Introduction
Alzheimer's disease (AD), the most common cause of
dementia in elderly individuals worldwide, is characterized by
progressive memory and cognitive impairments (1). The abnormal accumulation of
extracellular β-amyloid (Aβ) plaques in the brain is an important
molecular pathological feature of patients with AD (2). Aβ is produced by proteolytic cleavage
of the amyloid precursor protein (APP) by β-secretases and
γ-secretases (3,4), which generates soluble (s)APPβ,
amyloid-β1-40 (Aβ1-40) and Aβ1-42
fragments. The alternative proteolytic pathway of APP involves the
release of α-secretase, which prevents the formation of Aβ due to
its cleavage site and produces sAPPα that serves as a
neuroprotective player (5,6).
MicroRNAs (miRNAs/miRs) are a class of endogenous
22-nucleotide non-coding RNAs that base-pair with the
3'-untranslated region (UTR) of specific genes, thereby repressing
gene expression. Certain miRNAs serve as crucial players of
neuronal gene expression that contribute to neurogenesis, neuronal
maturation, brain development and neuroplasticity (7,8). The
involvement of miRNAs in the pathophysiology of AD has been
demonstrated in a number of previous studies. For example, miR-15b
inhibits Aβ accumulation by targeting NF-κB signaling and
β-secretase (BACE)1(9).
microRNA-219 downregulation promotes neurodegeneration via
post-transcriptional regulation of microtubule associated protein
tau (10). Recently, miR-149-5p was
reported to protect against high glucose-induced pancreatic β-cell
apoptosis (11). Furthermore,
miR-149-5p was also revealed to regulate pentraxin resistance in
breast cancer (12). However, the
role of miR-149-5p in regulating APP and Aβ production is not
completely understood. Epigenetic dysregulation serves a vital role
in the onset and progression of AD (13-21),
which include chromatin variation, DNA methylation and non-coding
RNAs (22). Among the widespread
epigenetic modifications, histone acetylation and deacetylation are
chromatin variations that regulate gene transcription (23). Histone acetylation is downregulated
at regulatory regions of memory genes in mouse models of AD
(24). Nonselective histone
deacetylase inhibitor treatments were reported to improve memory
impairments by restoring normal histone acetylation levels
(25,26). Among histone acetylation marks,
H4K16ac facilitates gene activation and damaged DNA repair by
influencing its chromatin structure (27). The largest meta-analytic genome-wide
association study identified branched chain keto acid dehydrogenase
kinase/KAT8 as a new AD-associated loci (28). KAT8 is the major lysine acetyl
transferase responsible for the acetylation of H4K16 in flies and
mammals (29). The present study
compared serum histone acetylation and KAT8 in patients with AD and
cognitively healthy individuals. In addition, the expression of
miR-149-5p in an AD cell model in vitro was investigated,
and the roles of miR-149-5p and its interaction with KAT8 in the AD
cell model were also explored. Collectively, the results suggested
that miR-149-5p levels were increased in patients with AD compared
with healthy subjects, which regulated H4K16ac in 293/APPsw cells
via targeting KAT8. Therefore, the present study indicated that
targeting miR-149-5p may display neuroprotective effects in AD,
suggesting that miR-149-5p may serve as a novel therapeutic
target.
