Introduction
Sepsis is a syndrome caused by infection that causes
systemic inflammation, which could cause cell apoptosis,
inflammation and oxidative stress (1-3).
The high mortality rates of sepsis has become a worldwide issue
(4), making it urgent to find key
targets for sepsis treatment. Lipopolysaccharide (LPS) is an
endotoxin that combines with toll-like receptor 4 (TLR4) on the
surface of the cell membrane to achieve cellular inflammatory
response (5). LPS induces NF-κB
signaling pathway activation in podocytes and promotes the levels
of pro-inflammatory cytokines, such as interleukin (IL)-6, tumor
necrosis factor-α (TNF-α) and IL-1β (5,6).
Therefore, LPS can be used to induce cell inflammatory response to
construct sepsis cell models.
Long non-coding RNAs (lncRNAs) are a class of
noncoding RNAs that are >200 nucleotides in length which are
involved in the regulation of various diseases (7,8). Nuclear
enriched abundant transcript 1 (NEAT1) is a cancer-related lncRNA
which exerts vital effects on innate immune responses (9,10). NEAT1
has been shown to be associated with immunosuppression (11), brain injury (12), prognosis (13) and kidney injury (14) in sepsis. Therefore, NEAT1 expression
could be used as a marker in sepsis diagnosis (15), but its specific mechanism remains to
be elucidated.
MicroRNAs (miRs/miRNAs) are small non-coding RNAs,
which plays a role in post-transcriptional regulation via binding
with target genes (16,17). At present, it has been proven that
numerous miRNAs can regulate the process of sepsis, such as
miR-145(18), miR-150(19) and miR-25(20). Studies have shown that miR-590-3p
plays a key role in the control of various cancers and associated
with the activation of multiple signaling pathways (21-23).
Zhao et al (24), reported
that miR-590-3p could target the NF-κB signaling pathway to
regulate the inflammation of myocarditis. Another study suggested
that miR-590-3p could relieve oxidative stress in
ischemia-reperfusion injury mice models by regulating the NF-κB
signaling pathway (25). Recent
studies have shown that the NF-κB signaling pathway is closely
associated with cell apoptosis and involved in the transcriptional
regulation of various apoptosis-related genes, playing a decisive
role in the process of apoptosis (26,27).
Therefore, NF-κB is one of the important downstream signaling
pathways regulated by miR-590-3p. However, to the best of our
knowledge, there are few studies investigating the role of
miR-590-3p in sepsis.
The present study aimed to investigate the mechanism
of NEAT1 in an LPS-induced sepsis cell model. Knockdown of NEAT1
reduced apoptosis and inflammatory response in LPS-induced H9c2
cells. The regulatory function of NEAT1 on sepsis may be achieved
partly by targeting miR-590-3p. The discovery of the
NEAT1/miR-590-3p axis may contribute to the development of new
targets for sepsis treatment.
Materials and methods
Serum sample collection
Blood from 22 patients with sepsis and 22 healthy
volunteers were collected at the First Affiliated Hospital of the
University of South China between March 2016 to June 2018. The
clinical characteristics of the enrolled subjects are shown in
Table I. All volunteers signed
informed consent to the study. The present study was approved by
the Ethics Committee of First Affiliated Hospital of the University
of South China.
 | Table IClinical characteristics of enrolled
subjects. |
Table I
Clinical characteristics of enrolled
subjects.
|
Characteristics | Healthy
volunteers | Sepsis | P-value |
|---|
| Number | 22 | 22 | - |
| Age (years) | 50.20±18.4 | 51.40±16.8 | >0.05 |
| Sex
(male/female) | 12/10 | 12/10 | >0.05 |
| WBC
(x109/l) | 6.35±1.27 | 14.12±5.35 | <0.01 |
| APACHEII score | - | 17.30±4.60 | <0.01 |
| Lac (mmol/l) | 0.49±0.02 | 4.02±1.98 | <0.01 |
| CRP (µg/l) | 1.98±1.21 | 18.12±4.33 | <0.01 |
Cell culture and LPS treatment
Cardiomyocytes (H9c2 cells) were purchased from
Shanghai Jining Shiye Co., Ltd., and maintained in DMEM (Beijing
Solarbio Science & Technology Co., Ltd.) supplemented with 10%
FBS (Thermo Fisher Scientific, Inc.) and 1% penicillin/streptomycin
(Thermo Fisher Scientific, Inc.) at 37˚C in an incubator containing
5% CO2. Cells were treated with 5 or 10 µg/ml LPS
(Beijing Solarbio Science & Technology Co., Ltd.) at 37˚C for
12 h to induce sepsis cell models as previously described (28,29).
Cell transfection
Small interfering RNA (siRNA) against NEAT1
(si-NEAT1; 5'-GAGCAATGACCCCGGTGACG-3') and its control (si-negative
control (NC); 5'-TAGATACCCCCA GGCCTACC-3'), pcDNA3.1 overexpressing
NEAT1 vector (pcDNA-NEAT1; forward (F), 5'-TTGGGACAGTGTGG-3' and
reverse (R), 5'-TCAGTCCAGCAGGCA-3') and its control (pcDNA-NC;
5'-TAGAAGGCACAGTCGAGG-3'), miR-590-3p mimic or inhibitor
(miR-590-3p; 5'-UAAUUU UAUGUAUAAGCUAGU-3' or anti-miR-590-3p;
5'-ACU AGCUUAUACAUAAAAUUA-3') and their controls (NC;
5'-CGAUCGCAUCAGCAUCGAUUGC-3' or anti-NC;
5'-CAGUACUUUUGUGUAGUACAA-3') were synthesized by Vigene
Biosciences. H9c2 cells transfection was performed using
Lipofectamine™3000 (Invitrogen; Thermo Fisher Scientific, Inc.).
