Introduction
Lung cancer is the most common cancer type with the
highest incidence and mortality among all cancers worldwide
(1). In 2018, >2 million new
lung cancer cases and 1.8 million lung cancer-associated deaths
were estimated to have occurred worldwide (1). Lung cancer is classified into small
cell lung carcinoma and non-small cell lung carcinoma (NSCLC),
which includes lung adenocarcinoma (LUAD), squamous cell carcinoma
and large cell carcinoma, and accounts for >80% of lung cancer
cases (2). Current treatment
options for NSCLC are surgery and adjuvant therapy, including
radiation, chemotherapy and targeted therapy (2). Targeted medicines, such as those
against epidermal growth factor receptor and anaplastic lymphoma
kinase, have been successfully applied in the clinic (3). However, it is still necessary to
discover more appropriate molecular targets in order to provide
personalized treatment or overcome potential drug resistance.
Sperm-associated antigen 5 (SPAG5, also known as
astrin) is involved in mitotic spindle formation and chromosome
segregation (4,5), and acts as an important regulator of
mitosis and the cell cycle (6).
Studies have revealed that SPAG5 has an oncogenic role during the
tumorigenesis of multiple cancer types. It was reported to inhibit
mTOR complex 1 (mTORC1) signaling to prevent apoptosis in HeLa
cells (7), interact with
centrosomal protein of 55 kDa to regulate PI3K/AKT signaling, and
downregulate scavenger receptor class A member 5 expression via the
Wnt/β-catenin pathway in hepatocellular carcinoma cells (8,9),
activate the AKT/mTOR pathway to upregulate Wnt3 expression in
bladder urothelial carcinoma cells (10), and upregulate survivin expression in
gastric cancer (11). It has been
indicated that SPAG5 may serve as a prognostic biomarker in
cervical cancer (12), breast
cancer (13) and bladder urothelial
carcinoma (10). A previous study
reported that p53 inhibition is essential for the upregulation of
SPAG5 in LUAD (14). However, the
role and the underlying mechanisms of SPAG5 in LUAD remain to be
fully elucidated.
In the present study, SPAG5 expression was assessed
in LUAD tissues and the association of SPAG5 expression with the
prognosis of patients with LUAD was analyzed. In addition, the role
of SPAG5 in cellular proliferation, apoptosis, autophagy and
motility of A549 cells was examined.
Materials and methods
Cell transfection and reagents
The LUAD cell lines A549, HCC827, HCC515, HCC1833,
NCI-H1975, NCI-H1993, NCI-H2347 and NCI-H3255 were purchased from
the Kunming Cell Bank of the Chinese Academy of Sciences. Cells
were maintained in RPMI-1640 medium (Gibco; Thermo Fisher
Scientific, Inc.) containing 10% FBS (Gibco; Thermo Fisher
Scientific, Inc.) at 37˚C in an incubator with 5% CO2.
A549 cells were grown to ~75% confluence and then transiently
transfected with SPAG5 small interfering RNA (siRNA; #1,
5'-GAGGAAAUUGUAGAGCAUTT-3', #2, 5'-CCCGACAACUCACAGAGAATT-3', #3,
5'-CCCGACAACUCACAGAGAATT-3') or scrambled siRNA
(5'-UUCUCCGAACGUGUCACGUTT-3') (purchased from Shanghai GenePharma
Co., Ltd.) at a concentration of 100 pM using
Lipofectamine® 3000 reagent (Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer's protocols.
Bafilomycin (used at a concentration of 100 nM) was purchased from
Cell Signaling Technology, Inc. (cat. no. 54645S).
Western blot analysis
At 48 h after transfection, total protein was
extracted from A549 cells using radioimmunoprecipitation assay
lysis buffer (150 mM NaCl; 50 M Tris-HCl; pH 7.5; 1% Triton X-100;
0.1% sodium deoxycholate and 0.1% SDS) containing 0.1% proteinase
and phosphatase inhibitors (Beijing Solarbio Science &
Technology Co., Ltd.). Immunoblot analysis was then performed as
described previously (15). The
primary antibodies used were as follows: Rabbit anti-SPAG5
polyclonal antibody [cat. no. 60940; 1:1,800; Cell Signaling
Technology, Inc. (CST)], rabbit anti-caspase-3 polyclonal antibody
(cat. no. 9662; 1:1,000; CST), rabbit anti-cleaved caspase-3
polyclonal antibody (cat. no. 9661; 1:500; CST), rabbit
anti-poly(ADP-ribose) polymerase (PARP; cat. no. 9542; 1:1,000;
CST), rabbit anti-S6 kinase (cat. no. 2708; 1:1,000; CST), mouse
anti-phosphorylated-S6 kinase (cat. no. 9206; 1:1,000; CST), rabbit
anti-light chain (LC) 3B polyclonal antibody (cat. no. NB100-2220;
1:1,000; Novus Biologicals, LLC), rabbit anti-p62 polyclonal
antibody (cat. no. 39749; 1:1,000; CST) and mouse anti-β-actin
monoclonal antibody (cat. no. TA811000; 1:1,000; OriGene
Technologies, Inc.). The secondary antibodies used were as follows:
Horseradish peroxidase (HRP)-conjugated anti-rabbit immunoglobulin
G (IgG; cat. no. 7074; 1:5,000; CST) and HRP-conjugated anti-mouse
IgG (cat. no. 7076; 1:5,000; CST). Densitometric analysis was
performed using FluorChem SA 6.0.0 (Informer Technologies,
Inc.).
