Introduction
Klebsiella pneumoniae is a Gram-negative
enteric bacillus that is a member of the Enterobacteriaceae family.
In immunocompromised patients, K. pneumoniae can
infect the lungs, blood, and urinary tract. In recent years, the
incidence of nosocomial infections of carbapenem-resistant
K. pneumoniae (CRKP) has continued to increase,
resulting in exceptionally high mortality rates (1,2). In
earlier studies, our group investigated the resistance mechanism
and conducted homology analysis and molecular epidemiological
studies of 88 clinical isolates of penicillin- and
carbapenem-resistant Enterobacteriaceae (25 CRKP) collected in
Ningxia, China, from 2011 to 2016. The results of these studies
revealed that 100% of carbapenem-resistant K.
pneumoniae were also resistant to imipenem. Furthermore, the
NDM-1 and KPC-2 genes were found to code for the carbapenemase
enzyme. Drug-resistant strains can pass resistant plasmids to
sensitive strains of Enterobacteriaceae, although the distribution
varied (3-6).
Since there have been relatively few studies of the
host innate immune response to K. pneumoniae
infection, the virulence and pathogenic mechanism of this species
remain unclear. Autophagy is an important component of natural
immunity and plays key roles in the host immune response, including
inflammation, to infections of pathogenic microbes.
Microorganism-mediated autophagy has been shown to clear the host
cells of Salmonella sp., Pseudomonas aeruginosa, and
group A streptococcus (7-11),
and may contribute to the long-term survival and replication in the
host cell of Brucella melitensis (12) and Mycobacterium tuberculosis
(13). Previous studies have
confirmed that autophagy also protects type II alveolar epithelial
cells (AECIIs) from M. tuberculosis infection
(14) and furthermore conveys
important immunomodulatory effects in the lung against microbial
infections (15). For example,
AECIIs can rapidly phagocytize and kill invading Aspergillus
fumigatus (16). The ability of
AECIIs to phagocytize M. tuberculosis is 60-70%
greater than that of macrophages (17), suggesting that autophagosomes may
play a role in the ability of K. pneumoniae to infect
AECIIs. In this study, human adenocarcinoma alveolar basal
epithelial (A549) cells were used to study the mechanism of
autophagy of K. pneumoniae in order to establish a
novel approach for effective intervention and treatment of
K. pneumoniae infection.
Materials and methods
Bacterial strain and cell type
The reference strain K. pneumoniae K6
(ATCC 700603) was obtained from the American Type Culture
Collection and preserved in our laboratory. A549 cells are
hypotriploid alveolar basal epithelial cells that are widely used
as a model of lung adenocarcinoma, as well as an in vitro
model of type II pulmonary epithelial cells, were purchased from
The Cell Bank of Type Culture Collection of the Chinese Academy of
Sciences (Shanghai, China).
Reagents and instruments
Dulbecco's modified Eagle's medium (DMEM), fetal
bovine serum, and trypsin were purchased from HyClone Laboratories,
Inc. Cell culture plates were obtained from Axygen Scientific, Inc.
Pierce™ Immunoprecipitation Lysis Buffer and the Pierce™
Bicinchoninic Acid (BCA) kit were purchased from Thermo Fisher
Scientific, Inc. An enhanced chemiluminescence kit was obtained
from Beyotime Institute of Biotechnology. Lysogeny broth (LB) solid
medium was obtained from Oxoid Ltd. Rapamycin and 3-methyladenine
(3-MA) were purchased from Sigma-Aldrich; Merck KGaA. Antibodies
against microtubule-associated protein 1A/1B-light chain 3 (LC3)
and β-actin were acquired from Abcam PLC. Alexa Fluor™ 488 goat
anti-rabbit secondary antibodies against immunoglobulin G were
purchased from Invitrogen; Thermo Fisher Scientific, Inc. An
apparatus for sodium dodecyl sulfate-polyacrylamide gel
electrophoresis and a wet electroblotting system were acquired from
Bio-Rad Laboratories, Inc. A laser confocal fluorescence microscope
was purchased from Olympus Corporation.
