Introduction
Circadian rhythm influences various behaviors and
physiological characteristics of the body, including movement,
sleep, body temperature, endocrine function and metabolic rhythms,
in both invertebrate and vertebrate species. Dysregulation of the
circadian rhythm is associated with numerous diseases, including
diabetes, obesity and cancer (1-4).
Circadian oscillations result from transcriptional-translational
negative feedback loops that are organized by circadian clock
components at the molecular level. Primarily, two core circadian
clock transcriptional factors [brain and muscle ARNT-like1 (BMAL1)
and clock circadian regulator (CLOCK)] form heterodimers and
rhythmically bind to enhancer boxes in the promoters of target
genes, such as the negative regulators periods 1-3 (PER 1-3) and
cryptochromes 1 and 2 (CRY 1 and 2). Subsequently, PERs and CRYs
suppress the transcriptional activity of BMAL1 and CLOCK (5-8).
The precision of the circadian clock is further influenced by clock
protein posttranslational modifications, including ubiquitination,
phosphorylation, acetylation and SUMOylation, that are critical for
the maintenance of normal physiological function (9-11).
Ubiquitination is one of the most important
post-translational modifications for proteins and the
ubiquitin-proteasome system (UPS) is responsible for the
degradation of various proteins (12). Dysfunction in the UPS can result in
multiple diseases, including metabolic diseases, cancer and
neurological diseases (13,14). The UPS is composed of three classes
of enzymes: E1 ubiquitin-activating enzymes, E2
ubiquitin-conjugating enzymes and E3 ubiquitin ligases. E3 ligases
are complementary to specific substrates (15,16).
Previous studies have indicated that ubiquitination is associated
with the regulation of the circadian clock (7,17).
Notably, two F-box E3 ligases, F-box and leucine rich repeat
protein 3 and F-box and leucine rich repeat protein 21 pseudogene,
cooperate to regulate the degradation of CRY proteins and the
oscillation of the circadian clock (18-20).
An additional two E3 ligases, ARF-BP1 and PAM, have been revealed
to form a complex that mediates degradation of the circadian heme
receptor REV-ERBα (also transcriptionally activated by BMAL1/CLOCK
heterodimers), which in turn inhibits BMAL1 transcription
(21-24).
Furthermore, BMAL1 is degraded via ubiquitination mediated by E3
ligase UBE3A (25). E3 ligase HRD1
(HRD1), an endoplasmic reticulum (ER) transmembrane protein, is an
E3 ubiquitin ligase encoded by the synoviolin 1 gene (26-28).
HRD has been suggested to influence ER-associated degradation
(ERAD), which is a protein quality control system that targets
misfolded ER-associated proteins for ubiquitination and subsequent
degradation (29). As HRD1 is a
well-established E3 ligase that mediates substrate ubiquitination
(26-28),
it was hypothesized that the UPS may influence HRD1-mediated
ubiquitination of BMAL1.
The results of the present study suggested that HRD1
enhanced the ubiquitination of BMAL1 and promoted its degradation
via the UPS. Furthermore, the results suggested that HRD1-dependent
degradation of BMAL1 protein regulated the expression of BMAL1
target genes and the amplitude of circadian oscillations in
mammalian cells, which indicated that HRD1 may influence circadian
rhythm.
Materials and methods
Plasmids
HA-Ub [wild-type (WT)], HA-Ub (K48R) and HA-Ub
(K63R) plasmids were kindly provided by Dr Hui Zheng (Soochow
University, Suzhou, China). The 3xFLAG-BMAL1,
PGL3-BMAL1-luciferase and PGL3-PER1-luciferase
plasmids were kindly provided by Dr Ying Xu (Soochow University).
The 3xFLAG-HRD1 plasmid was kindly provided by Dr Shengyun Fang
(University of Maryland Biotechnology Institute, Rockville, USA).
For pEGFP-N3-BMAL1 plasmid construction, the full length BMAL1 cDNA
was created by subcloning the PCR product with the primers
5'-CGGAATTCCATGGCGGACCAGAGAATG-3' and
5'-GCGTCGACCAGCGGCCATGGCAAGTC-3', which was then inserted into the
pEGFP-N3 (Takara Bio-USA, Inc.) vector at EcoRI/SalI
sites. A previously developed mutant FLAG-HRD1 vector that lost the
E3 ubiquitin ligase activity was used in the present study, in
which the RING finger domain was deleted (ΔRING) (30).
Cell culture, transfection and drug
treatment
293 T-cells or mouse neuroblastoma cells (Neuro-2a;
N2a) were obtained from the Cell Bank of Type Culture Collection of
the Chinese Academy of Sciences and cultured in DMEM (Gibco; Thermo
Fisher Scientific, Inc.) containing 10% FBS (Gibco; Thermo Fisher
Scientific, Inc.) with penicillin (100 mg/ml) and streptomycin (100
mg/ml) at 37˚C with 5% CO2. 30-50% seeding densities of
1x105 cultured cells/well in 12-wells were transfected
with 2 µg plasmids using Lipofectamine® 2000
(Invitrogen; Thermo Fisher Scientific, Inc.). After 24 h of
transfection at 37˚C with 5% CO2, 3x105 cells
were harvested for immunoblot analyses or treated with proteasome
inhibitor MG132 (10 µM) or control for 14 h for immunoprecipitation
assays. In order to analyze protein stability, transfected cells
were treated with cycloheximide (CHX; Sigma-Aldrich; Merck KGaA;100
µg/ml) for 0, 2, 4, 6 or 8 h before harvesting. Cells were
incubated with 100 nM bafilomycin A1 (Baf; Sigma-Aldrich; Merck
KGaA) for 24 h or 10 µM MG132 (Sigma-Aldrich; Merck KGaA) for 12 h
before harvesting. N2a cells were synchronized to the same stage of
cell cycle using horse serum treatment. N2a cells were shifted to a
DMEM containing 50% horse serum (Gibco; Thermo Fisher Scientific,
Inc.) and incubated for 2 h, after which the serum-rich DMEM was
replaced with serum-free medium.
Antibodies
The following primary antibodies were used: Mouse
monoclonal antibodies against FLAG (Sigma-Aldrich; Merck KGaA; cat.
no. F9291; 1:1,000), GAPDH (Chemicon; Merck KGaA; cat. no.