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA was isolated from A549 cells using
TRIzol® reagent (Thermo Fisher Scientific, Inc.)
according to the manufacturer's protocol. RNA concentration was
measured using a NanoDrop™ 2000 spectrophotometer
(Thermo Fisher Scientific, Inc.). A total of 2 µg RNA was reverse
transcribed into complementary DNA using a reverse transcription
kit (cat. no. KR103-04; Tiangen Biotech Co., Ltd.) according to the
manufacturer's protocol. qPCR was performed using SYBR®
Premix Ex Taq™ II (Takara Bio, Inc.) to detect SPAG5
gene expression. GAPDH was amplified in parallel as the internal
control. The gene-specific primers used were as follows: SPAG5
forward, 5'-CATCTCACAGTGGGATAACTAATAAAC-3' and reverse,
5'-CAGGGATAGGTGAAGCAAGGATA-3' and GAPDH forward,
5'-CAGGAGGCATTGCTGATGAT-3' and reverse, 5'-GAAGGCTGGGGCTCATTT-3'.
The following thermocycling conditions were used for the qPCR:
Pre-denaturation at 95˚C for 30 sec, followed by 40 cycles of
denaturation at 95˚C for 5 sec, annealing at 60˚C for 20 sec and
extension at 72˚C for 10 sec, and final extension at 72˚C for 30
sec. The 2-ΔΔCq method was used to calculate relative
gene expression (16).
Flow cytometric analysis
A549 cells (1x105 cells/well) were seeded
in 12-well plates and cultured for 17 h. Subsequently, the cells
were transfected with SPAG5 siRNA or control siRNA for 48 h. The
cells were harvested and incubated with 5 µl FITC Annexin V and 5
µl propidium iodide (PI) for 15 min in the dark, and 300 µl 1X
binding buffer was then added. Finally, the cells were analyzed by
flow cytometry (Muse™ Cell Analyzer; Merck Millipore).
Cells with positive FITC Annexin V and negative PI were considered
to be apoptotic.
Cell proliferation assay
A549 cells were transfected with SPAG5 siRNA or
control siRNA. At 48 h post-transfection, 5x103 cells
were seeded into 96-well plates in RPMI-1640 medium containing 2%
FBS. The media was replaced every day for 4 days. Cell
proliferation was determined with an MTT kit (cat. no. M1020;
Beijing Solarbio Science & Technology Co., Ltd.) according to
the manufacturer's protocol. In brief, MTT was dissolved in 0.1 M
PBS at a concentration of 5 mg/ml. Subsequently, 10 µl MTT solution
was added to each well, followed by incubation at 37˚C for 4 h.
Finally, RPMI-1640 culture medium was discarded and 200 µl dimethyl
sulfoxide was then added to each well to completely dissolve the
crystals. The absorbance at a wavelength of 490 nm was measured
with a microplate reader (SpectraMax® Plus 384;
Molecular Devices).
Cell migration and invasion assay
Following transfection with SPAG5 siRNA or negative
control for 48 h, 8-µm pore size culture inserts (Transwell;
Costar; Corning Inc.) were used for the migration assay and
Matrigel Invasion Chambers (Transwell; Costar; Corning Inc.) were
used for the invasion assay. The inserts were placed into the wells
of 24-well culture plates, separating the plates into upper and
lower chambers. A total of 600 µl RPMI-1640 medium containing 20%
FBS was added to the lower chambers. Furthermore, 3x104
cells in 200 µl serum-free RPMI-1640 medium were added to each
upper chamber. After incubation at 37˚C in an atmosphere with 5%
CO2 for 24 h, non-invading or non-migrating cells were
removed from the top wells with a cotton swab. Cells that had
transgressed to the bottom of the membrane were fixed with 4%
paraformaldehyde for 20 min at room temperature and stained with
0.1% crystal violet overnight at 4˚C. Randomly selected images of
three independent fields of each well were captured with a
phase-contrast microscope (magnification, x100; Olympus TH4-200;
Olympus Corporation) and the cells were manually counted.