Materials and methods
Plasma analyses
Blood samples were obtained from patients with AD
(n=30) and healthy volunteers (n=30) with typical cognitive
performance. Patients with AD were confirmed by screening cognitive
impairment by conducting a Mini-Mental State Examination (patients
with a score ≤24 were included in the present study) and a
neuropsychological test via Clinical Dementia Rating (an assay that
can identify very mild dementia; dementia scale: 0, none; 0.5, very
mild; 1, mild; 2, moderate; and 3, severe) (30). All patients with AD had an overall
CDR of ≥0.5. Patients with AD were diagnosed according to
NINCDS-ADRDA criteria (31). To
further diagnose AD based on imaging-based techniques, the magnetic
resonance image (MRI) relaxation time constant was examined in the
brains of patients with AD according to a previous report (32). Neuropsychological impairment was
measured according to the Global Deficit Score for classifying
Neuropsychological impairment (33). All healthy subjects had no brain
diseases, as assessed by MRI and computed tomography. In addition,
healthy subjects did not have a history or symptoms of ischemic or
hemorrhagic stroke. Exclusion criteria were as follows: i) history
of any vascular or systemic disease; ii) any other
neurodegenerative disease; iii) any psychiatric disease; iv)
epilepsy; and v) substance or drug abuse. The present study was
approved by the Institutional Ethics Committee of First Teaching
Hospital of Tianjin University of Traditional Chinese Medicine
(approval no. KY-E-2017-12-30). Written informed consent was
obtained from all patients and healthy subjects. Blood samples were
collected in EDTA-containing tubes and processed within 2 h. After
collection, blood samples were centrifuged at 1,000 x g for 10 min
at 4˚C. Plasma was isolated and stored 1 ml aliquots at -80˚C until
further analysis.
Cell Culture
Human embryonic kidney (293) cells carrying the
Swedish mutation of APP (293/APPsw), which were kindly provided by
the Centre for Translational Medicine, Tianjin University of
Traditional Chinese Medicine, were used in the present study. APPsw
is an established AD cell model. Cells were cultured in DMEM/F-12
supplemented with 10% FBS and 500 µg/ml G418 (all from Gibco;
Thermo Fisher Scientific, Inc.) at 37˚C in a humidified atmosphere
of 5% CO2.
Transfection
293/APPsw cells were plated at 5x104
cells per well in a 24-well plate and transfected with 100 nM
miR-149-5p inhibitor, inhibitor negative control (inhibitor NC),
short hairpin RNA (sh)-Scramble (sh-NC) orsh-KAT8, which were all
purchased from GenePharma. Lipofectamine® 3000
(Invitrogen; Thermo Fisher Scientific, Inc.) was used to transfect
cells according to the manufacturer's protocol. Subsequent in
vitro assays were performed 3 days post-transfection. Sequences
of the oligonucleotides were as follows: miR-149-5p inhibitor:
5'-GGGAGUGAAGACACGGAGCCAGA-3' and inhibitor NC:
5'-CGAACGUGUCACGUTT-3. For stable KAT8 overexpression, at 50%
confluence, 293/APPsw cells were infected with purified lentiviral
particles expressing KAT-8 (LV-KAT-8-puromycin; Shanghai GeneChem
Co., Ltd.) or NC lentiviral particles (LV-puromycin; Shanghai
GeneChem Co., Ltd.) at MOI=20. Subsequently, KAT8 overexpression
cells were selected using puromycin for 72 h at 37˚C in a
humidified atmosphere of 5% CO2. Reverse
transcription-quantitative PCR and western blotting were performed
to assess transfection efficiency.
ELISA assay
Aβ1-40 and Aβ1-42 levels in
the cell medium were determined using a specific sandwich ELISA kit
(Aβ1-40, cat. no. MBS263658; Aβ1-42, cat. no.
MBS703888; both MyBioSource, Inc.) according to the manufacturer's
protocol.
Western blotting
Total protein was extracted from cells using RIPA
buffer (Beyotime Institute of Biotechnology). Protein concentration
was determined using the Bicinchoninic Acid Protein Assay (Pierce;
Thermo Fisher Scientific, Inc.). Equal amounts of protein (20
µg/lane) were separated via 8-12% SDS-PAGE and transferred onto
PVDF membranes (EMD Millipore). After blocking with 5% skim milk in
TBS for 2 h at room temperature, the membranes were incubated with
the following primary antibodies at 4˚C overnight: Anti-H3K4ac
(Abcam; 1:500; cat. no. ab232931), anti-H3K27ac (Abcam; 1:500; cat.