SAB2108668; 1:5,000), GFP (Santa Cruz Biotechnology, Inc.; cat. no.
(B-2):SC-9996; 1:1,000), HA (Santa Cruz Biotechnology, Inc.; cat.
no. sc-7392; 1:500), BMAL1 (Santa Cruz Biotechnology, Inc.; cat.
no. SC-365645; 1:500), Ub (Santa Cruz Biotechnology, Inc.; cat. no.
sc-8017; 1:500); rabbit polyclonal antibodies against HRD1 (Abgent;
cat. no. ap2184a; 1:500) and Histone 2B (Abcam; cat. no. ab52599;
1:500).
The following secondary antibodies were used:
Horseradish peroxidase-conjugated sheep anti-mouse antibody (GE
Healthcare; cat. no. NA931; 1:5,000) and anti-rabbit antibody (GE
Healthcare; cat. no. NA934; 1:5,000) for 2 h at room temperature.
The proteins were visualized with an ECL detection kit (Thermo
Fisher Scientific, Inc.).
Immunoblot analysis
Cells were harvested and then lysed in cell lysis
buffer [50 mM Tris-HCl, pH 7.6; 150 mM NaCl; 0.5% sodium
deoxycholate; 1% Nonidet P-40; protease inhibitor cocktail (Roche
Diagnostics)]. The bicinchoninic acid method was used to determine
the protein concentration and the quantity of protein loaded in
each lane. 8% SDS-PAGE was used for the separation of FLAG, HA and
Ub proteins. 13.5% SDS-PAGE was used for the separation of BMAL1,
GAPDH and HRD1 proteins. SDS-PAGE were transferred onto a PVDF
membrane (EMD Millipore). The membranes were blocked with 5%
non-fat milk for 1 h at room temperature, followed by incubation
with the primary antibodies for 12 h at 4˚C and secondary
antibodies for 2 h at room temperature. Densitometric analyses of
immunoblots from three independent experiments were performed using
Adobe Photoshop CS5 (Adobe, Inc.), and the resulting data were
analyzed using Origin 6.0 (OriginLab Corporation). Graphs indicate
the quantification of the blots and were prepared according to a
previously reported method (31).
Immunocytochemistry
HEK293 cells transfected with EGFP-BMAL1 and
FLAG-HRD1 were washed with PBS twice and fixed with 4%
paraformaldehyde for 10 min at room temperature. After treatment
with 0.25% Triton X-100 for 15 min, the cells were blocked with 4%
FBS (Gibco; Thermo Fisher Scientific, Inc.) for 1 h at room
temperature. Cells were washed with PBS and incubated with
anti-FLAG antibody (Sigma-Aldrich; Merck KGaA; cat. no. F9291;
1:1,000) or GFP antibody (Santa Cruz Biotechnology, Inc.; cat. no.
(B-2):SC-9996; 1:1,000) in PBS overnight at 4˚C. Next, the cells
were incubated with Alexa Fluor 594 donkey anti-mouse secondary
antibodies (Thermo Fisher Scientific, Inc.; cat. no. A32744; 1:300)
for 2 h and then the nuclei were stained with DAPI (Sigma-Aldrich;
Merck KGaA; cat. no. D9542; 2 µg/ml) for 10 min. Finally, the cells
were observed with an inverted system microscope Ti2-E (Nikon,
Japan).
Immunoprecipitation assay
293 cells or N2a cells were co-transfected with
various combinations of tagged plasmids detailed in the figure
legends. After 24 h, the transfected cells were treated with 10
µM/ml MG132 (Sigma-Aldrich; Merck KGaA) for 14 h. The cells were
then harvested and lyzed in cell lysis buffer (50 mM Tris-HCl pH
7.5 buffer containing 150 mM NaCl, 1% NP-40 and 0.5% deoxycholate)
supplemented with the protease inhibitor cocktail (Roche
Diagnostics) at 4˚C. Cells were sonicated with 200 W amplitude and
duration of 10 sec on ice, and centrifuged at 3,000 x g for 30 min
at 4˚C. Meanwhile, protein G-Sepharose beads (Roche Diagnostics)
were incubated with anti-Flag antibody (Sigma-Aldrich; Merck KGaA;
cat. no. F9291; 1:1,000) or anti-GFP antibody (Santa Cruz
Biotechnology, Inc.; cat. no. (B-2):SC-9996; 1:1,000) and 0.01% BSA
(Sigma-Aldrich; Merck KGaA) at 4˚C for 12 h. After they were washed
with ice-cold PBS, the beads were incubated with the supernatants
for 6 h on ice. After incubation, the beads were washed with ice
cold PBS, six times, and then the bound proteins were eluted with
SDS sample buffer and analyzed by with immunoblotting.
RNA extraction and reverse
transcription-quantitative PCR (RT-qPCR)
Total RNA from N2a cells was extracted with
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc.), and then 500 ng of each RNA sample was reverse-transcribed
at 16˚C into cDNA using a PrimeScript RT Master Mix (Takara Bio,
Inc.). qPCR was performed with the following thermocycling
conditions: 95˚C for 10 min, 95˚C for 15 sec, 55˚C for 30 sec and
72˚C for 40 sec, for 40 cycles using Power SYBR Green PCR Master
Mix (Applied Biosystems; Thermo Fisher Scientific, Inc.) with their
relevant primers using a 7500 Real-Time PCR System (Applied
Biosystems; Thermo Fisher Scientific, Inc.). The following
sequences of primers were used: Mouse GAPDH: Forward,
5'-CATGGCCTTCCGTGTTCCTA-3' and reverse, 5'-CCTGCTTCACCACCTTCTT-3';
mouse HRD1: Forward, 5'-CGTGTGGACTTTATGGAACGC-3' and
reverse, 5'-CGGGTCAGGATGCTGTGATAAG-3'; mouse BMAL1: Forward,
5'-TCCAGTCTTGGCATCAATGAGT-3' and reverse,
5'-CCTAATTCTCAGGGCAGCAGAT-3'; mouse DBP: Forward,
5'-CGTGGAGGTGCTTAATGACCTTT-3' and reverse,
5'-CATGGCCTGGAATGCTTGA-3'; mouse PER1: Forward,
5'-TGGCTCAAGTGGCAATGAGTC-3' and reverse,
5'-GGCTCGAGCTGACTGTTCACT-3'. Relative gene expression was
calculated using the 2-ΔΔCq method (32).