Colony formation assay
Following transfection with SPAG5 siRNA or negative
control for 48 h, 6x102 cells were seeded into each well
of six-well plates, and 2 ml RPMI-1640 culture medium was added
into each well. Cells were cultured at 37˚C in a humidified
incubator with 5% CO2 for 10-15 days. Cells was washed
with PBS three times, fixed with 4% neutral paraformaldehyde
solution at room temperature for 30 min, followed with three PBS
washes, and 2 ml 1% crystal violet solution was added to each well
and incubated at room temperature for 2 h. The cells were washed
three times with PBS after the crystal violet solution was
discarded. Once the plates were dry, the colonies were counted and
images were captured.
Analysis of SPAG5 expression in LUAD
tissues
LUAD data from The Cancer Genome Atlas (TCGA) were
analyzed using the Gene Expression Profiling Interactive Analysis
(GEPIA; gepia.cancer-pku.cn) online tool
(17). The Okayama Lung cohort
(GSE31210 in Gene Expression Omnibus database) was analyzed using
ONCOMINE (www.oncomine.org) (18,19).
TCGA LUAD datasets (TCGA Pan-Cancer Atlas) were downloaded from
cbioportal (https://www.cbioportal.org/datasets) and SPAG5
expression at different clinical stages of TCGA LUAD was analyzed
using GraphPad Prism 5.0 software (GraphPad Software, Inc.)
(20). Comparisons among normal
groups and different clinical stages were performed using one-way
ANOVA followed by Tukey's post hoc test. SPAG5 expression data by
immunohistochemical (IHC) staining were obtained from The Human
Protein Atlas (HPA) database (www.proteinatlas.org) (21).
Prognosis analysis
The univariate analysis of SPAG5 expression on
overall survival (OS), first progression (FP) and post-progression
survival (PPS) was analyzed using the online tool Kaplan Meier
plotter (www.kmplot.com) (22). The cut-off values were as follows:
266 for OS, 216 for FP and 305 for PPS, which were the best
threshold values automatically determined by the software. Hazard
ratio (HR), 95% confidence intervals (CI) and log-rank P-values
were calculated using Cox regression. The log-rank test was used to
evaluate the differences between survival curves. Cox proportional
hazards model was used for multivariate analysis of sex, American
Joint Committee on Cancer (AJCC) stage T and stage N, smoking
history and SPAG5 expression on OS and FP. P<0.05 was considered
to indicate a statistically significant difference.
Statistical analysis
SPSS 13.0 software (SPSS, Inc.) was used to perform
statistical analysis. Values are expressed as the mean ± SD.
Comparisons between two groups were performed using Student's
t-test and comparisons among multiple groups were performed using
one-way ANOVA followed by Tukey's post hoc test. All experiments
were performed at least three times independently and
representative data are presented. The log-rank test was used to
evaluate the differences between survival curves. Cox proportional
hazards model was used for multivariate analysis. P<0.05 was
considered to indicate a statistically significant difference.
Results
SPAG5 expression is upregulated in
LUAD tissues and its high expression is associated with unfavorable
prognosis
To determine SPAG5 expression in LUAD tissues, the
LUAD data in TCGA were analyzed using the online tool GEPIA
(17). The results indicated that
SPAG5 expression was significantly upregulated in LUAD tissues
(n=483) compared with normal lung tissues (n=59) (P<0.05;
Fig. 1A). To confirm the TCGA
results, SPAG5 expression was analyzed in LUAD tissues (n=226) and
normal lung tissues (n=20) in the Okayama Lung cohort from the
ONCOMINE database (18,19). The results demonstrated that SPAG5
expression was significantly increased in LUAD compared with normal
lung tissues (P<0.001; Fig. 1B),
consistent with the results of the LUAD cohort from TCGA.
Furthermore, SPAG5 expression we analyzed in LUAD samples at
different clinical stages from TCGA. The results showed that SPAG5
expression at stages 1-4 (stage 1, n=277; stage 2, n=120; stage 3,
n=84; stage 4, n=26;) was significantly increased compared with
normal lung tissues (n=59) (P<0.001; Fig. 1C), while there was no significant
difference between any two stages of LUAD. Since the TCGA and
ONCOMINE data were obtained at the RNA level, alterations in SPAG5
expression were further analyzed at the protein level by searching
the HPA database (21). The results
of the HPA included two datasets for IHC staining of lung cancer
tissue (HPA022008 and HPA022479) comprising a total of 11 cases
stained with one of two different SPAG5 antibodies. The HPA022008
dataset comprised one LUAD sample with low and five with medium
SPAG5-positive staining, while the HPA022479 dataset comprised two
LUAD samples with negative, two with low and one with highly
positive SPAG5 staining. Representative images are provided in
Fig. 1D. The IHC data confirmed
SPAG5 expression in LUAD tissues at the protein level.