Briefly, H9c2 cells were seeded in six-well plates at a density of
1x105 cells/well. Oligonucleotides (20 nM) or vectors (2
mg/ml) were transfected into cells when cells reached 60%
confluence. Following incubation for 24 h at 37˚C, the medium was
changed and 10 µg/ml LPS was added to the cells for 12 h.
Subsequently, the cells were collected for functional assays. In
addition, cells that were not treated with LPS (control group) were
collected at 48 h after transfection and the transfection
efficiencies of miRNA mimics, inhibitors, siRNAs and overexpression
vectors were evaluated by detecting the expression of NEAT1 and
miR-590-3p.
Reverse transcription-quantitative PCR
(RT-qPCR)
TRIzol® reagent (Invitrogen; Thermo
Fisher Scientific, Inc.) was used to isolate total RNA from serum
samples and treated H9c2 cells. RNA was reverse transcribed into
cDNA using a cDNA reverse transcription kit (Invitrogen; Thermo
Fisher Scientific, Inc.) at the following conditions: 37˚C for 1 h
and 85˚C for 5 min. Gene amplifications was conducted on a
StepOnePlus thermocycler (Thermo Fisher Scientific, Inc.) using
SYBR Green (Invitrogen; Thermo Fisher Scientific, Inc.) according
to the manufacturer's instructions. The relative expression of
NEAT1 or miR-590-3p was normalized to endogenous GAPDH or U6
expression, respectively. The following primer pairs were used for
the qPCR: NEAT1 F, 5'-TTGGGACAGTGTGG-3' and R,
5'-TCAGTCCAGCAGGCA-3'; GAPDH F, 5'-AGGTCG GTGTGAACGGATTTG-3' and R,
5'-TGTAGACCA TGTAGTTGAGGTCA-3'; miR-590-3p F, 5'-GCCAGTCAG
AAATGAGCTTATTC-3' and R, 5'-GCTGCATGTTTC AATCAGAGAC-3' and U6 F,
5'-AAAGACCTGTACGCC AACAC-3' and R, 5'-GTCATACTCCTGCTTGCTGAT-3'.
Relative expression of genes was normalized by GAPDH or U6 and was
determined using the 2-ΔΔCq method (30). The thermocycling conditions used for
the qPCR were as follows: Initial denaturation at 95˚C for 30 sec;
followed by 40 cycles of 95˚C for 5 sec, 55˚C for 30 sec and 72˚C
for 30 sec.
Cell Counting Kit-8 (CCK-8) assay
H9c2 cells were cultured for 24 h after
transfection. According to the manufacturer's protocol, H9c2 cells
were washed with PBS and incubated with CCK-8 solution (Glp Bio
Technology) for 4 h in the dark. The absorbance of cells at 450 nm
was observed under a microplate reader (Molecular Devices,
LLC).
Cell apoptosis assay
The FITC-Annexin V Apoptosis Detection kit (BD
Biosciences) was used to measure cell apoptosis. Briefly, H9c2
cells were digested with trypsin (Beyotime Institute of
Biotechnology) and collected into centrifuge tubes. Following
washing with cold PBS, the cells were resuspended in 1X Binding
Buffer. Subsequently, the cells were stained with 5 µl FITC-Annexin
V and 5 µl propidium iodide for 15 min in the dark. Finally, Attune
NxT Flow Cytometer (Thermo Fisher Scientific, Inc.) was used to
detect fluorescence signals and evaluate cell apoptosis.
Western blot (WB) analysis
Total protein was extracted using RIPA lysis buffer
(Santa Cruz Biotechnology, Inc.) and quantified using a
bicinchoninic acid protein assay kit (Pierce; Thermo Fisher
Scientific, Inc.). Subsequently, 30 µg protein sample was separated
by 10% SDS-PAGE (Beyotime Institute of Biotechnology) and
transferred to PVDF membranes (EMD Millipore). Membranes were
blocked in 5% non-fat milk for 2 h at room temperature and
incubated with primary antibodies overnight at 4˚C. Primary
antibodies against Bcl-2 (1:500; cat. no. BA0412), Bax (1:1,000;
cat. no. BA0315-2), caspase 3 (1:1,000; cat. no. BA3257), TNF
receptor associated factor 6 (TRAF6; 1:1,500; cat. no. A00185),
phosphorylated (p)-p65 (1:1,000; cat. no. P00284), p65 (1:2,000;
cat. no. A00284) and GAPDH (1:2,000; cat. no. BA2913) were
purchased from Wuhan Boster Biological Technology, Ltd. Membranes
were then incubated with horseradish peroxidase-labeled secondary
antibody (1:5,000; cat no. BA1056; Wuhan Boster Biological
Technology, Ltd.) for 1 h at room temperature. Protein signals were
visualized using chemiluminescence reagent (EMD Millipore) and
captured using Invitrogen iBright 1500 (Thermo Fisher Scientific,
Inc.). Results were analyzed using ImageLab software (Version 5.0,
Bio-Rad Laboratories, Inc.).
ELISA
TNF-α (kt30484), IL-6 (BA23048), IL-9 (kt30445), and
IL-1β (kt30375) ELISA kits (Wuhan Merck Biotechnology Co., Ltd.)
were used to measure TNF-α, IL-6, IL-9, and IL-1β concentration in
cells according to the manufacturer's instructions.. In brief, the
cell medium was collected and the supernatant was collected after
being centrifuged with 2,000 x g for 10 min at 4˚C. Standards and
tested samples were configured and added to corresponding plates
according to the kit instructions. Following incubation at 37˚C for
30 min, the plates were washed with washing liquid. Subsequently,
50 µl enzyme-labeled reagent was added to each well for further
incubation for 30 min. Following washing, color developer was added
into each well and incubated for 15 min, and termination solution
was added into each well. The absorbance at a wavelength of 450 nm
was measured with a microplate reader, a standard curve was drawn,
and TNF-α, IL-6, IL-9, and IL-1β concentration in cells were
calculated separately.