Nuclear and cytoplasmic fractionation
assay
The 293 cells that had been transfected with
BMAL1-EGFP along with empty control vector or FLAG-tagged HRD1 for
24 h were lysed in fractionation buffer (320 mM sucrose; 3 mM
CaCl2; 2 mM MgAc; 0.1 mM EDT; 1 mM DTT; 0.5 mM
phenylmethylsulfonyl fluoride and 0.5% NP-40) for 20 min on ice.
After centrifugation at 600 x g for 15 min at 4˚C, the supernatant
was collected as the cytoplasmic fraction. The pellet was washed
once with fractionation buffer without NP-40 and lysed in cell
lysis buffer [150 mM NaCl, 50 mM Tris-HCl pH 7.5, 0.5%
deoxycholate, 1% NP-40 and protease inhibitor cocktail (Roche
Diagnostics)] as the nuclear fraction. Histone 2B (H2B) served as a
nuclear marker and GAPDH served as a cytoplasmic marker.
Luciferase reporter gene assay
293 cells were transfected using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) with 2 µg pGL3-BMAL1-Luciferase (500
ng/µl) or PGL3-PER1-Luciferase (500 ng/µl) construct and
co-transfected with various combinations of tagged plasmids (500
ng/µl). The Renilla luciferase-expressing plasmid pRL-CMV
(500 ng/µl) was co-transfected into cells to normalize the
variations in transfection efficiency. After 36 h of transfection,
the cells were harvested and treated with passive lysis buffer
(Promega Corporation). The activities of both firefly and
Renilla luciferase were measured using a dual luciferase
assay kit (Promega Corporation) through a Microplate reader
Infinite M1000 Pro (Tecan Group, Ltd.) according to the
manufacturer's instructions (Promega Corporation). The absolute
values of firefly luminescence were normalized to those of
Renilla and the ratios were presented as the median of three
transfected experiments, as described previously (31).
Statistical analysis
Quantitative data are presented as the mean ± SEM.
Statistical analysis of the data was performed by a paired
Student's t-test for two group comparisons and one-way ANOVA with
Tukey's test for multiple group comparisons. P<0.05 was
considered to be statistically significant.
Results
E3 ligase HRD1 decreases BMAL1 protein
levels
To investigate the degradation pathway of BMAL1, 293
cells were treated with the proteasome inhibitor, MG132, and the
autophagy inhibitor, Baf. It was revealed that treatment with
MG132, but not Baf, significantly increased the protein levels of
BMAL1 in comparison to vehicle (Fig.
1A). This suggests that BMAL1 protein is prone to degradation
via the UPS rather than lysosomes. To confirm whether other E3
ligases besides UBE3A were involved in BMAL1 degradation, 293 cells
were transfected with several E3 ligase plasmids. Among those E3
ligases, HRD1 reduced the protein levels of BMAL1 compared to empty
vector (Fig. 1B). Increased
expression of HRD1 was seen in cells transfected with FLAG-HRD1
compared with FLAG vector alone (Fig.
1C). In addition, the reduction of BMAL1 protein due to HRD1
overexpression could be rescued following treatment with the
proteasome inhibitor MG132 (Fig. 1D
and E), suggesting that
HRD1-mediated BMAL1 reduction by the proteasome system. To further
confirm the effect of HRD1 on BMAL1, short interfering RNA (siRNA)
was used to knock down HRD1 in different cell lines. Depletion of
HRD1 markedly increased endogenous BMAL1 levels in N2a cells
(Fig. 1F) as well as in 293 cells
(Fig. 1G). The current results
indicated that HRD1 may degrade BMAL1 protein.
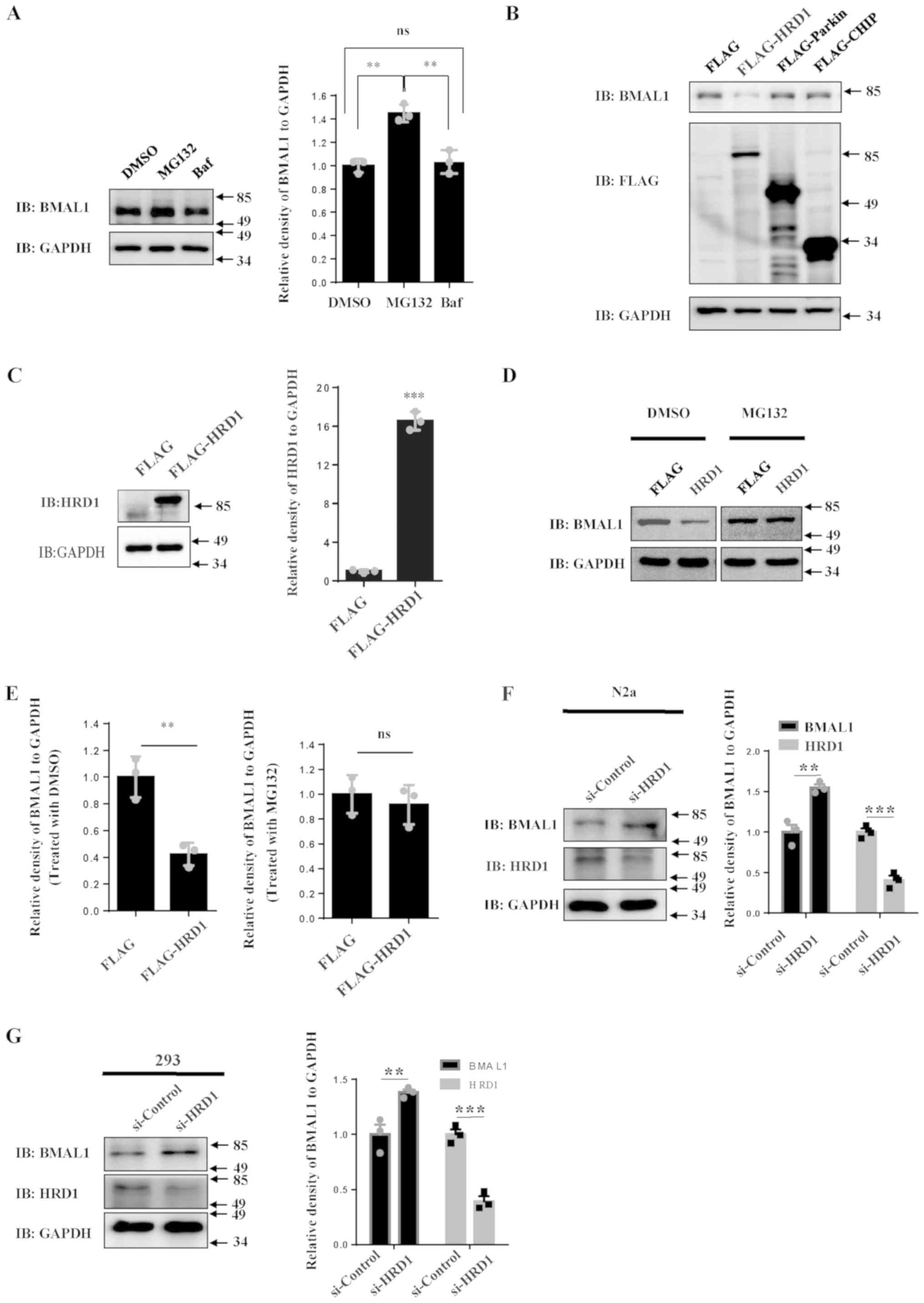 | Figure 1HRD1 decreases BMAL1 protein levels.