no. ab4729), anti-H4K12ac (EMD Millipore; 1:3,000, 04-119),
anti-H4K16ac (EMD Millipore; 1:3,000; cat. no. 07-329), anti-APP
(EMD Millipore; 1:1,500; cat. no. AB5300), anti-sAPPα (EMD
Millipore; 1:1,000; cat. no. JP11088), anti-sAPPβ (EMD Millipore;
1:1,000; cat. no. MABN640), anti-BACE1 (Epitomics; 1:2,000; cat.
no. ABCA0158537), anti-BACE2 (Epitomics; 1:2,000; cat. no.
ABCA0157823) and anti-GAPDH (Santa Cruz Biotechnology, Inc.;
1:1,000; cat. no. sc-47724). After washing with TBST (0.1% Tween
20), the membranes were incubated with horseradish
peroxidase-conjugated corresponding secondary antibodies (Santa
Cruz Biotechnology, Inc.; 1:5,000; cat. no. sc-2357 for anti-rabbit
IgG-HRP, and cat. no. sc-2005 for anti-mouse IgG-HRP) at 37˚C for 1
h. Protein bands were visualized using ECL substrate (Beyotime
Institue of Biotechnology). Protein expression levels were
semi-quantified using ImageJ software (v1.8.0, National Institutes
of Health) with GAPDH (KAT8, APP, sAPPα, sAPPβ, BACE1 and BACE2) or
H3 and H4 (H3K4ac, H3K27ac, H4K12ac and H4K16ac) as the loading
controls (30,31).
RT-qPCR
Total RNA was extracted from transfected 293/APPsw
cells using TRIzol® reagent (Invitrogen; Thermo Fisher
Scientific, Inc.) following the manufacturer's protocol. Total RNA
was reverse transcribed into cDNA using the PrimeScript™ RT reagent
Kit (Takara Biomedical Technology (Beijing) Co., Ltd.), and the
reverse transcription conditions were 37˚C, 15 min; 85˚C, 5 sec.
qPCR was performed using SYBR Premix Ex Taq kit (Takara Biomedical
Technology Co., Ltd.) according to the manufacturer's instructions.
The thermocycling conditions were 95˚C for 10 min (initial
denaturation) followed by 40 cycles of 95˚C, 15 sec (denaturation);
60˚C, 1 min (annealing). The final stage was 95˚C, 5 min; 60˚C, 1
min; 95˚C, 15 sec for 1 cycle to obtain a dissolution curve. The
following primers were used for qPCR: miR-149-5p forward,
5'-GGCTCTGGCTCCGTGTCTT-3' and reverse, 5'-CAGTGCAGGGTCCGAGGTATT-3';
KAT8 forward, 5'-GTCACGGTGGAGATCGGAGA-3' and reverse,
5'-CCCTCCTGGTCGTTCACTC-3'; U6 forward,
5'-GCTTCGGCAGCACATATACTAAAAT-3' and reverse,
5'-CGCTTCACGAATTTGCGTGTCAT-3'; and GAPDH forward,
5'-GCCTTCCGTGTCCCCACTGC-3' and reverse, 5'-CAATGCCAGCCCCAGCGTCA-3'.
miRNA and mRNA expression levels were quantified using the
2-∆∆Cq method (34) and
normalized to the internal reference genes U6 and GAPDH,
respectively.
Luciferase reporter assay
293/APPsw cells (2x104 cells/well) were
incubated in 24-well plates for 24 h. Subsequently, cells were
co-transfected with 50 ng pGL3 luciferase vector carrying wild-type
(WT) or mutated (MUT) 3'-UTR of KAT8 (Promega Corporation) and 50
nM miR-149-5p-mimic or mimic-NC using Lipofectamine®
3000 (Invitrogen; Thermo Fisher Scientific, Inc.). At 48 h
post-transfection, luciferase activities were detected using a
Dual-Luciferase Reporter Assay system (Guangzhou RiboBio Co., Ltd.)
according to the manufacturer's instructions. Relative luciferase
activity was normalized to Renilla luciferase activity.