Dual-luciferase reporter assay
The StarBase v2.0 tool (http://starbase.sysu.edu.cn/) was used to predict the
binding sites between NEAT1 and miR-590-3p. NEAT1 wild-type
(NEAT1-wt) or NEAT1 mutated type (NEAT1-mut) reporter vectors were
synthesized by General Biosystems. H9c2 cells were inoculated in
24-well plates (8x104 cells/well) and co-transfected
with miR-590-3p mimic or miR-NC and NEAT1-wt or NEAT1-mut using
LipofectamineÔ 3000 (Invitrogen; Thermo Fisher Scientific, Inc.).
Using the dual-luciferase reporter assay kit (Shanghai Genomeditech
Biotechnology Co., Ltd.), the Firefly luciferase reaction intensity
(RLU1) and Renilla luciferase reaction intensity (RLU2) were
determined following transfection for 48 h. The luciferase activity
of cells was the ratio of the two groups of data (RLU1/RLU2).
RNA immunoprecipitation (RIP)
assay
A Magna RIP kit (EMD Millipore) was used to evaluate
the binding degree of NEAT1 and miR-590-3p to protein argonaute-2
(Ago2). Cells were washed with pre-cooled PBS and collected into an
Eppendorf tube. Following centrifugation at 377 x g at 4˚C for 5
min, the cells were lysed with RIP buffer (EMD Millipore) for 5
min. The magnetic beads were pre-incubated with Ago2 antibodies
(anti-Ago2; 1:50; cat. no. ab186733; Abcam) and immunoglobulin G
(IgG) antibody (anti-IgG; 1:100; cat. no. ab48386; Abcam).
Subsequently, cell lysates were added into the magnetic beads
mixture and incubated overnight at 4˚C. RT-qPCR was used to measure
the enrichment of NEAT1 and miR-590-3p to anti-Ago2 and
anti-IgG.
Biotin-labeled RNA pull-down
assay
Biotin-labeled miR-590-3p (Bio-miR-590-3p) and
control probe Bio-NC were purchased from Sangon Biotech Co., Ltd.
H9c2 cells were transfected with these probes for 24 h and then
lysed to collect the cell lysates. The cell lysates were incubated
with streptavidin-labeled magnetic beads (Purimag Biotech Ltd.)
overnight at 4˚C. Relative NEAT1 enrichment was detected using
RT-qPCR.
Statistical analysis
Data are presented as the mean ± SD. Experiments
were repeated three times independently. Statistical analysis was
performed using GraphPad Prism v7.0 (GraphPad Software, Inc.).
Student's t-test or one-way ANOVA followed by Tukey's post-hoc test
was used for data comparison. P<0.05 was considered to indicate
a statistically significant difference.
Results
NEAT1 is highly expressed in patients
with sepsis and LPS-induced H9c2 cells
First, NEAT1 expression in the serum of patients
with sepsis and LPS-induced sepsis cell models was examined.
Compared with normal controls, NEAT1 was significantly upregulated
in patients with sepsis (Fig. 1A).
Similarly, NEAT1 was highly expressed in H9c2 cells treated with 5
and 10 μg/ml LPS compared with in the control group, as detected
using RT-qPCR (Fig. 1B). These data
indicated that NEAT1 might play a role in sepsis progression.
LPS can induce H9c2 cell apoptosis and
inflammation
To verify the success of the sepsis cell model, the
biological function of cells was assessed. Compared with controls,
CCK-8 assay results showed that LPS significantly reduced H9c2 cell
viability (Fig. 2A), and flow
cytometry results proved that the number of apoptotic cells
significantly increased in LPS-induced H9c2 cells (Fig. 2B). Moreover, WB analysis was
performed to measure the levels of apoptosis-related proteins. It
was found that Bax and cleaved caspase-3 levels significantly
increased, while Bcl-2 levels significantly decreased in
LPS-induced H9c2 cells compared with controls, suggesting that LPS
could accelerate H9c2 cell apoptosis (Fig. 2C). In addition, ELISA was performed
to detect the concentrations of TNF-α, IL-6, IL-8 and IL-1β in
LPS-induced H9c2 cells. The results showed that the concentrations
of TNF-α, IL-6, IL-8 and IL-1β significantly increased in H9c2
cells treated with 5 and 10 µg/ml LPS compared with controls
(Fig. 2D-G). These results suggested
that LPS could induce cell apoptosis and inflammation, confirming
the successful establishment of a sepsis cell model. In addition,
the impact 10 µg/ml LPS caused a greater degree of damage to the
cells than that of 5 µg/ml; therefore, 10 µg/ml was adopted for
subsequent tests.
 | Figure 2LPS induces H9c2 cell damage. H9c2
cells were treated with 0, 5 and 10 μg/ml LPS. Cell viability,
apoptosis and apoptosis-related protein expression were measured
using (A) Cell Counting Kit-8 assay, (B) flow cytometry and (C)
western blot analysis, respectively. ELISA was performed to
determine the concentrations of (D) TNF-α, (E) IL-6, (F) IL-8 and
(G) IL-1β in H9c2 cells. *P<0.05. LPS,
lipopolysaccharide; TNF, tumor necrosis factor; IL, interleukin;
PI, propidium iodide. |
Knockdown of NEAT1 relieves
LPS-induced H9c2 cell damage
Since high NEAT1 expression was observed in the
LPS-induced sepsis model, a NEAT1 loss-of-function experiment was
conducted to verify the effect of NEAT1 on sepsis. The low
expression of NEAT1 in H9c2 cells confirmed the transfection
efficiency of si-NEAT1 (Fig. S1A).