(A) 293 cells were treated with MG132 (10 µM) or Baf (100 µM) for
14 h and the levels of endogenous immunoblotting BMAL1 were
determined by western blot analysis. The relative levels of BMAL1
to GAPDH were quantified. (B) 293 cells were transfected with empty
control vector or FLAG-tagged HRD1, Parkin or CHIP, respectively.
After 24 h of transfection, cell lysates were subjected to western
blotting. 293 cells were transfected with empty control vector or
FLAG-tagged HRD1. After 24 h of transfection, the cells were
treated with MG132 (10 µM) or vehicle. The levels of (C) HRD1 and
(D) BMAL1 were determined by western blotting. (E) The relative
levels of BMAL1 to GAPDH were analyzed. (F) N2a cells and (G) 293
cells were transfected with si-control or si-HRD1. After 72 h, the
cell lysates were subjected to western blotting. The relative
levels of BMAL1 to GAPDH were analyzed. **P<0.01,
***P<0.001. Data were collected from 3 repeats,
indicated by the squares and circles. Baf, bafilomycin A1; BMAL1,
brain and muscle ARNT-like 1; DMSO, dimethyl sulfoxide; HRD1,
endoplasmic reticulum transmembrane E3 ubiquitin ligase HRD1; ns,
no statistical significance; si, small interfering. |
HRD1 regulates the stability of BMAL1
protein
As HRD1 is an E3 ligase, it was hypothesized that
HRD1-mediated reduction of BMAL1 protein was relevant to its
ubiquitin ligase activity. The C-terminal RING finger domain of
HRD1 is essential for its activity as an E3 ubiquitin ligase
(28). Hence, the effect of a RING
finger domain deletion mutant of HRD1 (ΔRING) on BMAL1 degradation
was investigated. By contrast to wild-type HRD1, overexpression of
the mutant HRD1 did not result in degradation of BMAL1 (Fig. 2A). To further confirm the finding
that HRD1 promoted the degradation of BMAL1, 293 cells were
transfected with different doses of FLAG-tagged HRD1. The abundance
of BMAL1 decreased in association with an increase in FLAG-HRD1
(Fig. 2B). The increased expression
of BMAL1 in cells transfected with BMAL1-EGFP compared with EGFP
vector only is shown in Fig. 2C. It
was revealed that the degradation rate of BMAL1 protein was faster
in cells transfected with HRD1, compared with the control cells, in
a pulse-chase experiment (Fig. 2D
and E), indicating that HRD1 may
accelerate BMAL1 protein degradation. A subcellular fractionation
assay was performed to investigate whether HRD1 differentially
mediated subcellular BMAL1 protein degradation. Notably, HRD1
significantly reduced BMAL1 protein levels in both the nucleus and
the cytoplasm (Fig. 2F).
 | Figure 2HRD1 regulates the stability of BMAL1
protein. (A) 293 cells were transfected with empty control vector,
FLAG-tagged HRD1 or FLAG-tagged mutant HRD1 (ΔRING), respectively.
After 24 h of transfection, cell lysates were subjected to western
blotting. The relative levels of BMAL1 to GAPDH were analyzed. (B)
293 cells were transfected with a mixture of empty control vector
and quantities of FLAG-tagged HRD1 as indicated. The levels of
endogenous BMAL1 protein and FLAG-HRD1 were determined by western
blotting. (C) 293 cells were transfected with empty control vector
or EGFP-tagged BMAL1 for 24 h. The protein levels of BMAL1 were
determined by western blotting. (D) 293 cells were co-transfected
with BMAL1-EGFP along with empty control vector or FLAG-tagged
HRD1. After transfection for 24 h, the cells were treated with 125
µg/ml CHX and harvested at the indicated timepoints. The protein
levels of BMAL1-EGFP and FLAG-HRD1 were determined by western
blotting. (E) The relative levels of BMAL1 to GAPDH at the
different CHX treated times in (D) were analyzed. (F) 293 cells
were co-transfected with BMAL1-EGFP along with empty control vector
or FLAG-tagged HRD1. After 24 h of transfection, the cells were
subjected to nuclear and cytoplasmic fractionation assay.