Bacterial count
Activated single K. pneumoniae
colonies were inoculated on conventional LB solid medium and
incubated at 37˚C for 24 h. Then a typical single colony was
cultivated in LB liquid medium to the logarithmic phase at 37˚C
while rotating at 250 rpm. The bacterial culture was diluted in
sterile physiological saline to a concentration of
1.2x1011 bacteria/ml.
Cell culture
A549 cells were inoculated into a Petri dish and
cultured in a DMEM supplemented with 10% fetal bovine serum, 100
U/ml of penicillin, and 100 µg/ml of streptomycin under an
atmosphere of 5% CO2/95% air at 37˚C. At approximately
80% confluence, the medium was discarded and the cells were washed
one or two times with phosphate-buffered saline (PBS). After the
PBS was removed, the cells were digested for 1-3 min by the
addition of 1 ml of trypsin. Then, 3 ml of complete medium was
added to terminate the digestion and the cells were transferred to
a 15-ml centrifuge tube and centrifuged 300 x g for 5 min at room
temperature. After the supernatant was discarded, the cells were
resuspended in 3 ml of medium and passaged (1:3) in a petri
dish.
Establishment of a cellular infection
model
A549 cells were seeded in T25 flasks and grown to a
confluence of 80-90%. Next, the cells were digested with trypsin
and plated in the wells of 6-well plates at a concentration of
2x106 cells, as determined with a hemocytometer. After
culturing overnight, the cells were infected with bacteria at a
multiplicity of infection (MOI) of 100:1, 50:1, 10:1, 5:1, or 1:1
and incubated for 3 h. Then, the medium was removed and the cells
were washed three times with PBS and then incubated in complete
DMEM containing gentamycin for 24 h. The resulting cells were used
for immunofluorescence staining and western blot analysis.
Detection of autophagic changes to LC3
by confocal microscopy
A549 cells were cultured in DMEM containing 10%
fetal bovine serum. At 80 to 90% confluence, the culture medium was
removed and the cells were washed twice with PBS, fixed with 4%
paraformaldehyde at room temperature for 10 min, and then washed
three times with pre-chilled PBS. Following the addition of PBS
containing 0.5% Triton X-100, the cell membranes were lysed on ice
for 10 min. The cells were then washed three times with pre-chilled
PBS-Triton X-100. PBS containing 3% bovine serum albumin (BSA) was
added and blocked at room temperature for 30 min. LC3 antibody was
diluted to 1:100 in PBS-1% BSA solution. After removal of the
blocking solution, the primary antibody was added and incubated at
room temperature for 2 h or at 4˚C overnight. The primary antibody
was removed by washing three times with PBS-0.35% Tween-20.
Fluorescent secondary antibodies were diluted to 1:400 in PBS-1%
BSA solution. After removal of PBS-T, the secondary antibody was
added and the samples were incubated for 1 h at room temperature in
the dark. For removal of the secondary antibody, the samples were
washed three times with PBS-0.35% Tween-20. Following the addition
of 100 ng/ml of 4',6-diamidino-2-phenylindole (DAPI) solution, the
samples were incubated at room temperature in the dark for 10 min.
Then, the DAPI solution was removed and the samples were washed
three times with PBS-0.35% Tween-20. Following the addition of
anti-fluorescence quenching solution, the samples were placed in
the dark at 4˚C or imaged using a laser confocal fluorescence
microscope.
Western blot detection of the
autophagic protein LC3-II
Triplicate samples of A549 cells were infected with
K. pneumoniae at multiple MOIs for various times.
Then, the expression levels of LC3-II and LC3-II/LC3-I ratios were
detected. The BCA assay was used to detect the concentrations of
proteins extracted from K. pneumoniae-infected A549
cells. After separation by vertical electrophoresis on 10% sodium
dodecyl sulfate-polyacrylamide gels, the proteins were transferred
to polyvinylidene fluoride membranes by semi-dry electroporation.
The membranes were sealed and incubated overnight with the primary
antibody. The next day, the membranes were washed and then
incubated with the secondary antibody. After a final washing, the
protein bands on the membranes were visualized using a gel imager.