Subsequently, H9c2 cells were transfected with si-NEAT1 or si-NC
for 12 h, followed by treatment with 10 µg/ml LPS for 12 h. RT-qPCR
results showed that si-NEAT1 significantly inhibited NEAT1
expression compared with si-NC transfection (Fig. 3A). CCK-8 assay results showed that
NEAT1 knockdown significantly increased the viability of
LPS-induced H9c2 cells compared with si-NC transfection (Fig. 3B). Flow cytometry results revealed
that NEAT1 silencing significantly suppressed apoptosis in
LPS-induced H9c2 cells compared with si-NC transfection (Fig. 3C). Moreover, compared with si-NC
groups, silencing of NEAT1 significantly downregulated the levels
of Bax and cleaved caspase-3 and significantly upregulated Bcl-2
levels in LPS-induced H9c2 cells (Fig.
3D). In addition, the concentrations of TNF-α, IL-6, IL-8 and
IL-1β were significantly decreased by NEAT1 knockdown in
LPS-induced H9c2 cells compared with si-NC groups (Fig. 3E-H). These results suggested that
NEAT1 may play a regulatory role in sepsis progression.
 | Figure 3Knockdown of NEAT1 relieves
LPS-induced H9c2 cell damage. H9c2 cells were transfected with
si-NEAT1 or si-NC for 24 h, followed by stimulation with LPS (10
µg/ml) for 12 h. (A) NEAT1 expression was measured using reverse
transcription-quantitative PCR to evaluate the transfection
efficiency of si-NEAT1. (B) Cell Counting Kit-8 assay, (C) flow
cytometry and (D) western blot analysis were performed to detect
cell viability, apoptosis and apoptosis-related protein expression
in H9c2 cells, respectively. The concentrations of (E) TNF-α, (F)
IL-6, (G) IL-8 and (H) IL-1β were determined using ELISA.
*P<0.05. LPS, lipopolysaccharide; TNF, tumor necrosis
factor; IL, interleukin; PI, propidium iodide; NEAT1, nuclear
enriched abundant transcript 1; si-NEAT1, small interfering RNA
targeting NEAT1; NC, negative control; lncRNA, long non-coding
RNA. |
Overexpression of miR-590-3p
attenuates LPS-induced H9c2 cell damage
By detecting the expression of miR-590-3p, it was
found that miR-590-3p was significantly decreased in patients with
sepsis and LPS-induced H9c2 cells compared with controls (Fig. 4A and B). Subsequently, gain-of-function
experiments were performed using miR-590-3p mimic. Increased
miR-590-3p expression confirmed the transfection efficiency of
miR-590-3p mimic (Fig. S1B).
Subsequently, miR-590-3p mimic was transfected into H9c2 cells,
followed by LPS treatment. Compared to the LPS + miR-NC group, LPS
+ miR-590-3p group also markedly enhanced miR-590-3p expression,
which further confirmed the transfection efficiency of the
miR-590-3p mimic (Fig. 4C). CCK-8
assay results showed that miR-590-3p overexpression significantly
enhanced the viability of LPS-induced H9c2 cells compared with the
NC group (Fig. 4D). By measuring the
number of apoptotic cells and the levels of apoptosis-related
proteins, it was found that miR-590-3p mimic significantly
suppressed the apoptosis; inhibited the levels of Bax and cleaved
caspase-3; and increased Bcl-2 level in LPS-induced H9c2 cells
(Fig. 4E and F). In addition, the detection results of
inflammatory cytokines showed that overexpression of miR-590-3p
significantly decreased the concentrations of TNF-α, IL-6, IL-8 and
IL-1β in LPS-stimulated H9c2 cells compared with NC groups
(Fig. 4G-J). It was found that LPS
promoted the expression of TRAF6, while miR-590-3p also inhibited
TRAF6 expression in LPS-induced H9c2 cells compared with the NC
group (Fig. S2). Therefore, the
data indicated that miR-590-3p might play an inhibitory role in
sepsis progression.
 | Figure 4miR-590-3p overexpression attenuates
LPS-induced H9c2 cell damage. (A) miR-590-3p expression in patients
with sepsis and healthy volunteers was detected by RT-qPCR. (B)
RT-qPCR was employed to measure miR-590-3p expression in H9c2 cells
treated with LPS at different concentrations. H9c2 cells were
transfected with miR-590-3p mimic or NC for 24 h, followed by
stimulation with LPS (10 µg/ml) for 12 h. (C) miR-590-3p levels
were detected using RT-qPCR. Cell viability, apoptosis and
apoptosis-related protein expression of cells were determined using
(D) CCK-8 assay, (E) flow cytometry and (F) western blot analysis,
respectively. ELISA was performed to measure the concentrations of
(G) TNF-α, (H) IL-6, (I) IL-8 and (J) IL-1β in H9c2 cells.
*P<0.05. miR, microRNA; LPS, lipopolysaccharide; TNF,
tumor necrosis factor; IL, interleukin; PI, propidium iodide; NC,
negative control; RT-qPCR, reverse transcription-quantitative
PCR. |
NEAT1 directly interacts with
miR-590-3p
To investigate the association between NEAT1 and
miR-590-3p, bioinformatics analysis was performed. The StarBase
v2.0 tool predicted that miR-590-3p had binding sites with NEAT1
(Fig. 5A NEAT1-wt and NEAT1-mut
plasmids were constructed for dual-luciferase reporter assays. The
results revealed that miR-590-3p mimic significantly inhibited the
luciferase activity of NEAT1-wt compared with the NC group, while
showing no effects on NEAT1-mut (Fig.
