28
patients with genotype DD were identified.
The exposure to cardiovascular risk factors of DD
patients with severe coronary lesions: in the 3C group 82.4% of
patients had the genotype DD, 82.1% were male, 57.1% were >65
years of age, all with dyslipidemia, 92.9% diabetics and 57.1% were
obese, half were smokers (Table
III).
 | Table IIIGenotype-phenotype correlations for
each study group. |
Table III
Genotype-phenotype correlations for
each study group.
| | Group 0C (n=24) | Group 1C (n=48) |
|---|
| Risk factor | II (n=21) | ID (n=3) | DD (n=0) | P-value | II (n=8) | ID (n=40) | DD (n=0) | P-value |
|---|
| Male | 76.2 | - | - | 0.006 | 75.0 | 80.0 | - | 0.755 |
| >65 years | 57.1 | 100.0 | - | 0.080 | 50.0 | 65.0 | - | 0.430 |
| Diabetes | 19.0 | - | - | 0.278 | 100.0 | 55.0 | - | 0.004 |
| Dyslipidemia | 81.0 | 100.0 | - | 0.278 | 100.0 | 80.0 | - | 0.073 |
| Obesity | 71.4 | 100.0 | - | 0.028 | 100.0 | 50.0 | - | 0.001 |
| Smoking | 23.8 | - | - | 0.219 | 37.5 | 55.0 | - | 0.364 |
| | Group 2C (n=48) | Group 3C (n=34) |
| Risk factor | II (n=4) | ID (n=36) | DD (n=8) | P-value | II (n=0) | ID (n=6) | DD (n=28) | P-value |
| Male | 100.0 | 61.1 | 100.0 | 0.007 | - | 50.0 | 82.1 | 0.113 |
| >65 years | 100.0 | 41.7 | 100.0 | 0.001 | - | 33.3 | 57.1 | 0.287 |
| Diabetes | - | 44.4 | 100.0 | 0.001 | - | 100.0 | 92.9 | 0.370 |
| Dyslipidemia | 100.0 | 88.9 | 100.0 | 0.298 | - | 100.0 | 100.0 | 1.000 |
| Obesity | - | 61.1 | 100.0 | 0.001 | - | 100.0 | 57.1 | 0.012 |
| Smoking | 100.0 | 47.2 | 25.0 | 0.022 | - | - | 50.0 | 0.007 |
Discussion
Coronary artery disease is a polygenic pathology.
Its onset and severity depend on the interaction between genetic
and environmental factors. The DD, ID and II genotypes are
associated with high, intermediate and low levels of ACE. Our study
shows that the three genotypes influence the severity of CAD,
interacting with conventional risk factors. The relationship
between CAD and genetic polymorphism (genotype DD) was first
discussed by Cambien et al (6). Since then, the topic has long been
debated, the results are still unclear, with some positive studies
(7), and others negative (8,9).
Polymorphism I/D is responsible for 20-50% of the differences in
plasma level of ACE, which assumes that 50-80% come from the
influence of environmental factors or from the interaction between
these polymorphisms and environmental factors (6). Our results in this direction are
indisputable, since we demonstrated a growing precentage of the
genotype DD proportional with the growing severity of the coronary
disease, from absent genotype DD in the no-vessel disease, to 82.5%
DD carriers among the three-vessel disease patients. Also, II
genotype, which was predominant in the control group, was absent
among the 3C patients, who were all D-allele carriers (17.64%
genotype ID and 82.35% genotype DD). This was similar to the
results of Nakai et al (10),
Vargas-Alarcón et al (11)
Jamil et al (12) and
Acarturk et al (13), who
demonstrated an association between CAD and ACE gene I/D
polymorphism. By contrast, Ragia et al (14), Qiu et al (15), and Ramakrishnan et al
(16) found no association between
those two parameters.
Traditional risk factors such as smoking, high blood
pressure, diabetes mellitus, family history of cardiovascular
disease at a young age, obesity influence RAAS by stimulating ACE
synthesis, leading to excessive vasoconstriction, enhancing
ischemic risk. Freitas et al (17) concluded that genotype DD induces
onset and severity of coronary lesions only when exposure to common
risk factors is present, like hypertension and diabetes. Also, the
case control study of Niemec et al (18) demonstrated correlations between ACE
gene polymorphisms and CAD in the presence of hypercholesterolemia.
The present study revealed that three-vessel disease genotype DD
patients were diabetic and dyslipidemic and half of them were obese
and heavy smokers.
The present study has some limitations. First, the
sample size of included patients is small, that may reduce the
statistical power. Second, this is a single-center clinical study,
which might cause some selection bias.
In conclusion, by analysing all the study groups,
significant associations were obtained between the D-allele and
male sex, age over 65 years, smoking, diabetes mellitus, obesity,
dyslipidemia, which illustrates the strong interaction between ACE
polymorphism and environmental factors (genetic-epigenetic
interaction) in initiation and worsening of coronary heart disease.
Based on this study it is concluded that the D-allele is associated
with a greater risk for acute coronary events and severe coronary
stenosis, especially when risk genotype and risk phenotype
interact.
Acknowledgements
Not applicable.
Funding
The present study was supported by the European
Social Fund, Human Resources Development Operational Programme
2007-2013 (project no. POSDRU/159/1.5/S/133377).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
MCV and IBB acquired the data. MCV, IBB, OVB and AB
analyzed the data and drafted the manuscript. MB, MC and CAG
designed the study and supervised data analysis. MCV, MCB and DI
were involved in the design of the study and analyzed the data. All
authors read and approved the final manuscript.