**P<0.01. n=3 for all experiments. BMAL1, brain and
muscle ARNT-like 1; CHX, cycloheximide; EGFP, enhanced green
fluorescent protein; HRD1, endoplasmic reticulum transmembrane E3
ubiquitin ligase HRD1; ns, no statistical significance. |
HRD1 interacts with BMAL1 and
regulates ubiquitination of BMAL1
Subsequently, whether HRD1 interacts with BMAL1 in
cells was investigated. Immunoprecipitation using a BMAL1 antibody
indicated that there were interactions between endogenous HRD1 and
BMAL1 in 293 cells (Fig. 3A). In
addition, the interactions between HRD1 and BMAL1 in FLAG-HRD1
overexpressing cells were confirmed using anti-FLAG antibody;
however, CLOCK was not determined to bind to HRD1 (Fig. 3B). As displayed in Fig. 3C, it was revealed that overexpression
of FLAG-HRD1 significantly increased the polyubiquitination of
BMAL1-EGFP. Immunocytochemistry to detect the distribution of HRD1
and BMAL1 was also performed. Although EGFP-BMAL1 was mainly
distributed in the nucleus, it was partly localized to the
cytoplasm and co-localized with FLAG-HRD1 (Fig. 3D).
 | Figure 3HRD1 interacts with and increases
BMAL1 ubiquitination. (A) 293 cells were treated with 10 µM MG132
for 14 h, and the cell lysates were subjected to
immunoprecipitation using an anti-BMAL1 antibody. The levels of
proteins in the cell lysates (input) and the eluted bound proteins
(IP) were assessed by western blotting. Input lane represents whole
cell lysate and IP lane represents immunoprecipitation of bound
eluted proteins. GAPDH proteins were consider to indicate equal
loading for all figures. (B) 293 cells were transfected with empty
control vector or FLAG-tagged HRD1 for 24 h and then were treated
with 10 µM MG132 for 14 h. Then cell lysates were subjected to
immunoprecipitation assay using an anti-FLAG antibody. The cell
lysates and the bound proteins were assessed by western blotting.
(C) 293 cells were co-transfected with BMAL1-EGFP-N3 along with
empty control vector or FLAG-tagged HRD1 for 24 h and then treated
with 10 µM MG132 for 14 h. The cell lysates were then subjected to
immunoprecipitation assay using an anti-EGFP antibody. The cell
lysates and the bound proteins were assessed by western blotting.
(D) 293 cells were co-transfected with FLAG-HRD1, EGFP or
BMAL1-EGFP and 48 h after transfection, immunocytochemistry was
performed with anti-FLAG antibodies. (E) 293 cells were transfected
with HA-Ub (WT), its mutant types HA-Ub (K48R or K63R) or empty HA
vector for 36 h. The protein levels of Ub were determined by
western blotting. (F) 293 cells were co-transfected with
BMAL1-EGFP, FLAG or FLAG-HRD1 along with HA-Ub (WT) or its mutant
types of HA-Ub (K48R or K63R) as indicated for 24 h and then were
treated with MG132 (10 µM) for 14 h. The cell lysates were then
subjected to immunoprecipitation assay using an anti-GFP antibody.
The cell lysates and the bound proteins were analyzed by western
blotting. ***P<0.001 vs. HA; n=3 for all experiments.
BMAL1, brain and muscle ARNT-like 1; CHX, cycloheximide; EGFP,
enhanced green fluorescent protein; HRD1, endoplasmic reticulum
transmembrane E3 ubiquitin ligase HRD1; ns, no statistical
significance; Ub, ubiquitin; WT, wild-type. |
The type of ubiquitin chains conjugated to BMAL1 by
HRD1 was investigated. Cells were co-transfected with GFP-tagged
BMAL1 and FLAG-tagged HRD1 with either HA-Ub (WT), or Lys site
mutant of Ub (K48R or K63R) and underwent a ubiquitination assay.
Expression levels of the various HA-Ub vectors are presented in
Fig. 3E. Both wild-type and K63R
mutant Ub were able to form polyubiquitin chains conjugated to
BMAL1 and the numbers of polyubiquitin chains conjugated to BMAL1
were significantly increased by HRD1 overexpression(Fig. 3F). By contrast, HRD1 did not induce
K48R Ub to form polyubiquitin chains conjugated to BMAL1. The
present results indicate that the E3 ligase HRD1 promotes
K48-linked polyubiquitination of BMAL1 and mediates its degradation
via the UPS.
HRD1 regulates CLOCK gene expression
and circadian rhythm via targeting of BMAL1
BMAL1 and CLOCK form heterodimers to bind to the
E-box motifs in promoters of target genes, such as PERs,
CRYs and DBP, and thus activate their transcription
(5,6). It was hypothesized that overexpression
of HRD1, which promotes the degradation of BMAL1, may influence the
transcription of those genes. Although BMAL1 protein levels were
markedly reduced in FLAG-HRD1 transfected cells, BMAL1 mRNA
levels were not significantly altered by overexpression of HRD1,
according to qPCR analysis (Fig.
4A), indicating that HRD1 does not influence the mRNA level of
BMAL1. However, the mRNA levels of PER1 and DBP were
significantly decreased (Fig. 4B and
C) with FLAG-HRD1 transfection in
comparison to a control, indicating that HRD1 may suppress the
transcription of BMAL1 downstream genes. To further investigate
these findings, the activity of BMAL1 and PER1
promoters were measured using a dual-luciferase reporter system.
Overexpression of HRD1 did not affect the activity of BMAL1
promoter (Fig. 4D), but it strongly
repressed the activity of PER1 promoter (Fig. 4E and F) when co-transfected with BMAL-EGFP and
FLAG-CLOCK. The expression of FLAG-CLOCK is shown in Fig. 4E. The results indicated that HRD1 may
regulate the expression of clock genes such as PER1 and
DBP via regulation of BMAL1 protein levels. In addition,
BMAL1 is a core clock transcription factor, which is critical for
circadian rhythms (3,6). Therefore, whether HRD1 influences the
circadian rhythm of clock gene transcription was investigated. To
study the effect of HRD1 on the circadian rhythm of clock gene
transcription, N2a cells, which are frequently used in circadian
studies, were used (33-35).