5B). In addition, RIP assay showed that the enrichment of NEAT1
and miR-590-3p significantly increased in anti-Ago2 groups compared
with anti-IgG groups (Fig. 5C). RNA
pull-down assay results showed that NEAT1 was significantly
enriched in Bio-miR-590-3p compared with Bi-NC (Fig. 5D), indicating that NEAT1 could
interact with miR-590-3p. To further confirm this, pcDNA3.1
overexpressing NEAT1 vector was constructed and its transfection
efficiency was confirmed (Fig.
S1A). miR-590-3p expression was detected and it was found that
NEAT1 overexpression significantly decreased miR-590-3p expression,
while NEAT1 inhibition resulted in the opposite effects (Fig. 5E). Hence, the results revealed that
NEAT1 could sponge miR-590-3p.
miR-590-3p inhibitor reverses the
inhibitory effect of NEAT1 knockdown on LPS-induced H9c2 cells
To further investigate whether NEAT1 regulated
sepsis progression by targeting miR-590-3p, anti-miR-590-3p was
used to perform rescue experiments. Decreased miR-590-3p expression
confirmed that miR-590-5p transfection was successful (Fig. S1B). Subsequently, si-NEAT1 and
anti-miR-590-3p were co-transfected into H9c2 cells. Through
measuring the expression of miR-590-3p, it was found that
anti-miR-590-3p could reverse the promotive effects of NEAT1
silencing on miR-590-3p expression, indicating that the
transfection efficiency of si-NEAT1 and anti-miR-590-3p was
successful (Fig. 6A). CCK-8 assay
results revealed that miR-590-3p inhibitor reversed the increasing
effect of NEAT1 knockdown on the viability of LPS-induced H9c2
cells (Fig. 6B). Additionally, the
suppressive effects of NEAT1 silencing on the apoptosis of
LPS-induced H9c2 cells could be reversed by miR-590-3p inhibitor,
as demonstrated by detection of apoptotic cells and the levels of
Bax, Bcl-2 and cleaved caspase 3 (Fig.
6C and D). Similarly, miR-590-3p
inhibitor also recovered the inhibitory effects of silenced NEAT1
on the concentrations of TNF-α, IL-6, IL-8 and IL-1β in LPS-induced
H9c2 cells (Fig. 6E-H). These
results revealed that NEAT1 regulated sepsis progression by
sponging miR-590-3p.
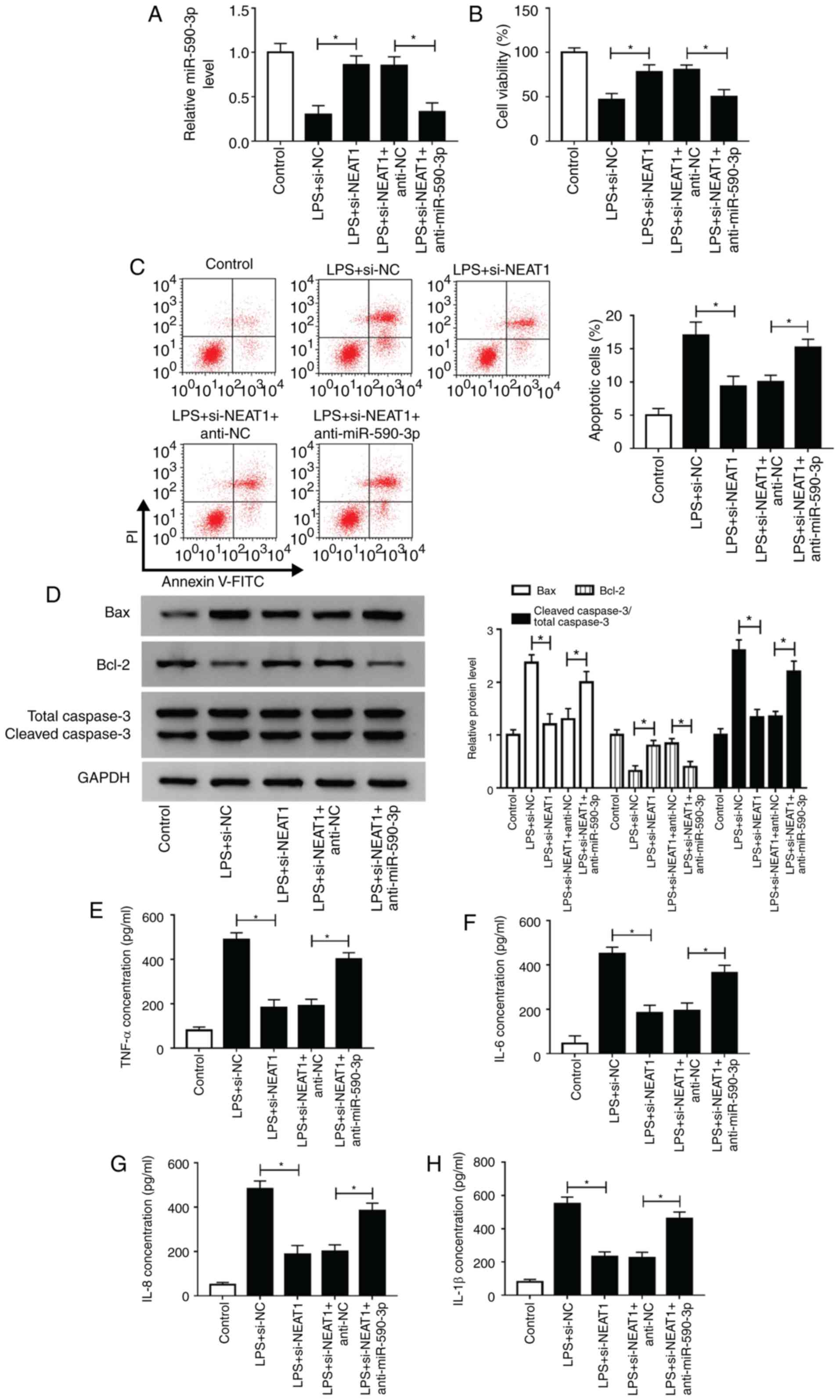 | Figure 6miR-590-3p inhibitor reverses the
inhibitory effect of NEAT1 knockdown on LPS-induced H9c2 cells.