Quantity One 1-D Analysis Software (Bio-Rad Laboratories) was used
to quantify the protein bands with β-actin as a reference.
Induction and inhibition of
autophagy
Induction and inhibition of autophagy were assessed
using four experimental groups: An infection group (Pneu),
autophagy inhibition group (Pneu + 3-MA), autophagy induction group
(Pneu + rapamycin), and negative control group. At a confluence of
80-90%, the A549 cells were infected at a bacteria:cell ratio of
100:1 and incubated for 3 h. After the medium was removed, the
cells were washed three times with PBS and then cultivated in
medium containing gentamicin. At the same time, trimethylpurine
(3-MA), rapamycin, purine (3-MA), and rapamycin were added to the
medium at final concentrations of 5 and 10 µM, respectively. At 24,
48, and 72 h, the cells were stained with immunofluorescent markers
and subjected to western blot analysis.
Statistical analysis
Statistical analysis was performed using SPSS
software for Windows, version 15.0. (SPSS, Inc.). The data of
experiments repeated three times were used for analysis and are
presented as the mean ± standard deviation. One-way analysis of
variance was used for comparisons between groups. The Tukey test
was used to identify differences among three or more groups. At
α=0. 05, a probability (p) value of <0.05 was considered
statistically significant.
Results
Morphologies of K. pneumoniae-infected
cells
The bacteria were added to the cell cultures at
ratios of 100:1, 50:1, 10:1, 5:1, and 1:1. As shown in Fig. 1, there were no significant changes to
the morphology of A549 cells.
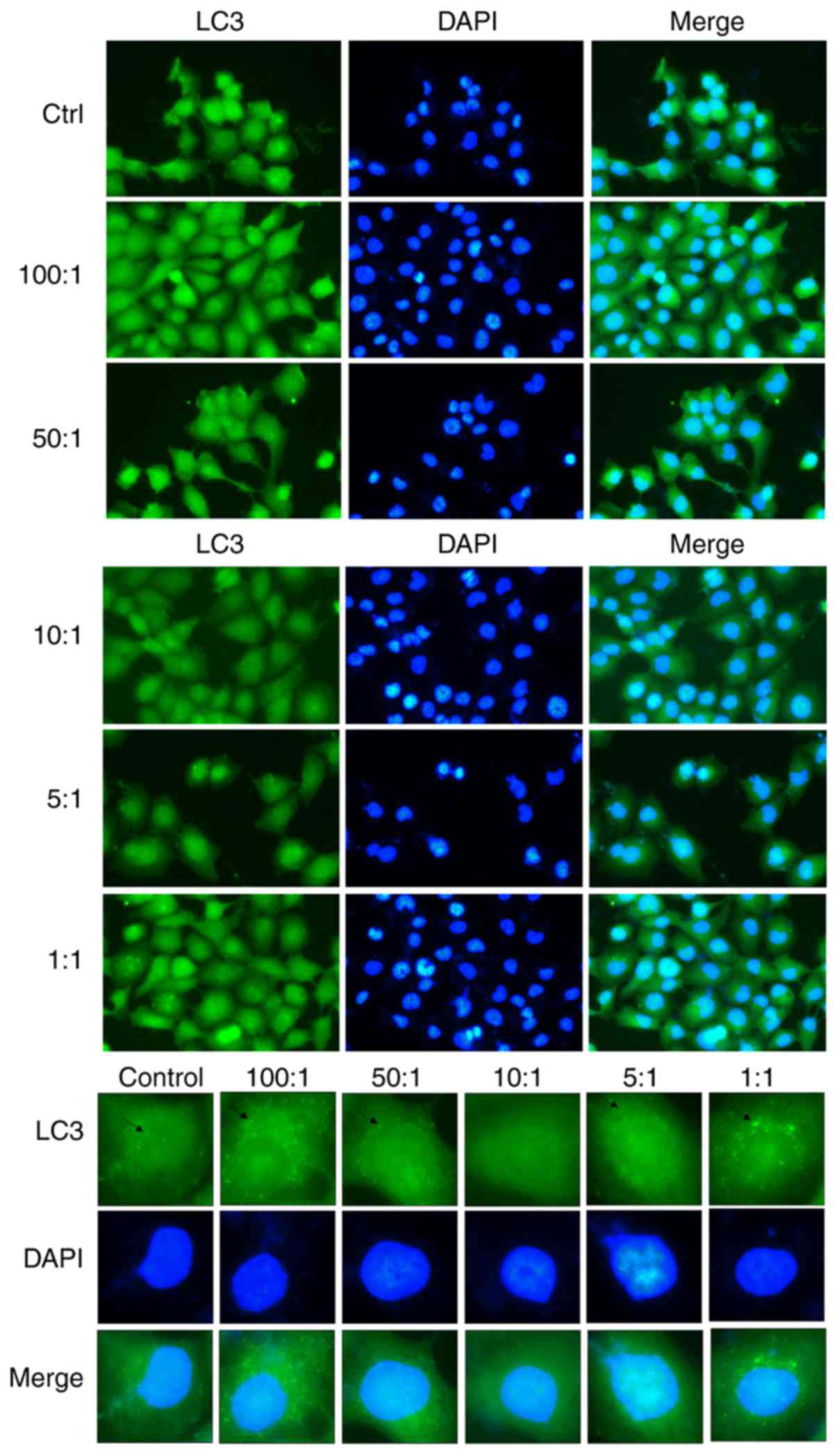
Confocal microscopy of autophagic
changes to LC3 in A549 cells
As compared with the control group of cells that
were not infected with K. pneumoniae, the
autophagosomes of LC3 in the infected group were significantly
changed. The immunofluorescence results showed that LC3 content in
autophagosomes was lowest at a bacteria:cell ratio of 10:1. As
shown in Fig. 2, K.
pneumoniae inhibited autophagy and weakened the resistance
of A549 cells, resulting in increased necrosis and proliferation of
K. pneumoniae.
Western blot detection of the
autophagic protein LC3-II