Ethics approval and consent to
participate
The study was approved by the Ethics Committee of
the ‘Grigore T. Popa’ University of Medicine and Pharmacy (Iasi,
Romania) on 09.06.2015, according to the law of medical research,
no. 206 from 27.05.2004. Informed consent was obtained from all
patients included in the study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Chen YH, Liu JM, Hsu RJ, Hu SC, Harn HJ,
Chen SP, Jeng JR, Wu CL, Ho JY and Yu CP: Angiotensin converting
enzyme DD genotype is associated with acute coronary syndrome
severity and sudden cardiac death in Taiwan: A case-control
emergency room study. BMC Cardiovasc Disord. 12(6)2012.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Sayed-Tabatabaei FA, Oostra BA, Isaacs A,
van Duijn CM and Witteman JCM: ACE polymorphisms. Circ Res.
98:1123–1133. 2006.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Pfohl M, Koch M, Prescod S, Haase KK,
Häring HU and Karsch KR: Angiotensin I-converting enzyme gene
polymorphism, coronary artery disease and myocardial infarction. An
angiographically controlled study. Eur Heart J. 20:1318–1325.
1999.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Hubert C, Houot AM, Corvol P and Soubrier
F: Structure of the angiotensin I-converting enzyme gene. Two
alternate promoters correspond to evolutionary steps of a
duplicated gene. J Biol Chem. 266:15377–15383. 1991.PubMed/NCBI
|
|
5
|
Windecker S, Kolh P, Alfonso F, Collet JP,
Cremer J, Falk V, Filippatos G, Hamm C, Head SJ, Jüni P, et al:
Authors/Task Force members: 2014 ESC/EACTS Guidelines on myocardial
revascularization: The Task Force on Myocardial Revascularization
of the European Society of Cardiology (ESC) and the European
Association for Cardio-Thoracic Surgery (EACTS)Developed with the
special contribution of the European Association of Percutaneous
Cardiovascular Interventions (EAPCI). Eur Heart J. 35:2541–2619.
2014.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Cambien F, Costerousse O, Tiret L, Poirier
O, Lecerf L, Gonzales MF, Evans A, Arveiler D, Cambou JP and Luc G:
Plasma level and gene polymorphism of angiotensin-converting enzyme
in relation to myocardial infarction. Circulation. 90:669–676.
1994.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Gardemann A, Fink M, Stricker J, Nguyen
QD, Humme J, Katz N, Tillmanns H, Hehrlein FW, Rau M and Haberbosch
W: ACE I/D gene polymorphism: Presence of the ACE D allele
increases the risk of coronary artery disease in younger
individuals. Atherosclerosis. 139:153–159. 1998.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Ferrières J, Ruidavets JB, Fauvel J,
Perret B, Taraszkiewicz D, Fourcade J, Niéto M, Chap H and Puel J:
Angiotensin I-converting enzyme gene polymorphism in a low-risk
European population for coronary artery disease. Atherosclerosis.
142:211–216. 1999.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Canavy I, Henry M, Morange PE, Tiret L,
Poirier O, Ebagosti A, Bory M and Juhan-Vague I: Genetic
polymorphisms and coronary artery disease in the south of France.
Thromb Haemost. 83:212–216. 2000.PubMed/NCBI
|
|
10
|
Nakai K, Itoh C, Miura Y, Hotta K, Musha
T, Itoh T, Miyakawa T, Iwasaki R and Hiramori K: Deletion
polymorphism of the angiotensin I-converting enzyme gene is
associated with serum ACE concentration and increased risk for CAD
in the Japanese. Circulation. 90:2199–2202. 1994.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Vargas-Alarcón G, Zamora J, Sánchez-García
S, Rodríguez-Pérez JM, Cardoso G and Posadas-Romero C:
Angiotensin-I-converting enzyme (ACE) insertion/deletion
polymorphism in Mexican patients with coronary artery disease.
Association with the disease but not with lipid levels. Exp Mol
Pathol. 81:131–135. 2006.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Jamil K, Syed R and Rao H: Implications of
I/D (rs4340) polymorphism in CAD among South Indian population. Int
J Med Med Sci. 1:151–157. 2009.
|
|
13
|
Acarturk E, Attila G, Bozkurt A, Akpinar
O, Matyar S and Seydaoglu G: Insertion/deletion polymorphism of the
angiotensin converting enzyme gene in coronary artery disease in
southern Turkey. J Biochem Mol Biol. 38:486–490. 2005.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Ragia G, Nikolaidis E, Tavridou A,
Arvanitidis KI, Kanoni S, Dedoussis GV, Bougioukas G and
Manolopoulos VG: Renin-angiotensin-aldosterone system gene
polymorphisms in coronary artery bypass graft surgery patients. J
Renin Angiotensin Aldosterone Syst. 11:136–145. 2010.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Qiu C, Han Z, Lu W and Zhang C:
Association of polymorphisms in angiotensin-converting enzyme and
type 1 angiotensin II receptor genes with coronary heart disease
and the severity of coronary artery stenosis. J Huazhong Univ Sci
Technolog Med Sci. 27:660–663. 2007.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Ramakrishnan V, Jaikumar V, Gowtham Kumar
S, Thiyagarajan G and Vincent S: Angiotensin-converting enzyme gene
polymorphism in patients with coronary artery disease. J Adv Lab
Res Biol. 1:36–40. 2010.
|
|
17
|
Freitas AI, Mendonça I, Brión M, Sequeira
MM, Reis RP, Carracedo A and Brehm A: RAS gene polymorphisms,
classical risk factors and the advent of coronary artery disease in
the Portuguese population. BMC Cardiovasc Disord.
8(15)2008.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Niemiec P, Zak I and Wita K: Modification
of the coronary artery disease risk associated with the presence of
traditional risk factors by insertion/deletion polymorphism of the
ACE gene. Genet Test. 11:353–359. 2007.PubMed/NCBI View Article : Google Scholar
|