H9c2 cells were transfected with si-NEAT1 and anti-miR-590-3p or
their corresponding negative controls (si-NC and anti-NC), followed
by treatment with LPS. (A) miR-590-3p expression was detected using
reverse transcription-quantitative PCR to evaluate the transfection
efficiency of si-NEAT1 and anti-miR-590-3p. (B) Cell Counting Kit-8
assay, (C) flow cytometry and (D) western blot analysis were
employed to assess the viability, apoptosis and apoptosis-related
protein expression of H9c2 cells, respectively. The concentrations
of (E) TNF-α, (F) IL-6, (G) IL-8 and (H) IL-1β were assessed using
ELISA. *P<0.05. miR, microRNA; LPS,
lipopolysaccharide; TNF, tumor necrosis factor; IL, interleukin;
PI, propidium iodide; NC, negative control; si-NEAT1, small
interfering RNA targeting NEAT1. |
Inhibition of NEAT1 suppresses the
NF-κB signaling pathway via targeting miR-590-3p
It was reported that NF-κB signaling pathway plays
an important role in mediating inflammatory response and cell
apoptosis (31). The present study
investigated the effects of NEAT1 and miR-590-3p on the activity of
the NF-κB signaling pathway in LPS-induced H9c2 cells. Compared
with the si-NC group, NEAT1 knockdown inhibited p-p65/p65 levels in
LPS-induced H9c2 cells. In addition, miR-590-3p inhibitor also
reversed the inhibitory effects of NEAT1 knockdown on p-p65/p65
levels (Fig. 7A and B). These results indicated that the
NEAT1/miR-590-3p axis could regulate the activity of the NF-κB
signaling pathway in LPS-induced H9c2 cells.
Discussion
Sepsis is a common complication resulting from
severe trauma, burns, infection and major surgical operations,
which can lead to multiple organ dysfunction syndromes in severe
cases (1-3).
Therefore, it is urgent to elucidate the molecular mechanism that
affects the sepsis progression. The present study found that NEAT1
was highly expressed in sepsis. In addition, the sepsis cell model
was constructed using LPS, and it was found that NEAT1 deficiency
alleviated apoptosis and inflammatory response of cardiomyocytes
induced by LPS, which provided a way to inhibit the progress of
sepsis. Consistent with the results of previous studies, the
present study showed that NEAT1 might be an effective target for
the treatment of sepsis (12-15).
The involvement of miR-590-3p in inflammatory
responses has been previously reported. He et al (32), showed that miR-590 decreased the
levels of pro-inflammatory cytokines TNF-α, MCP-1, IL-1b and IL-6
via targeting lipoprotein lipase. Moreover, Li et al
(33), showed that inhibition of
miR-590-3p improved the levels of IL-18 in osteoblasts. The present
study found that miR-590-3p was downregulated in sepsis, and
miR-590-3p could relieve LPS-induced cardiomyocyte damage.
Additionally, miR-590-3p also decreased TRAF6 expression in
LPS-induced cardiomyocytes. Ma et al (34), discovered that miR-590-3p could
suppress LPS-induced acute kidney injury by targeting TRAF6.
Therefore, it was hypothesized that miR-590-3p might also regulate
the progression of sepsis by targeting TRAF6, which requires
further investigation. The present study also found that NEAT1
could serve as a sponge of miR-590-3p, and rescue experiments
confirmed that NEAT1 mediated the regulation of cardiomyocyte
damage by regulating miR-590-3p. Considering the important role of
the NF-κB signaling pathway in inflammatory response (35), the activity of the NF-κB signaling
pathway was assessed and it was confirmed that NEAT1/miR-590-3p
could regulate the activity of the NF-κB signaling pathway. Similar
to previous studies, the regulatory role of miR-590-3p and the
NF-κB signaling pathway has also been confirmed in our study
(24-25).
However, studies have also shown that miR-590-3p inhibited the
activity of the NF-κB signaling pathway in peritoneal macrophages
(24) and mice primary
cardiomyocytes (25). Therefore, the
role of the miR-590-3p/NF-κB signaling pathway in other heart or
myocardial cell lines will be the focus of further research.
LncRNAs regulates gene expression via multiple
mechanisms in various biological processes, including apoptosis,
inflammatory response, cell differentiation and development
(36-38).
It was reported that lncRNA metastasis associated lung
adenocarcinoma transcript 1 could be used as a therapeutic target
for patients with sepsis (39). Chen
et al (40), showed that
lncRNAs urothelial carcinoma associated 1 and highly up-regulated
in liver cancer were involved in the inflammatory response of
LPS-induced endothelial cells. NEAT1 is a noncoding transcript with
a length of ~4 kb and is also involved in the regulation of sepsis
(11-15).
In the present study, the NEAT1 knockdown and miR-590-3p mimic
transfection showed promising effects in alleviating LPS-induced
cardiomyocyte damage. This showed that NEAT1 could be used as a
biomarker lncRNA to evaluate sepsis progression, and NEAT1
inhibition or miR-590-3p mimic could provide a method for the
treatment of sepsis.
In summary, the present study suggested that NEAT1
plays a promotive role in sepsis progression by sponging
miR-590-3p. This is a novel mechanism by which NEAT1 acts on the
progression of sepsis. The inhibitory effects of NEAT1 knockdown
and miR-590-3p overexpression on LPS-induced cardiomyocyte damage
can provide new strategies for the clinical treatment of sepsis.
Hence, the discovery of the NEAT1/miR-590-3p axis may contribute to
the development of new treatments for sepsis.