The western blot results showed that LC3-II protein
expression was significantly higher in infected A549 cells than in
the control uninfected group and the LC3-II/LC3-I ratio was
significantly increased (P<0.05), indicating that K.
pneumoniae promoted autophagy. As the proportion of
bacterial cells (MOI) increased, autophagy also increased. However,
as shown in Fig. 3, at a
bacteria:cell ratio of 10:1, the level of autophagic cells had
relatively decreased as compared with that of the other infection
groups.
Immunofluorescence results
The immunofluorescence results revealed significant
morphological changes to the LC3 autophagosomes of infected cells
as compared with those of the uninfected control cells. With the
extension of time and under the action of gentamycin, the invasive
abilities of K. pneumoniae were weakened (autophagy
increased). As shown in Fig. 4, 3-MA
inhibited autophagy, while rapamycin promoted autophagy.
Western blot results
The western blot results (Fig. 5) showed that as compared with the
control uninfected group, autophagy was increased in cells infected
with K. pneumoniae. Moreover, with time, LC3-II
protein expression gradually increased. The addition of 100 nM
rapamycin to induce autophagy further upregulated LC3-II protein
expression. Moreover, the addition of 10 mM 3-MA to inhibit
autophagy significantly downregulated LC3-II protein expression.
LC3-II protein expression levels were significantly higher in the
infection and autophagy induction groups as compared with the
control group (P<0.05). In addition, LC3-II protein expression
was significantly lower in the infection group treated with 3-MA
(P<0.05).
Discussion
Over the past two decades, K.
pneumoniae has surpassed Escherichia coli as the
important pathogen isolated from patients with purulent liver
abscesses, and has tended to spread globally (18). With the large number and irrational
abuse of antibacterial drugs, the drug resistance rate of K.
pneumoniae continues to increase (19). The US Centers for Disease Control and
Prevention listed Krk-resistant pneumococcus pneumoniae (CRKP) as
the highest urgent grade in 2013(20). Moreover, the World Health
Organization mentioned CRKP in the global drug resistance
surveillance report (21).