 | Figure 1SPAG5 expression is upregulated in
LUAD. (A) SPAG5 expression in LUAD tissues (n=483) and normal lung
tissues (n=59) from TCGA database was analyzed with the Gene
Expression Profiling Interactive Analysis online software. The
results demonstrated that SPAG5 expression was significantly
increased in LUAD. *P<0.05. (B) SPAG5 expression in
LUAD tissues (n=226) and normal lung tissues (n=20) from the
ONCOMINE database (Okayama Lung) was analyzed online. The results
demonstrated that SPAG5 expression was significantly increased in
LUAD. ***P<0.001. (C) SPAG5 expression in different
stages of LUAD was significantly increased compared with in normal
tissues. However, there was no significant difference between any
stages. SPAG5 expression at different stages (stage 1, n=277; stage
2, n=120; stage 3, n=84; stage 4, n=26) and normal lung tissues
(n=59) from TCGA database was analyzed. One-way ANOVAs and Tukey's
post hoc test were used for the comparison between the groups.
***P<0.001. (D) Immunohistochemistry staining results
from The Human Protein Atlas database demonstrated that SPAG5
expression was upregulated in LUAD tissues. The patient ID for the
sample with low expression was 2585 and that for high expression
was 3391. Scale bar, 100 µm in low-magnification images and 25 µm
in high-magnification images. TCGA, The Cancer Genome Atlas; LUAD,
lung adenocarcinoma; SPAG5, sperm-associated antigen 5; TPM,
transcripts per million; FPKM, fragments per kilobase of transcript
per million mapped reads. |
To reveal the potential prognostic value of SPAG5 in
patients with LUAD, the association between SPAG5 expression and
OS, FP and PPS was analyzed using the online tool Kaplan Meier
plotter (22). The results of the
univariate analysis demonstrated that high expression of SPAG5 is
associated with unfavorable OS and FP (Fig. 2A and B), while there was no significant
association with PPS (Fig. 2C),
suggesting that SPAG5 expression may serve as a predictor of OS and
FP. The results of the multivariate analysis of AJCC stage, sex and
smoking history suggested that high expression of SPAG5 is an
independent prognostic indicator for OS and FP (Table I).
 | Table ICox regression multivariate analysis
on OS and FP. |
Table I
Cox regression multivariate analysis
on OS and FP.
| Rate | Variable | Hazard ratio (95%
CI) | P-value |
|---|
| OS | Sex | 1.55 (0.86-2.79) | 0.1410 |
| | AJCC stage T | 2.64 (1.34-5.21) | 0.0051 |
| | AJCC stage N | 2.83 (1.69-4.75) | 0.0001 |
| | Smoking history | 0.82 (0.38-1.77) | 0.6138 |
| | SPAG5 | 1.89
(1.01-3.53) | 0.0469 |
| FP | Sex | 1.22
(0.66-2.26) | 0.5333 |
| | AJCC stage T | 2.87
(1.34-6.17) | 0.0068 |
| | AJCC stage N | 2.48
(1.41-4.37) | 0.0017 |
| | Smoking
history | 1.41
(0.7-2.84) | 0.3412 |
| | SPAG5 | 2.05 (1.05-4) | 0.0357 |
Knockdown of SPAG5 suppresses the
proliferation, migration and invasion of A549 cells
To investigate the role of SPAG5 in lung cancer
in vitro, SPAG5 protein expression was examined in multiple
LUAD cell lines by western blot analysis. The results indicated
SPAG5 was expressed in all cell lines tested and its expression in
A549 cells was the highest (Fig.
3A). Therefore, the A549 cell line was selected for subsequent
experiments. A total of three siRNAs against SPAG5 were designed
and their effects to suppress SPAG5 expression were assessed by
RT-qPCR and western blot analysis. A549 cells were individually
transfected with SPAG5 siRNAs#1-3 and control siRNA, and SPAG5
expression was examined 48 h after transfection. The results
indicated that all SPAG5 siRNAs significantly reduced SPAG5
expression at the RNA and protein level compared with control siRNA
transfection, and SPAG5 siRNA#2 had the most potent inhibitory
effect (Fig. 3B). Thus, SPAG5
siRNA#2 was used in further experiments.