When N2a cells were synchronized by horse serum treatment,
PER1 was expressed in a manner consistent with the circadian
rhythm. Notably, the oscillation amplitude of PER1 mRNA
levels was lower in N2a cells harboring FLAG-HRD1 compared with in
control cells (Fig. 4G). In
conclusion, the present results indicate that HRD1, a novel E3
ubiquitin ligase, interacted with BMAL1 and enhanced the
ubiquitination modification of BMAL1 protein (Fig. 4H).
 | Figure 4HRD1 influences CLOCK gene expression
by degrading BMAL1 protein. (A) N2a cells were transfected with
empty control vector or FLAG-tagged HRD1 for 24 h. The levels of
endogenous BMAL1 and FLAG-HRD1 protein were determined by western
blot analysis and the mRNA levels of BMAL1 were determined
by RT-qPCR. The mRNA levels of (B) DBP and (C) PER1
were determined using RT-qPCR. **P<0.01. (D) 293
cells were co-transfected with BMAL1-Luc or the PGL3-Basic
negative control along with empty control vector or FLAG-tagged
HRD1 for 24 h. The activity of the Bmal1 promoter was
analyzed. (E) 293 cells were co-transfected with BAML1-EGFP and
FLAG-CLOCK, a PGL3-PER1-Luc or the PGL3-Basic negative
control along with empty control vector or FLAG-tagged HRD1. The
activity of the PER1 promoter was analyzed.
**P<0.01. (F) 293 cells were transfected with empty
control vector or FLAG-tagged CLOCK for 24 h. The levels of CLOCK
protein were determined by immunoblot assay **P<0.01
vs. FLAG. (G) N2a cells were transfected with empty control vector
or FLAG-tagged HRD1. The mRNA levels of PER1 were determined
by RT-qPCR. *P<0.05, **P<0.01,
***P<0.001 vs. FLAG-tagged HRD1. (H) HRD1 promoted
K48-linked poly-ubiquitination of BMAL1 and subsequently mediated
its degradation by the ubiquitin-proteasome system. n=3 for all
experiments. BMAL1, brain and muscle ARNT-like 1; CHX,
cycloheximide; EGFP, enhanced green fluorescent protein; HRD1,
endoplasmic reticulum transmembrane E3 ubiquitin ligase HRD1; RT-q,
reverse transcription quantitative; ns, no statistical
significance; Luc, luciferase reporter; ss, synchronized with 50%
horse serum. |
Discussion
Circadian oscillation relies on the molecular
transcriptional-translational feedback loop (36,37). The
positive components of this system, BMAL1 and CLOCK, are necessary
for promoting the expression of clock genes (5,21,36,37).
Post-translational modification has been revealed to regulate the
stability of clock proteins and, thus, serves an essential role in
the maintenance of circadian rhythm. Ubiquitin-conjugating enzyme
E2O, an E3-independent E2 ubiquitin-conjugating enzyme, reduces
BMAL1 levels by promoting its ubiquitination and degradation
(38). E3 ubiquitin ligase, tumor
necrosis factor receptor-associated factor 2 (TRAF2), interacts
with BMAL1 to reduce its stability through ubiquitination (39). BMAL1 preferentially interacts with
the zinc finger domain, but not the conventional substrate
recognition domain in TRAF2(39).
Additionally, ubiquitin carboxyl-terminal hydrolase FAF-X, a
deubiquitinating enzyme, was found to modulate the ubiquitination
and degradation of BMAL1(40).
Although several ubiquitinating and deubiquitinating enzymes have
previously been identified to mediate BMAL1 degradation, it is
still of interest to investigate novel E3s that influence BMAL1
protein degradation. In the current study, it was revealed that an
ERAD-associated E3 ligase HRD1 influenced BMAL1 degradation.
BMAL1 is a well-established transcriptional factor
and a nucleocytoplasmic shuttling protein (41-44).
A nuclear localization signal and nuclear export signal have been
identified at the N-terminus of BMAL1, which suggests it may serve
cytoplasmic functions (43).
Previously, it was reported that BMAL1 influences the regulation of
translation (45). BMAL1 regulates
protein translation by interacting with a number of
translation-associated factors and ribosomal proteins in the
cytoplasm (45). The present results
also suggested that HRD1, an ER transmembrane protein may interact
with BMAL1 whereas other E3 ligases, such as Parkin and CHIP, did
not influence BMAL1 degradation. The specificity of HRD1 in the
regulation of BMAL1 degradation provides further evidence that HRD1
is a BMAL1-specific E3 ligase. Furthermore, nuclear and cytoplasmic
fractionation assays indicated that both nuclear and cytoplasmic
BMAL1 levels were decreased by the overexpression of HRD1. It is
possible that degradation of BMAL1 by HRD1 in the cytoplasm
decreases the BMAL1 protein level, resulting in a reduction in
BMAL1 transport into the nucleus.
Polyubiquitin chains are formed at different lysine
residues of ubiquitin molecules (46,47).
Different topologies of polyubiquitin chains have different
three-dimensional structures and functions. Two well-characterized
forms are K48-linked and K63-linked polyubiquitin chains.
K48-linked polyubiquitin chains are typically considered to target
substrates to the proteasome, while K63-linked polyubiquitin chains
function in numerous cellular pathways, such as vesicle
trafficking, aggresome formation, nuclear transport and NF-κB
signaling (46,47). In the current study, it was revealed
that BMAL1 was rarely ubiquitinated with the K48R mutant of
ubiquitin when HRD1 was overexpressed. By contrast, the K63R mutant
of ubiquitin did not influence BMAL1 ubiquitination via
overexpression of HRD1 compared with the wild-type ubiquitin. The
current data demonstrate that the K48 site, but not the K63 site,
of ubiquitin influences HRD1-mediated BMAL1 polyubiquitination.
BMAL1 and CLOCK form heterodimers to activate the
transcription of key clock genes such as PERs and CRYs, as well as
many clock-mediated genes (2,4,6,37). The
present study indicated that immunoprecipitation of endogenous HRD1
was able to detect BMAL1. Immunoprecipitation was also performed to
immunoprecipitate endogenous HRD1 to detect BMAL1. The HRD1
antibody used was not available for the immunoprecipitation
experiment. It was also revealed that, HRD1 may bind to BMAL1, but
not CLOCK, which indicates the specificity of HRD1 to certain
circadian clock proteins. Meanwhile, overexpression of HRD1
significantly suppressed the expression of PER1 and DBP1, which
indicates that a decrease in BMAL1 levels found in BMAL1/CLOCK
heterodimers formed by HRD1 results in the dysfunction of circadian
rhythm regulation. The current data indicate that HRD1 modulates
molecular circadian rhythm via regulation of BMAL1 protein
stability.