Therefore, elucidating the pathogenic mechanism underlying
K. pneumoniae infection and finding effective control
measures is of great value for the treatment and prevention of
K. pneumoniae infection.
The role of autophagy in bacteria-infected cells has
attracted increasing attention and has been shown to play an
important role in host defense, especially in immune cells
(22,23). Autophagy can directly affect the
immune and inflammatory responses throughout the body. For example,
autophagy participates in the clearance of invading bacteria via
degradation of autophagic lysosomes and also plays an effective
regulatory role in the immune response against pathogen invasion
(24). Autophagy and bacterial
infection restrict and promote one another. In addition,
microbe-mediated autophagy can help the body to clear
Salmonella sp., P. aeruginosa, and group A
streptococcus (7-11),
thereby promoting long-term survival and replication in host cells.
For example, M. tuberculosis can survive in
macrophages in latent infections (25). Recently, Ato et al found that
miR-129-3p can inhibit autophagy through Atg4b, thus contributing
to the survival of M. tuberculosis (26). Some Shigella and
Listeria species have evolved various mechanisms to disrupt
the growth and survival of autophagy systems, and to destroy
autophagosomes (27), while others,
such as Legionella pneumophila, regulate intracellular
transport and inhibit autophagosome formation (28).
LC3 is an autophagosomal membrane protein that is
considered to be a specific autophagosome marker. The expression
and transformation of LC3-II (LC3-II/LC3-I) are important
indicators to evaluate the level of intracellular autophagy. AECIIs
serve as the first line of defense against pulmonary exposure to
exogenous pathogens and play an extremely important defensive
function in lung infections. Hence, in-depth studies of the
molecular mechanism underlying the interactions of AECIIs,
autophagosomes, and K. pneumoniae are warranted to
further elucidate the mechanism underlying the unique immune
function of AECIIs for the prevention and treatment of K.
pneumoniae infection.
In the present study, human alveolar type II
epithelial (A549) cells were cultured in vitro and infected
with K. pneumoniae at ratios of 100:1, 50:1, 10:1,
5:1, and 1:1 to establish an infection model. The cells were then
collected at 0, 24, 48, and 72 h after infection for detection of
relevant indicators. At the same time, A549 cells infected with
K. pneumoniae were treated with the autophagy
inhibitor 3-MA and the autophagy inducer rapamycin. The
immunofluorescence and western blot results showed that in A549
cells, LC3 autophagosome expression, LC3-II protein expression, and
the LC3-II/LC3-I ratio were significantly increased relative to the
control uninfected cells, indicating that K.
pneumoniae promotes autophagy by A549 cells. As the
proportion of K. pneumoniae-infected A549 cells had
increased, autophagy also increased. However, at a bacteria:cell
ratio of 10:1, autophagy relatively decreased. At this ratio,
K. pneumoniae can inhibit autophagy and weaken the
resistance of A549 cells, resulting in increased necrosis, thereby
promoting the proliferation of K. pneumoniae. With
prolonged infection, cell autophagy gradually increased. In
response to the addition of the autophagy inducer rapamycin, LC3-II
protein expression was further upregulated, which promoted the
autophagic effect of bacteria in the infected cells. On the
contrary, in response to the addition of the autophagy inhibitor
3-MA, LC3-II protein expression was downregulated, which inhibited
the autophagic effect of bacteria. These results further confirm
that K. pneumoniae can induce autophagy in A549 cells
in vitro.
There were some limitations to this study that
should be addressed. Due to the COVID-19 pandemic, we were unable
to perform proliferation/viability assays to quantitatively assess
the responses of A549 cells or to repeat the experiments in
vitro and in vivo with other cell lines in order to
illustrate the hypothesis of the article.
Autophagy and inflammation are hot topics in the
study of infectious diseases. Recent studies have shown that Notch
signaling can serve as a substrate and participates in the process
of autophagy (29). Autophagy is
involved in the regulation of the inflammatory response, since the
upregulation and absence of autophagy are closely related to the
development of infectious diseases. In future studies, our group
plans to use molecular biology techniques, both in vitro and
in vivo, to further study autophagy-related targets and
related molecular signaling pathways involved with inflammation.
The results of the present study provide theoretical and
experimental evidence of the pathogenic mechanism for the
prevention and treatment of K. pneumoniae
infection.
Acknowledgements
Not applicable.