Since SPAG5 was indicated to be associated with
unfavorable OS and FP of patients with LUAD, it was hypothesized
that SPAG5 may have a role in the regulation of proliferation and
motility of LUAD cells. To test this hypothesis, the proliferation
and motility of A549 cells transfected with SPAG5 siRNA were
analyzed using MTT and Transwell assays, respectively. The results
suggested that, compared with that in the control siRNA group, the
proliferation (Fig. 4A), migration
(Fig. 4B) and invasion (Fig. 4C) of A549 cells transfected with
SPAG5 siRNA was significantly attenuated.
Knockdown of SPAG5 induces apoptosis
in A549 cells
The effects of SPAG5 knockdown on A549 cell
apoptosis were then examined by flow cytometry and western blot
analysis. Flow cytometric analysis demonstrated that SPAG5 siRNA
led to significant increase in apoptotic cells compared with
control siRNA (Fig. 5A). To confirm
the flow cytometry results, the activation of the apoptotic markers
caspase-3 and PARP was detected by western blot analysis. The
result suggested that, compared with the control siRNA group,
transfection of SPAG5 siRNA resulted in a decrease of caspase-3 and
PARP protein and a simultaneous increase of cleaved caspase-3 and
cleaved PARP protein (Fig. 5B),
indicating that SPAG5 knockdown induced apoptosis in A549
cells.
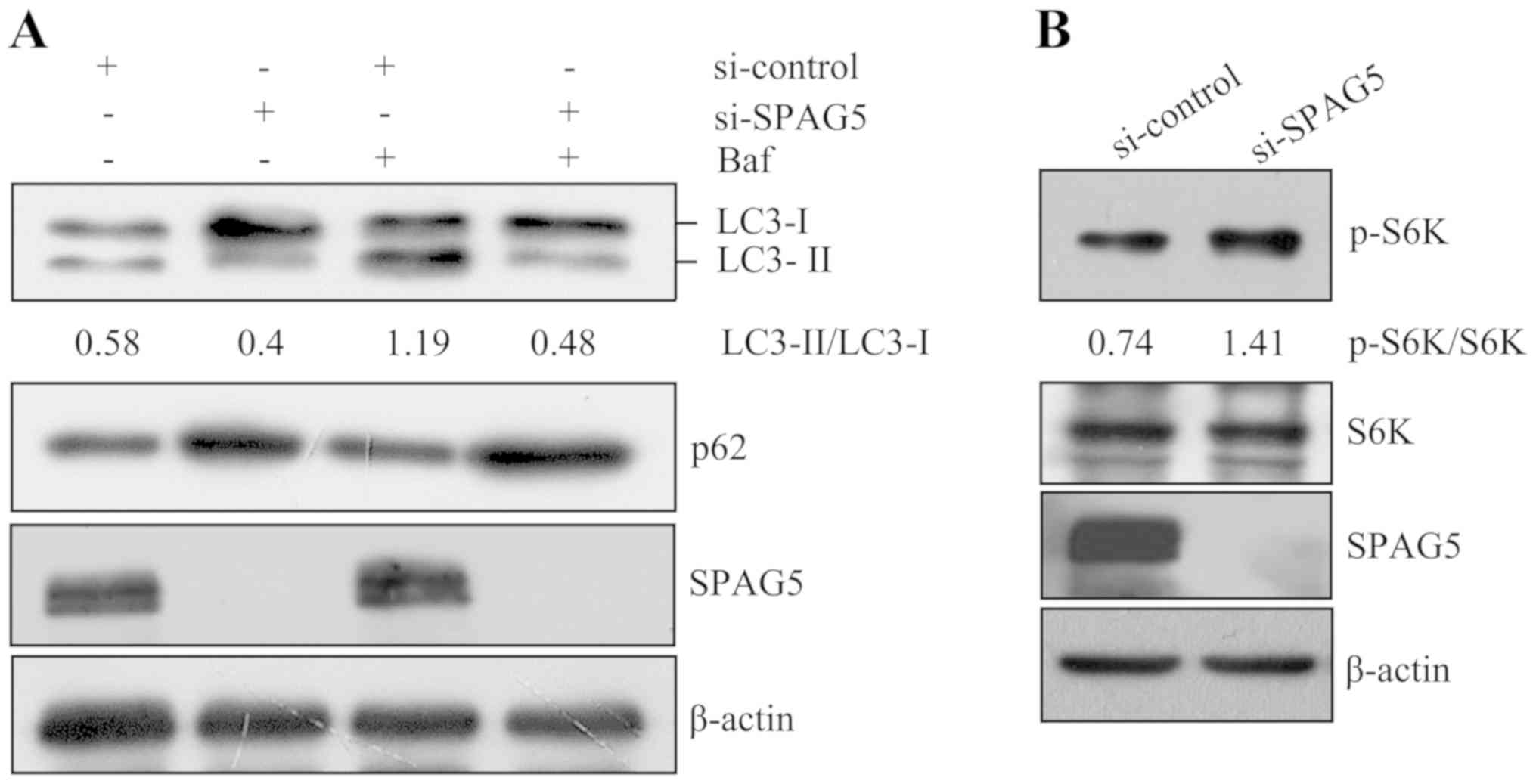
Knockdown of SPAG5 inhibits autophagy
in A549 cells
As a previous study indicated that in HeLa cells,
SPAG5 inhibits mTORC1, which is a vital upstream negative regulator
of autophagy (7), it was
hypothesized that SPAG5 may enhance autophagy. To assess this
hypothesis, A549 cells were transfected with SPAG5 siRNA for 48 h
and then treated with the autophagy inhibitor bafilomycin for 1 h.