In conclusion, the results of the present study
indicate that the E3 ligase HRD1 is an endogenous regulator of the
circadian clock that serves its function via modulation of BMAL1
stability.
Acknowledgements
Not applicable.
Availability of data and materials
The datasets generated and/or analyzed during the
current study are available from the corresponding author on
reasonable request.
Funding
This work was supported by the National Natural
Sciences Foundation of China (grant nos. 31970966 and 31871023),
Natural Science Foundation of Jiangsu Province (SBK2020040753), The
National Key Scientific R&D Program of China (grant nos.
2016YFC1306000 and 2012CB947602), The Science and Technology
Project of Jiangsu Traditional Chinese Medicine Bureau (grant no.
YB2017063), Suzhou Society of Integrated Chinese and Western
medicine (grant no. SYSD2016169), Suzhou Science and Technology
Bureau (grant no. SYSD2019172), The Medical and Health Technology
Project of Suzhou National New and Hi-Tech Industrial Development
Zone (grant no. 2019Q001) and Suzhou Science and Technology Town
Hospital (grant no. 2019D01).
Authors' contributions
DG performed the majority of the experiments,
analyzed the data, and drafted the manuscript. YZ and HW performed
immunoblot assays and analyzed the data. GW analyzed the data and
revised manuscript, CW analyzed and interpreted the data, and
revised manuscript. HR designed experiments, analyzed the data and
revised manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Cao Y and Wang RH: Associations among
metabolism, circadian rhythm and age-associated diseases. Aging
Dis. 8:314–333. 2017.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Mitsumoto Y and Mori A: Acute restraint
stress augments 1-Methyl-4-phenyl-1,2,3,6-tetrahydropyridine
neurotoxicity via increased toxin uptake into the brain in C57BL/6
mice. Neurosci Bull. 34:849–853. 2018.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Li X and Li X: The antidepressant effect
of light therapy from retinal projections. Neurosci Bull.
34:359–368. 2018.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Poggiogalle E, Jamshed H and Peterson CM:
Circadian regulation of glucose, lipid, and energy metabolism in
humans. Metabolism. 84:11–27. 2018.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Brown SA: Circadian metabolism: From
mechanisms to metabolomics and medicine. Trends Endocrinol Metab.
27:415–426. 2016.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Mendoza-Viveros L, Bouchard-Cannon P,
Hegazi S, Cheng AH, Pastore S and Cheng HM: Molecular modulators of
the circadian clock: Lessons from flies and mice. Cell Mol Life
Sci. 74:1035–1059. 2017.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Papazyan R, Zhang Y and Lazar MA: Genetic
and epigenomic mechanisms of mammalian circadian transcription. Nat
Struct Mol Biol. 23:1045–1052. 2016.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Chen D, Li YP, Yu YX, Zhou T, Liu C, Fei
EK, Gao F, Mu CC, Ren HG and Wang GH: Dendritic cell nuclear
protein-1 regulates melatonin biosynthesis by binding to BMAL1 and
inhibiting the transcription of N-acetyltransferase in C6 cells.
Acta Pharmacol Sin. 39:597–606. 2018.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Lim C and Allada R: Emerging roles for
post-transcriptional regulation in circadian clocks. Nat Neurosci.
16:1544–1550. 2013.PubMed/NCBI View
Article : Google Scholar
|
|
10
|
Preußner M and Heyd F:
Post-transcriptional control of the mammalian circadian clock:
Implications for health and disease. Pflugers Arch. 468:983–991.
2016.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Beckwith EJ and Yanovsky MJ: Circadian
regulation of gene expression: At the crossroads of transcriptional
and post-transcriptional regulatory networks. Curr Opin Genet Dev.
27:35–42. 2014.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Swatek KN and Komander D: Ubiquitin
modifications. Cell Res. 26:399–422. 2016.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Covill-Cooke C, Howden JH, Birsa N and
Kittler JT: Ubiquitination at the mitochondria in neuronal health
and disease. Neurochem Int. 117:55–64. 2018.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Rape M: Ubiquitylation at the crossroads
of development and disease. Nat Rev Mol Cell Biol. 19:59–70.
2018.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Lecker SH, Goldberg AL and Mitch WE:
Protein degradation by the ubiquitin-proteasome pathway in normal
and disease states. J Am Soc Nephrol. 17:1807–1819. 2006.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Myung J, Kim KB and Crews CM: The
ubiquitin-proteasome pathway and proteasome inhibitors. Med Res
Rev. 21:245–273. 2001.PubMed/NCBI View
Article : Google Scholar
|
|
17
|
Stojkovic K, Wing SS and Cermakian N: A
central role for ubiquitination within a circadian clock protein
modification code. Front Mol Neurosci. 7(69)2014.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Busino L, Bassermann F, Maiolica A, Lee C,
Nolan PM, Godinho SI, Draetta GF and Pagano M: SCFFbxl3 controls
the oscillation of the circadian clock by directing the degradation
of cryptochrome proteins. Science. 316:900–904. 2007.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Xing W, Busino L, Hinds TR, Marionni ST,
Saifee NH, Bush MF, Pagano M and Zheng N: SCF(FBXL3) ubiquitin
ligase targets cryptochromes at their cofactor pocket. Nature.
496:64–68. 2013.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Yoo SH, Mohawk JA, Siepka SM, Shan Y, Huh
SK, Hong HK, Kornblum I, Kumar V, Koike N, Xu M, et al: Competing
E3 ubiquitin ligases govern circadian periodicity by degradation of
CRY in nucleus and cytoplasm. Cell. 152:1091–1105. 2013.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Cho H, Zhao X, Hatori M, Yu RT, Barish GD,
Lam MT, Chong LW, DiTacchio L, Atkins AR, Glass CK, et al:
Regulation of circadian behaviour and metabolism by REV-ERB-α and
REV-ERB-β. Nature. 485:123–127. 2012.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Zhang Y, Fang B, Emmett MJ, Damle M, Sun
Z, Feng D, Armour SM, Remsberg JR, Jager J, Soccio RE, et al: GENE
REGULATION Discrete functions of nuclear receptor Rev-erbα couple
metabolism to the clock. Science. 348:1488–1492. 2015.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Preitner N, Damiola F, Lopez-Molina L,
Zakany J, Duboule D, Albrecht U and Schibler U: The orphan nuclear
receptor REV-ERBalpha controls circadian transcription within the
positive limb of the mammalian circadian oscillator. Cell.