Funding
The study was supported by grants from the
First-Class Discipline Construction Founded Project of NingXia
Medical University and the School of Clinical Medicine (grant no.
NXYLXK2017A05), and the Ningxia Natural Science Foundation Project
(grant no. 2019AAC03216).
Availability of data and materials
The datasets used and/or analyzed in the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
ZS wrote the manuscript and performed western blot
analysis. GL and LZ were responsible for the cell culture and
transfection. MM and WJ contributed to the analysis of the
observation indexes. All authors read and approved the final
version of the manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Hoxha A, Kärki T, Giambi C, Montano C,
Sisto A, Bella A and D'Ancona F: Study Working Group. Attributable
mortality of carbapenem-resistant klebsiella pneumoniae
infections in a prospective matched cohort study in Italy,
2012-2013. J Hosp Infect. 92:61–66. 2016.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Zhang X, Gu B, Mei Y, Wen Y and Xia W:
Increasing resistance rate to carbapenem among blood culture
isolates of Klebsiella pneumoniae, Acinetobacter
baumannii and Pseudomonas aeruginosa in a
university-affiliated hospital in China, 2004-2011. J Antibiot
(Tokyo). 68:115–120. 2015.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Shi Z, Zhao H, Li G and Jia W: Molecular
characteristics of carbapenem resistant enterobacter cloacae in
ningxia province, China. Front Microbiol. 8(94)2017.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Shi ZY, Zhao HJ, Li CY, Li G, Tao J, Li
SS, Yao Y and Jia W: Bacterial resistance and genotypic
distribution of enterobacteriaceae in burn patients. Chin J
Nosocomiol. 26:2660–2663. 2016.
|
|
5
|
Li G, Zhao HD, Jia W, Zhao M, Zhou XY, Ma
H, Wang LL, Li SS, Dong H and Shi ZY: Analysis of clinical
distribution and drug resistance of 7 157 strains of
Enterobacteriaceae. Int Test Med J. 3:494–497. 2015.
|
|
6
|
Tängdén T and Giske CG: Global
dissemination of extensively drug-resistant carbapenemase-producing
enterobacteriaceae: Clinical perspectives on detection, treatment
and infection control. J Intern Med. 277:501–512. 2015.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Noad J, von der Malsburg A, Pathe C,
Michel MA, Komander D and Randow F: LUBAC-synthesized linear
ubiquitin chains restrict cytosol-invading bacteria by activating
autophagy and NF-κB. Nat Microbiol. 2(17063)2017.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Negroni A, Colantoni E, Vitali R, Palone
F, Pierdomenico M, Costanzo M, Cesi V, Cucchiara S and Stronati L:
NOD2 induces autophagy to control AIEC bacteria infectiveness in
intestinal epithelial cells. Inflamm Res. 65:803–813.
2016.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Yuan K, Huang C, Fox J, Laturnus D,
Carlson E, Zhang B, Yin Q, Gao H and Wu M: Autophagy plays an
essential role in the clearance of Pseudomonas aeruginosa by
alveolar macrophages. J Cell Sci. 125:507–515. 2012.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Cemma M, Kim PK and Brumell JH: The
ubiquitin-binding adaptor proteins p62/SQSTM1 and NDP52 are
recruited independently to bacteria-associated microdomains to
target Salmonella to the autophagy pathway. Autophagy.
7:341–345. 2011.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Nakagawa I, Amano A, Mizushima N, Yamamoto
A, Yamaguchi H, Kamimoto T, Nara A, Funao J, Nakata M, Tsuda K, et
al: Autophagy defends cells against invading group A streptococcus.
Science. 306:1037–1040. 2004.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Guo F, Zhang H, Chen C, Hu S, Wang Y, Qiao
J, Ren Y, Zhang K, Wang Y and Du G: Autophagy favors Brucella
melitensis survival in infected macrophages. Cell Mol Biol
Lett. 17:249–257. 2012.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Romagnoli A, Etna MP, Giacomini E, Pardini
M, Remoli ME, Corazzari M, Falasca L, Goletti D, Gafa V, Simeone R,
et al: ESX-1 dependent impairment of autophagic flux by
Mycobacterium tuberculosis in human dendritic cells.