Subsequently, the autophagic markers LC3 and p62 were examined by
western blot analysis. The results suggested that transfection of
SPAG5 siRNA led to a decrease of LC3-II and a simultaneous increase
of p62 compared with control siRNA (Fig. 6A), indicating that SPAG5 knockdown
inhibited autophagy. In addition, mTORC1 activity was examined by
detecting phosphorylation of its substrate S6K by western blot
analysis. The results indicated that compared with the control
siRNA group, S6K phosphorylation increased following SPAG5
knockdown (Fig. 6B), implying
increased mTORC1 activity.
Discussion
Patients with lung cancer have a poor prognosis and
the 5-year survival is <18% (2).
Therefore, it is urgent to discover novel biomolecules that may be
used as diagnostic markers and/or therapeutic targets. Välk et
al (23) analyzed the gene
expression profiles of 81 tissue samples of patients with NSCLC,
revealing that SPAG5 is upregulated in NSCLC and therefore
suggested that SPAG5 may be a biomarker in this cancer type.
However, the underlying mechanisms of SPAG5 in LUAD have remained
elusive. The present study demonstrated that SPAG5 expression was
upregulated in LUAD by analyzing its expression in two different
large LUAD cohorts from two databases. However, SPAG5 expression
did not change significantly from LUAD stage 1 to stage 4. These
results indicated that SPAG5 upregulation occurs early during the
tumorigenesis of LUAD, implying that SPAG5 may be a potential
diagnostic marker for LUAD. In addition, high expression of SPAG5
was indicated to be associated with unfavorable OS and FP, which
may also suggest that SPAG5 may be used as a prognostic marker in
LUAD.
Dysregulation of proliferation is a hallmark of
malignant tumor growth (24). It
was hypothesized that inhibiting SPAG5 expression may suppress
certain cancerous features of LUAD cells. Thus, this possibility
was tested in vitro using LUAD A549 cells, which exhibited
high endogenous SPAG5 expression. The present study demonstrated
that, in A549 cells, knockdown of SPAG5 inhibited cell
proliferation and induced apoptosis, indicating that SPAG5 may have
a role in the regulation of cell proliferation and death. These
results are in agreement with those of previous studies on cervical
cancer (12), bladder urothelial
carcinoma (10) and gastric cancer
(11). Although the apoptosis of
A549 cells was analyzed upon SPAG5 knockdown by flow cytometry to
detect annexin and by western blotting to examine the apoptotic
markers caspase-3 and PARP, it would be useful to confirm the
results in cells treated with caspase inhibitor when performing
further experiments. Certain carcinomas arise from epithelial
tissues and may subsequently obtain the capability of migration and
invasion to progress to higher pathological grades of malignancy
due to aberrations in the expression of relevant genes associated
with invasion and metastasis, such as N-cadherin (24). The present study indicated that
knockdown of SPAG5 in A549 cells inhibited cell migration and
invasion, suggesting that SPAG5 may have a role in tumor
metastasis. As SPAG5 may act via the Wnt/β-catenin and AKT/mTOR
pathways (8,10), both of which are crucial pathways in
a variety of cancers, it is necessary to explore which mechanism
enables LUAD cells to obtain those features in the future. As
epidermal growth factor receptor (EGFR) acts upstream of the
AKT/mTOR pathway, it was questioned whether there is any
association between EGFR and SPAG5. To address this, the
correlation between EGFR and SPAG5 expression in LUAD tissues was
analyzed with GEPIA. However, a weak but not significant
correlation was observed (data not shown), indicating they do not
significantly affect the expression of each other.
Autophagy is a lysosomal degradation pathway through
which cells are able to recycle unwanted or damaged proteins or
organelles to maintain cellular homeostasis or in response to
microenvironmental stress (25).