Supplementary Material
Confirmation of transfection
efficiency. H9c2 cells were transfected with si-NC, si-NEAT1,
pcDNA-NC, pcDNA-NEAT1, NC, miR-590-5p, anti-NC or anti-miR-590-5p
for 48 h. The expression levels of (A) NEAT1 and (B) miR-590-3p
were determined using reverse transcription-quantitative PCR.
*P<0.05. NEAT1, nuclear enriched abundant transcript
1; miR, microRNA; si-NEAT1, small interfering RNA targeting NEAT1;
NC, negative control; lncRNA, long non-coding RNA.
miR-590-5p suppresses TRAF6 expression
in LPS-induced H9c2 cells. H9c2 cells were transfected with NC or
miR-590-5p for 24 h, followed by stimulation with LPS (10
μg/ml) for 12 h. TRAF6 protein levels were measured and
quantified by western blot analysis. *P<0.05. LPS,
lipopolysaccharide; miR, microRNA; NC, negative control; TRAF6, TNF
receptor associated factor 6.
Acknowledgements
Not applicable.
Funding
This study was supported by the Hunan Provincial
Department of Education Science Research Fund (grant no. 18C0426) -
Effect of hydrogen sulfide on mitochondrial damage of myocardial
cells in septic rats.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
LL and ZY conceived, designed and revised the
present study. FL and ZS analyzed the data and wrote the
manuscript. ZP and TY analyzed the data. All authors read and
approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of The First Affiliated Hospital of the University of
South China. Written informed consent forms was obtained from each
patient.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Napolitano LM: Sepsis 2018: Definitions
and Guideline Changes. Surg Infect (Larchmt). 19:117–125.
2018.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Delano MJ and Ward PA: Sepsis-induced
immune dysfunction: Can immune therapies reduce mortality? J Clin
Invest. 126:23–31. 2016.PubMed/NCBI View
Article : Google Scholar
|
|
3
|
Hotchkiss RS, Moldawer LL, Opal SM,
Reinhart K, Turnbull IR and Vincent JL: Sepsis and septic shock.
Nat Rev Dis Primers. 2(16045)2016.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Stevenson EK, Rubenstein AR, Radin GT,
Wiener RS and Walkey AJ: Two decades of mortality trends among
patients with severe sepsis: A comparative
meta-analysis*. Crit Care Med. 42:625–631.
2014.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Sheng X, Zuo X, Liu X, Zhou Y and Sun X:
Crosstalk between TLR4 and Notch1 signaling in the IgA nephropathy
during inflammatory response. Int Urol Nephrol. 50:779–785.
2018.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Wang S, Li Z, Chen Q, Wang L, Zheng J, Lin
Z and Li W: NF-kappaB-induced microRNA-211 inhibits interleukin-10
in macrophages of rats with lipopolysaccharide-induced acute
respiratory distress syndrome. Cell Physiol Biochem. 45:332–342.
2018.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Boon RA, Jaé N, Holdt L and Dimmeler S:
Long noncoding RNAs: From clinical genetics to therapeutic targets?
J Am Coll Cardiol. 67:1214–1226. 2016.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Mohanty V, Gökmen-Polar Y, Badve S and
Janga SC: Role of lncRNAs in health and disease-size and shape
matter. Brief Funct Genomics. 14:115–129. 2015.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Qian K, Liu G, Tang Z, Hu Y, Fang Y, Chen
Z and Xu X: The long non-coding RNA NEAT1 interacted with miR-101
modulates breast cancer growth by targeting EZH2. Arch Biochem
Biophys. 615:1–9. 2017.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Chakravarty D, Sboner A, Nair SS,
Giannopoulou E, Li R, Hennig S, Mosquera JM, Pauwels J, Park K,
Kossai M, et al: The oestrogen receptor alpha-regulated lncRNA
NEAT1 is a critical modulator of prostate cancer. Nat Commun.
5(5383)2014.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Chen JX, Xu X and Zhang S: Silence of long
noncoding RNA NEAT1 exerts suppressive effects on immunity during
sepsis by promoting microRNA-125-dependent MCEMP1 downregulation.
IUBMB Life. 71:956–968. 2019.PubMed/NCBI View
Article : Google Scholar
|
|
12
|
Liu WQ, Wang YJ, Zheng Y and Chen X:
Effects of long non-coding RNA NEAT1 on sepsis-induced brain injury
in mice via NF-κB. Eur Rev Med Pharmacol Sci. 23:3933–3939.
2019.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Huang Q, Huang C, Luo Y, He F and Zhang R:
Circulating lncRNA NEAT1 correlates with increased risk, elevated
severity and unfavorable prognosis in sepsis patients. Am J Emerg
Med. 36:1659–1663. 2018.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Chen Y, Qiu J, Chen B, Lin Y, Chen Y, Xie
G, Qiu J, Tong H and Jiang D: Long non-coding RNA NEAT1 plays an
important role in sepsis-induced acute kidney injury by targeting
miR-204 and modulating the NF-κB pathway. Int Immunopharmacol.
59:252–260. 2018.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Huang S, Qian K, Zhu Y, Huang Z, Luo Q and
Qing C: Diagnostic value of the lncRNA NEAT1 in peripheral blood
mononuclear cells of patients with sepsis. Dis Markers.
2017(7962836)2017.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Bartel DP: MicroRNAs: Target recognition
and regulatory functions. Cell. 136:215–233. 2009.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Stefani G and Slack FJ: Small non-coding
RNAs in animal development. Nat Rev Mol Cell Biol. 9:219–230.
2008.PubMed/NCBI View
Article : Google Scholar
|
|
18
|
Cao X, Zhang C, Zhang X, Chen Y and Zhang
H: miR-145 negatively regulates TGFBR2 signaling responsible for
sepsis-induced acute lung injury. Biomed Pharmacother. 111:852–858.