Statistical analysis
All experiments were repeated at least three times
independently. Data are presented as the mean ± SEM. Statistical
analyses were conducted using SPSS software (version 20.0; IBM
Corp.). The figures and graphs were prepared using GraphPad Prism 7
(GraphPad Software, Inc.). The unpaired Student's t-test was used
to compare two groups, and comparisons among multiple groups were
analyzed using one-way ANOVA followed by Tukey's post hoc test.
Pearson's correlation coefficient analysis was used to analyze the
correlation between miR-149-5p and KAT8. P<0.05 was considered
to indicate a statistically significant difference.
Results
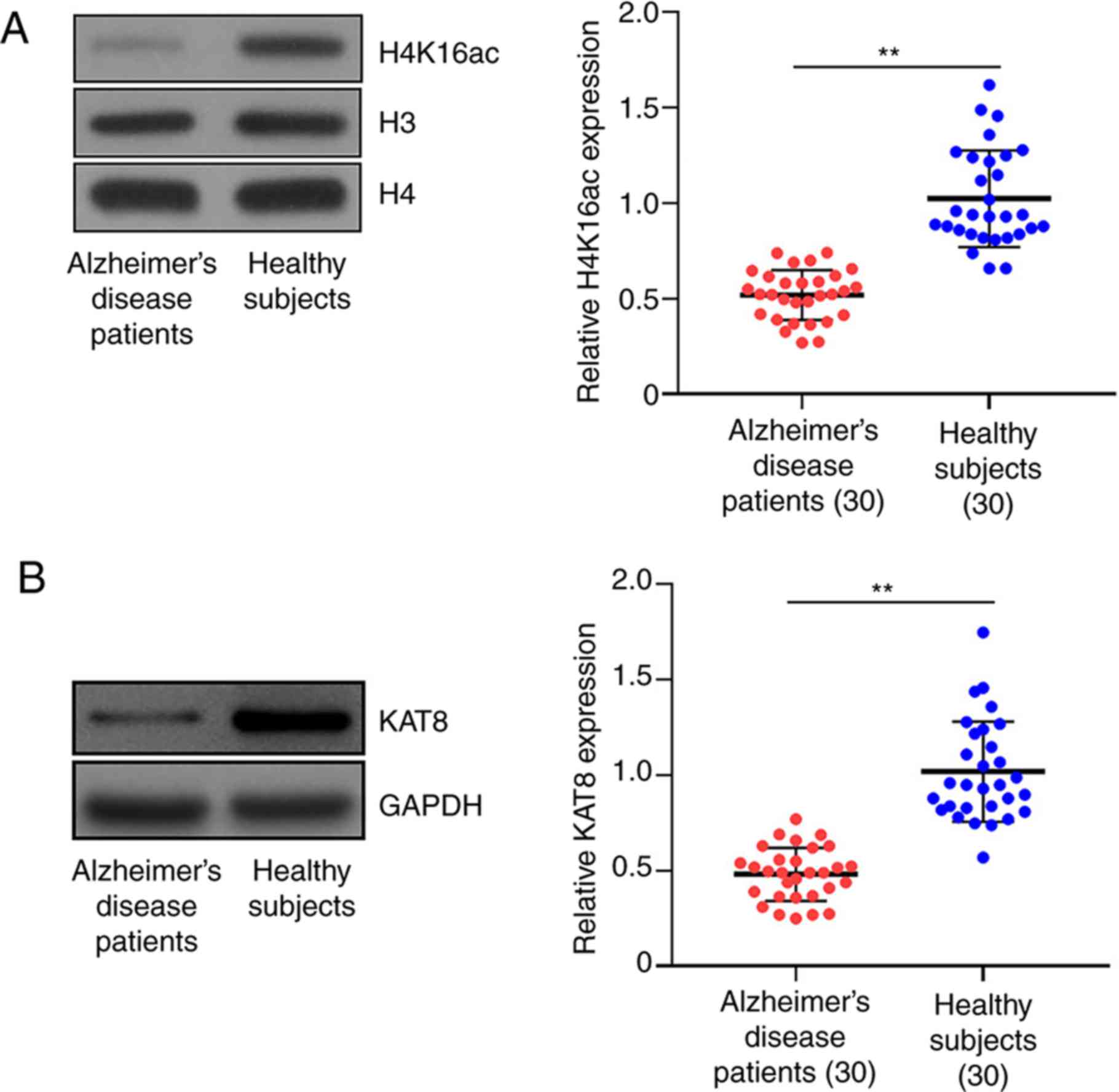
H4K16ac and KAT8 were downregulated in
the plasma of patients with AD
First, primary acetylated histone protein (H3K4ac,
H3K27ac, H4K12ac and H4K16ac) levels in the plasma of healthy
subjects and patients with AD were analyzed via western blotting.