Accumulating evidence has indicated that autophagy may serve as a
tumor-suppressing or tumor-promoting process, depending on the
context or tumor type (26). A
previous study reported that SPAG5 inhibits mTORC1 activity by
binding to Raptor to prevent mTORC1 assembly in HeLa cells
(7). Given that mTORC1 acts as a
suppressor of autophagy, it was hypothesized that SPAG5 may have a
role in the regulation of autophagy. Thus, the effect of SPAG5
knockdown on autophagy was examined in the present study. The
results indicated that knockdown of SPAG5 inhibited autophagy
accompanied by increased S6K phosphorylation, implying SPAG5
promoted autophagy through regulating the activity of mTORC1 in
A549 cells. Therefore, SPAG5 may promote tumor cell survival
through enhancing autophagy to recycle cellular components in the
microenvironment lacking oxygen and nutrients. To the best of our
knowledge, the present study was the first to reveal the role of
SPAG5 in the regulation of autophagy.
Since high SPAG5 expression was observed in LUAD
tissues and may contribute to cancerous features, including
excessive proliferation, evasion of apoptosis and enhancement of
autophagy, it is important to uncover how SPAG5 expression is
regulated in LUAD. Wang et al (14) observed that SPAG5 expression in LUAD
cells is suppressed by p53. Song et al (27) reported that microRNA (miR)-1179
inhibited SPAG5 expression in NSCLC cells. Wang et al
(28) revealed that long non-coding
RNA LINC00875 may sponge miR-1179 to regulate SPAG5 expression in
LUAD cells. As the in vitro experiments in the present study
were only performed in A549 cells with siRNA to knock down SPAG5,
further experiments including knockdown and overexpression of SPAG5
should be performed in other LUAD cell lines in order to confirm
the conclusions.
In summary, the present study demonstrated that the
expression of SPAG5 was upregulated in LUAD tissues and its high
expression is associated with unfavorable clinical outcomes. It may
promote the proliferation, motility and autophagy, and inhibit
apoptosis of LUAD cells. The present results indicated that SPAG5
has an oncogenic role in LUAD and may be a potential prognostic
predictor and therapeutic target for LUAD.
Acknowledgements
Not applicable.
Funding
This work was supported in part by the Research
Enhancement Project for Junior Faculty in Higher Education
Institutes of Guangxi (grant no. 2019KY0522), the Scientific
Research Project for Junior Faculty in Guilin Medical College
(grant no. 2018glmcy055) and the Open Research Fund from Guangxi
Key Laboratory of Liver Injury and Repair Molecular Medicine (grant
no. GXLIRMMKL-201816).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
AL conceived the study. RH performed the
bioinformatical analysis and AL performed the experiments. RH and
AL analyzed and interpreted the data. AL and RH wrote the
manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Zappa C and Mousa SA: Non-small cell lung
cancer: Current treatment and future advances. Transl Lung Cancer
Res. 5:288–300. 2016.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Herbst RS, Morgensztern D and Boshoff C:
The biology and management of non-small cell lung cancer. Nature.
553(446)2018.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Mack GJ and Compton DA: Analysis of
mitotic microtubule-associated proteins using mass spectrometry
identifies astrin, a spindle-associated protein. Proc Natl Acad Sci
USA. 98:14434–14439. 2001.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Thein KH, Kleylein-Sohn J, Nigg EA and
Gruneberg U: Astrin is required for the maintenance of sister
chromatid cohesion and centrosome integrity. J Cell Biol.
178:345–354. 2007.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Liu L, Akhter S, Bae JB, Mukhopadhyay SS,
Richie CT, Liu X and Legerski R: SNM1B/Apollo interacts with astrin
and is required for the prophase cell cycle checkpoint. Cell Cycle.
8:628–368. 2009.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Thedieck K, Holzwarth B, Prentzell MT,
Boehlke C, Kläsener K, Ruf S, Sonntag AG, Maerz L, Grellscheid SN,
Kremmer E, et al: Inhibition of mTORC1 by astrin and stress
granules prevents apoptosis in cancer cells. Cell. 154:859–874.
2013.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Yang YF, Zhang MF, Tian QH, Fu J, Yang X,
Zhang CZ and Yang H: SPAG5 interacts with CEP55 and exerts
oncogenic activities via PI3K/AKT pathway in hepatocellular
carcinoma. Mol Cancer. 17(117)2018.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Liu H, Hu J, Wei R, Zhou L, Pan H, Zhu H,
Huang M, Luo J and Xu W: SPAG5 promotes hepatocellular carcinoma
progression by downregulating SCARA5 through modifying β-catenin
degradation. J Exp Clin Cancer Res. 37(229)2018.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Liu JY, Zeng QH, Cao PG, Xie D, Yang F, He
LY, Dai YB, Li JJ, Liu XM, Zeng HL, et al: SPAG5 promotes
proliferation and suppresses apoptosis in bladder urothelial
carcinoma by upregulating Wnt3 via activating the AKT/mTOR pathway
and predicts poorer survival. Oncogene. 37:3937–3952.