Autophagy. 8:1357–1370. 2012.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Crotzer VL and Blum JS: Autophagy and
intracellular surveillance: Modulating MHC class II antigen
presentation with stress. Proc Natl Acad Sci USA. 102:7779–7780.
2005.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Stegemann-Koniszewski S, Jeron A, Gereke
M, Geffers R, Kröger A, Gunzer M and Bruder D: Alveolar type II
epithelial cells contribute to the anti-influenza A virus response
in the lung by integrating pathogen- and microenvironment-derived
signals. mBio. 7:e00276–e00216. 2016.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Zhao YX, Li G and Yu RJ: The effect of
retinoic acid on C3 and factor B secretion of human alveolar type
II epithelial cells induced with cytokines. Xi Bao Yu Fen Zi Mian
Yi Xue Za Zhi. 21:442–444. 2005.PubMed/NCBI(In Chinese).
|
|
17
|
Gutierrez MG, Master SS, Singh SB, Taylor
GA, Colombo MI and Deretic V: Autophagy is a defense mechanism
inhibiting BCG and Mycobacterium tuberculosis survival in
infected macrophages. Cell. 119:753–766. 2004.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Liu Y, Wang JY and Jiang W: An increasing
prominent disease of klebsiella pneumoniae liver abscess:
Etiology, diagnosis, and treatment. Gastroenterol Res Pract.
2013(258514)2013.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Baraniak A, Izdebski R, Fiett J,
Gawryszewska I, Bojarska K, Herda M, Literacka E, Żabicka D,
Tomczak H, Pewińska N, et al: NDM-producing enterobacteriaceae in
Poland, 2012-14: Inter-regional outbreak of klebsiella
pneumoniae ST11 and sporadic cases. J Antimicrob Chemother.
71:85–91. 2016.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Schuchat A, Tappero J and Blandford J:
Global health and the US centers for disease control and
prevention. Lancet. 384:98–101. 2014.PubMed/NCBI View Article : Google Scholar
|
|
21
|
World Health Organization (WHO):
Antimicrobial resistance: global report on surveillance. WHO,
Geneva, 2014. https://www.who.int/antimicrobial-resistance/publications/surveillancereport/en/.
|
|
22
|
Deretic V: Autophagy in infection. Curr
Opin Cell Biol. 22:252–262. 2010.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Deretic V: Autophagy in immunity and
cell-autonomous defense against intracellular microbes. Immunol
Rev. 240:92–104. 2011.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Deretic V and Levine B: Autophagy,
immunity, and microbial adaptations. Cell Host Microbe. 5:527–549.
2009.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Mizushima N, Yoshimori T and Levine B:
Methods in mammalian autophagy. Cell. 2:313–326. 2010.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Ato K, Akaki T, Shimizu T, Sano C,
Ogasawara K and Tomioka H: Invasion and intracellular growth of
Mycobacterium tuberculosis and myco-bacterium avium complex
adapted to intramacrophagic environment within macrophages and type
II alveolar epithelial cells. Kekkaku. 76:53–57. 2001.PubMed/NCBI(In Japanese).
|
|
27
|
Orsi GB, García-Fernández A, Giordano A,
Venditti C, Bencardino A, Gianfreda R, Falcone M, Carattoli A and
Venditti M: Risk factors and clinical significance of ertapenem-
resistant Klebsiella pneumoniae in hospitalised patients. J
Hosp Infect. 78:54–58. 2011.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Won S, Munoz-Price LS, Lolans K, Hota B,
Weinstein RA and Hayden MK: Centers for Disease Control and
Prevention Epicenter Program. Emergence and rapid regional spread
of Klebsiella pneumoniae carbapenemase-producing
enterobacteriaceae. Clin Infect Dis. 53:532–540. 2011.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Wu X, Fleming A, Ricketts T, Pavel M,
Virgin H, Menzies FM and Rubinsztein DC: Autophagy regulates notch
degradation and modulates stem cell development and neurogenesis.
Nat Commun. 7:10533–10550. 2016.PubMed/NCBI View Article : Google Scholar
|