Table I summarizes the
characteristics of patients with AD and cognitively healthy
volunteers. Although no difference was identified in H3K4ac and
H3K27ac levels between healthy subjects and patients with AD, the
H4K12ac level in patients with AD was notably downregulated
compared with healthy subjects (Fig.
S1). H4K16ac protein levels were notably decreased in patients
with AD compared with healthy controls (Fig. 1A), which was further indicated at
the mRNA level via RT-qPCR (Fig.
1A). The expression profile of KAT8, the major lysine
acetyltransferase responsible for the acetylation of H4K16, was
also evaluated. The RT-qPCR results demonstrated a reduction in
KAT8 mRNA and protein expression in patients with AD compared with
healthy subjects (Fig. 1B).
Collectively, the results demonstrated that aberrant expression
patterns of H4K16ac and KAT8 could be related to the progression of
AD.
 | Table ICharacteristics of patients with AD
and healthy subjects. |
Table I
Characteristics of patients with AD
and healthy subjects.
| Variable | Patients with AD
(n=30) | Healthy subjects
(n=30) | Statistical
value | P-value |
|---|
| Sex
(male/female) | 17/13 | 14/16 | 0.601a | 0.438 |
| Age (years) | 63.4±18.0 | 64.1±16 | 0.341b | 0.899 |
| BMI (kg/m²) | 24.5±3.4 | 24.7±4.4 | 0.413b | 0.931 |
|
Diabetes/non-diabetes | 8/22 | 5/25 | 0.884a | 0.347 |
| High blood
pressure/normal blood pressure | 10/20 | 7/23 | 0.739a | 0.390 |
| Coronary heart
disease/non-coronary heart disease | 6/24 | 3/27 | 1.176a | 0.278 |
|
Smoker/non-smoker | 11/19 | 10/20 | 0.073a | 0.787 |
|
Drinker/non-drinker | 15/15 | 16/14 | 0.067a | 0.796 |
| Educational
years | 11.3±3.4 | 11.7±3.7 | 0.968b | 0.867 |
| Left handed/right
handed | 3/27 | 5/25 | 0.577a | 0.448 |
| Duration of illness
(years) | 8.5±4.7 | - | - | - |
| Pathological MRI
(%) | 75 (60%) | - | - | - |
| Neuropsychological
impairment | 90 (64%) | | | |
| (2 T-scores 35; %)
(51,52) | | - | - | - |
| MMSE scores | 18.9±3.6 | 25.4±2.8 | 5.65b | 0.001 |
| CDR scale | | | | |
|
CDR 0 | 0 | 30 | - | - |
|
CDR 0.5 | 7 | - | - | - |
|
CDR 1 | 11 | - | - | - |
|
CDR
2+ | 12 | - | - | - |
KAT8 overexpression increases H4K16ac
expression, reduces APP processing and decreases the
Aβ1-40/Aβ1-42 ratio in 293/APPsw cells
Histone-associated heterochromatin structural
alterations are involved in the formation of long-term memory
(35). Certain studies reported
that increased levels of H3 and H4 acetylation are closely
associated with memory consolidation (36,37).