2018.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Liu G, Liu S, Cao G, Luo W, Li P, Wang S
and Chen Y: SPAG5 contributes to the progression of gastric cancer
by upregulation of Survivin depend on activating the wnt/β-catenin
pathway. Exp Cell Res. 379:83–91. 2019.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Yuan LJ, Li JD, Zhang L, Wang JH, Wan T,
Zhou Y, Tu H, Yun JP, Luo RZ, Jia WH and Zheng M: SPAG5
upregulation predicts poor prognosis in cervical cancer patients
and alters sensitivity to taxol treatment via the mTOR signaling
pathway. Cell Death Dis. 5(e1247)2014.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Abdel-Fatah TMA, Agarwal D, Liu DX,
Russell R, Rueda OM, Liu K, Xu B, Moseley PM, Green AR, Pockley AG,
et al: SPAG5 as a prognostic biomarker and chemotherapy sensitivity
predictor in breast cancer: A retrospective, integrated genomic,
transcriptomic, and protein analysis. Lancet Oncol. 17:1004–1018.
2016.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Wang T, Li K, Song H, Xu D, Liao Y, Jing
B, Guo W, Hu M, Kuang Y, Sun B, et al: p53 suppression is essential
for oncogenic SPAG5 upregulation in lung adenocarcinoma. Biochem
Biophys Res Commun. 513:319–325. 2019.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Li A, Wang Q, He G, Jin J and Huang G: DEP
domain containing 1 suppresses apoptosis via inhibition of A20
expression, which activates the nuclear factor κB signaling pathway
in HepG2 cells. Oncol Lett. 16:949–955. 2018.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Tang Z, Li C, Kang B, Gao G, Li C and
Zhang Z: GEPIA: A web server for cancer and normal gene expression
profiling and interactive analyses. Nucleic Acids Res. 45:W98–W102.
2017.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Rhodes DR, Yu J, Shanker K, Deshpande N,
Varambally R, Ghosh D, Barrette T, Pandey A and Chinnaiyan AM:
ONCOMINE: A cancer microarray database and integrated data-mining
platform. Neoplasia. 6:1–6. 2004.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Okayama H, Kohno T, Ishii Y, Shimada Y,
Shiraishi K, Iwakawa R, Furuta K, Tsuta K, Shibata T, Yamamoto S,
et al: Identification of genes upregulated in ALK-positive and
EGFR/KRAS/ALK-negative lung adenocarcinomas. Cancer Res.
72:100–111. 2012.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Sanchez-Vega F, Mina M, Armenia J, Chatila
WK, Luna A, La KC, Dimitriadoy S, Liu DL, Kantheti HS, Saghafinia
S, et al: Oncogenic signaling pathways in the cancer genome atlas.
Cell. 173:321–37.e10. 2018.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Uhlen M, Zhang C, Lee S, Sjöstedt E,
Fagerberg L, Bidkhori G, Benfeitas R, Arif M, Liu Z, Edfors F, et
al: A pathology atlas of the human cancer transcriptome. Science.
357(eaan2507)2017.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Győrffy B, Surowiak P, Budczies J and
Lánczky A: Online survival analysis software to assess the
prognostic value of biomarkers using transcriptomic data in
non-small-cell lung cancer. PLoS One. 8(e82241)2013.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Välk K, Vooder T, Kolde R, Reintam MA,
Petzold C, Vilo J and Metspalu A: Gene expression profiles of
non-small cell lung cancer: Survival prediction and new biomarkers.
Oncology. 79:283–292. 2010.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Hanahan D and Weinberg RA: Hallmarks of
cancer: The next generation. Cell. 144:646–674. 2011.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Levine B and Kroemer G: Biological
functions of autophagy genes: A disease perspective. Cell.
176:11–42. 2019.PubMed/NCBI View Article : Google Scholar
|
|
26
|
White E: The role for autophagy in cancer.
J Clin Invest. 125:42–46. 2015.PubMed/NCBI View
Article : Google Scholar
|
|
27
|
Song L, Dai Z, Zhang S, Zhang H, Liu C, Ma
X, Liu D, Zan Y and Yin X: MicroRNA-1179 suppresses cell growth and
invasion by targeting sperm-associated antigen 5-mediated Akt
signaling in human non-small cell lung cancer. Biochem Biophys Res
Commun. 504:164–170. 2018.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Wang L, Cao L, Wen C, Li J, Yu G and Liu
C: LncRNA LINC00857 regulates lung adenocarcinoma progression,
apoptosis and glycolysis by targeting miR-1179/SPAG5 axis. Hum
Cell. 33:195–204. 2020.PubMed/NCBI View Article : Google Scholar
|