110:251–260. 2002.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Yin L, Joshi S, Wu N, Tong X and Lazar MA:
E3 ligases Arf-bp1 and Pam mediate lithium-stimulated degradation
of the circadian heme receptor Rev-erb alpha. Proc Natl Acad Sci
USA. 107:11614–11619. 2010.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Gossan NC, Zhang F, Guo B, Jin D,
Yoshitane H, Yao A, Glossop N, Zhang YQ, Fukada Y and Meng QJ: The
E3 ubiquitin ligase UBE3A is an integral component of the molecular
circadian clock through regulating the BMAL1 transcription factor.
Nucleic Acids Res. 42:5765–5775. 2014.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Bordallo J, Plemper RK, Finger A and Wolf
DH: Der3p/Hrd1p is required for endoplasmic reticulum-associated
degradation of misfolded lumenal and integral membrane proteins.
Mol Biol Cell. 9:209–222. 1998.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Kaneko M, Ishiguro M, Niinuma Y, Uesugi M
and Nomura Y: Human HRD1 protects against ER stress-induced
apoptosis through ER-associated degradation. FEBS Lett.
532:147–152. 2002.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Kikkert M, Doolman R, Dai M, Avner R,
Hassink G, van Voorden S, Thanedar S, Roitelman J, Chau V and
Wiertz E: Human HRD1 is an E3 ubiquitin ligase involved in
degradation of proteins from the endoplasmic reticulum. J Biol
Chem. 279:3525–3534. 2004.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Nomura J, Hosoi T, Kaneko M, Ozawa K,
Nishi A and Nomura Y: Neuroprotection by endoplasmic reticulum
stress-induced HRD1 and chaperones: Possible therapeutic targets
for Alzheimer's and Parkinson's disease. Med Sci (Basel).
4(14)2016.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Mao J, Xia Q, Liu C, Ying Z, Wang H and
Wang G: A critical role of Hrd1 in the regulation of optineurin
degradation and aggresome formation. Hum Mol Genet. 26:1877–1889.
2017.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Guo DK, Zhu Y, Sun HY, Xu XY, Zhang S, Hao
ZB, Wang GH, Mu CC and Ren HG: Pharmacological activation of
REV-ERBα represses LPS-induced microglial activation through the
NF-κB pathway. Acta Pharmacologica Sinica. 40:26–34.
2019.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Repouskou A, Sourlingas TG,
Sekeri-Pataryas KE and Prombona A: The circadian expression of
c-MYC is modulated by the histone deacetylase inhibitor
trichostatin A in synchronized murine neuroblastoma cells.
Chronobiol Int. 27:722–741. 2010.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Chang HC and Guarente L: SIRT1 mediates
central circadian control in the SCN by a mechanism that decays
with aging. Cell. 153:1448–1460. 2013.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Chilov D, Hofer T, Bauer C, Wenger RH and
Gassmann M: Hypoxia affects expression of circadian genes PER1 and
CLOCK in mouse brain. FASEB J. 15:2613–2622. 2001.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Buhr ED and Takahashi JS: Molecular
components of the mammalian circadian clock. Handb Exp Pharmacol.
3–27. 2013.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Musiek ES and Holtzman DM: Mechanisms
linking circadian clocks, sleep, and neurodegeneration. Science.
354:1004–1008. 2016.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Chen S, Yang J, Zhang Y, Duan C, Liu Q,
Huang Z, Xu Y, Zhou L and Xu G: Ubiquitin-conjugating enzyme UBE2O
regulates cellular clock function by promoting the degradation of
the transcription factor BMAL1. J Biol Chem. 293:11296–11309.
2018.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Chen S, Yang J, Yang L, Zhang Y, Zhou L,
Liu Q, Duan C, Mieres CA, Zhou G and Xu G: Ubiquitin ligase TRAF2
attenuates the transcriptional activity of the core clock protein
BMAL1 and affects the maximal Per1 mRNA level of the circadian
clock in cells. FEBS J. 285:2987–3001. 2018.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Zhang Y, Duan C, Yang J, Chen S, Liu Q,
Zhou L, Huang Z, Xu Y and Xu G: Deubiquitinating enzyme USP9X
regulates cellular clock function by modulating the ubiquitination
and degradation of a core circadian protein BMAL1. Biochem J.
475:1507–1522. 2018.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Lee Y, Lee J, Kwon I, Nakajima Y, Ohmiya
Y, Son GH, Lee KH and Kim K: Coactivation of the CLOCK-BMAL1
complex by CBP mediates resetting of the circadian clock. J Cell
Sci. 123:3547–3557. 2010.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Kondratov RV, Chernov MV, Kondratova AA,
Gorbacheva VY, Gudkov AV and Antoch MP: BMAL1-dependent circadian
oscillation of nuclear CLOCK: Posttranslational events induced by
dimerization of transcriptional activators of the mammalian clock
system. Genes Dev. 17:1921–1932. 2003.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Kwon I, Lee J, Chang SH, Jung NC, Lee BJ,
Son GH, Kim K and Lee KH: BMAL1 shuttling controls transactivation
and degradation of the CLOCK/BMAL1 heterodimer. Mol Cell Biol.
26:7318–7330. 2006.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Miki T, Zhao Z and Lee CC: Interactive
organization of the circadian core regulators PER2, BMAL1, CLOCK
and PML. Sci Rep. 6(29174)2016.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Lipton JO, Yuan ED, Boyle LM,
Ebrahimi-Fakhari D, Kwiatkowski E, Nathan A, Güttler T, Davis F,
Asara JM and Sahin M: The circadian protein BMAL1 regulates
translation in response to S6K1-mediated phosphorylation. Cell.
161:1138–1151. 2015.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Behrends C and Harper JW: Constructing and
decoding unconventional ubiquitin chains. Nat Struct Mol Biol.
18:520–528. 2011.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Hochstrasser M: Lingering mysteries of
ubiquitin-chain assembly. Cell. 124:27–34. 2006.PubMed/NCBI View Article : Google Scholar
|


