2019.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Ma Y, Liu Y, Hou H, Yao Y and Meng H:
miR-150 predicts survival in patients with sepsis and inhibits
LPS-induced inflammatory factors and apoptosis by targeting NF-κB1
in human umbilical vein endothelial cells. Biochem Biophys Res
Commun. 500:828–837. 2018.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Yao Y, Sun F and Lei M: miR-25 inhibits
sepsis-induced cardiomyocyte apoptosis by targetting PTEN. Biosci
Rep. 38(38)2018.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Salem M, O'Brien JA, Bernaudo S, Shawer H,
Ye G, Brkić J, Amleh A, Vanderhyden BC, Refky B, Yang BB, et al:
miR-590-3p promotes ovarian cancer growth and metastasis via a
novel FOXA2-versican pathway. Cancer Res. 78:4175–4190.
2018.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Rohini M, Gokulnath M, Miranda PJ and
Selvamurugan N: miR-590-3p inhibits proliferation and promotes
apoptosis by targeting activating transcription factor 3 in human
breast cancer cells. Biochimie. 154:10–18. 2018.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Du B, Wang T, Yang X, Wang J, Shi X, Wang
X, Wu D, Feng L, Chen L and Zhang W: SOX9, miR-495, miR-590-3p, and
miR-320d were identified as chemoradiotherapy-sensitive genes and
miRNAs in colorectal cancer patients based on a microarray dataset.
Neoplasma. 66:8–19. 2019.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Zhao S, Yang G, Liu PN, Deng YY, Zhao Z,
Sun T, Zhuo XZ, Liu JH, Tian Y, Zhou J, et al: miR-590-3p Is a
novel microRNA in myocarditis by targeting nuclear factor kappa-b
in vivo. Cardiology. 132:182–188. 2015.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Zhao C, Jiang J, Wang YL and Wu YQ:
Overexpression of microRNA-590-3p promotes the proliferation of and
inhibits the apoptosis of myocardial cells through inhibition of
the NF-κB signaling pathway by binding to RIPK1. J Cell Biochem.
120:3559–3573. 2019.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Xu L, Wu Q, Zhou X, Wu Q and Fang M:
TRIM13 inhibited cell proliferation and induced cell apoptosis by
regulating NF-κB pathway in non-small-cell lung carcinoma cells.
Gene. 715(144015)2019.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Qi R, Huang J, Wang Q, Liu H, Wang R, Wang
J and Yang F: MicroRNA-224-5p regulates adipocyte apoptosis induced
by TNFα via controlling NF-κB activation. J Cell Physiol.
233:1236–1246. 2018.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Zhang H, Li H, Ge A, Guo E, Liu S and
Zhang L: Long non-coding RNA TUG1 inhibits apoptosis and
inflammatory response in LPS-treated H9c2 cells by down-regulation
of miR-29b. Biomed Pharmacother. 101:663–669. 2018.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Yao Y, Xu K, Sun Y, Tian T, Shen W, Sun F,
Yuan W, Wu H, Chen G, Yuan L, et al: MiR-215-5p inhibits the
inflammation injury in septic H9c2 by regulating ILF3 and LRRFIP1.
Int Immunopharmacol. 78(106000)2020.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) Method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Song B, Wang Z, Liu Y, Xu S, Huang G,
Xiong Y, Zhang S, Xu L, Deng X and Guan S: Immunosuppressive
activity of daphnetin, one of coumarin derivatives, is mediated
through suppression of NF-κB and NFAT signaling pathways in mouse T
cells. PLoS One. 9(e96502)2014.PubMed/NCBI View Article : Google Scholar
|
|
32
|
He PP, Ouyang XP, Tang YY, Liao L, Wang
ZB, Lv YC, Tian GP, Zhao GJ, Huang L, Yao F, et al: MicroRNA-590
attenuates lipid accumulation and pro-inflammatory cytokine
secretion by targeting lipoprotein lipase gene in human THP-1
macrophages. Biochimie. 106:81–90. 2014.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Li TM, Liu SC, Huang YH, Huang CC, Hsu CJ,
Tsai CH, Wang SW and Tang CH: YKL-40-Induced Inhibition of
miR-590-3p promotes interleukin-18 expression and angiogenesis of
endothelial progenitor cells. Int J Mol Sci. 18(18)2017.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Ma J, Li YT, Zhang SX, Fu SZ and Ye XZ:
miR-590-3p attenuates acute kidney injury by inhibiting tumor
necrosis factor receptor-associated factor 6 in septic mice.
Inflammation. 42:637–649. 2019.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Karin M: NF-kappaB as a critical link
between inflammation and cancer. Cold Spring Harb Perspect Biol.
1(a000141)2009.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Rossi MN and Antonangeli F: LncRNAs: New
players in spoptosis control. Int J Cell Biol.
2014(473857)2014.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Mirza AH, Berthelsen CH, Seemann SE, Pan
X, Frederiksen KS, Vilien M, Gorodkin J and Pociot F:
Transcriptomic landscape of lncRNAs in inflammatory bowel disease.
Genome Med. 7(39)2015.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Fatica A and Bozzoni I: Long non-coding
RNAs: New players in cell differentiation and development. Nat Rev
Genet. 15:7–21. 2014.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Geng F, Liu W and Yu L: Potential role of
circulating long noncoding RNA MALAT1 in predicting disease risk,
severity, and patients' survival in sepsis. J Clin Lab Anal.
33(e22968)2019.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Chen Y, Fu Y, Song YF and Li N: Increased
expression of lncRNA UCA1 and HULC is required for pro-inflammatory
response during LPS induced sepsis in endothelial cells. Front
Physiol. 10(608)2019.PubMed/NCBI View Article : Google Scholar
|





