In the present study, KAT8-overexpression 293/APPsw cells were
constructed to explore the effect of KAT8 on the acetylation status
of core histones in vitro. KAT8 mRNA and protein expression
profiles were assessed via RT-qPCR and western blotting (Fig. 2A and B). Cellular H4K16ac was markedly higher in
KAT8-overexpression 293/APPsw cells compared with NC and vector
cells (Fig. 2C). Furthermore,
cellular H4K12ac was slightly increased in KAT8-overexpression
cells compared with NC and vector cells; however, there was no
notable difference in H3K4ac and H3K27ac levels between
KAT8-overexpression and control cells (Fig. S2). The results indicated that KAT8
overexpression in 293/APPsw cells induced H416K acetylation.
Subsequently, the effects of KAT8 on APP processing in 293/APPsw
cells were assessed. KAT8-overexpression 293/APPsw cells displayed
markedly increased neuroprotective sAPPα expression levels, and
decreased expression levels of sAPPβ and APP expression compared
with NC and vector 293/APPsw cells (Fig. 2D). The
Aβ1-40/Aβ1-42 ratio was significantly
increased in KAT8-overexpression 293/APPsw cells compared with
vector cells (Fig. 2E). The
expression level of enzymes involved in proteolytic cleavage of
APP, BACE1 (a key enzyme in sAPPβ generation) and BACE2 (a homolog
of BACE1, which functions as an antagonist of BACE1 and blocks Aβ
production), were assessed (38,39).
The western blotting results indicated that BACE2 expression was
markedly upregulated in KAT8-overexpression cells compared with NC
and vector cells. Likewise, BACE1 expression was downregulated in
KAT8-overexpression cells compared with NC and vector cells
(Fig. 2D), which suggested that
KAT8 was associated with the proteolytic processing of APP
protein.
KAT8 is a direct target of
miR-149-5p
Since miRNAs inhibit transcription of specific mRNAs
by binding to their 3'-UTRs (40),
starBase (version 2.0, http://starbase.sysu.edu.cn/starbase2/) computational
analyses were conducted to predict potential miRNA interactions in
the 3'-UTR of KAT8. A putative miR-149-5p target site in the 3'-UTR
of KAT8 was identified (Fig. 3A).
To validate the interaction between miR-149-5p and KAT8, a
luciferase reporter assay was conducted. The luciferase activity of
293/APPsw cells co-transfected with miR-149-5p mimic and WT 3'-UTR
was significantly reduced compared with cells co-transfected with
miR-NC and WT 3'UTR (Fig. 3B);
however, the luciferase activity was not significantly different
between cells co-transfected with miR-149-5p mimic and MUT 3'-UTR,
and cells co-transfected with miR-NC and MUT 3'-UTR (Fig. 3B). The results suggested that KAT8
may serve as a potential target of miR-149-5p. Since miRNAs can
circulate in the blood and cerebrospinal fluid, they have been
identified as biomarkers of various diseases, including AD
(41). Subsequently, the expression
of miR-149-5p in the plasma of patients with AD and healthy
controls was assessed. miR-149-5p expression levels were
significantly upregulated in the plasma of patients with AD
compared with healthy subjects (Fig.
3C). Pearson's correlation coefficient analysis revealed a
negative linear correlation between the expression level of
miR-149-5p and KAT8 (Fig. 3D),
which further indicated that KAT8 may serve as a potential target
of miR-149-5p. To further clarify the negative relationship between
miR-149-5p and KAT8, miR-149-5p expression was assessed in
KAT8-overexpression 293/APPsw cells. The RT-qPCR results suggested
that miR-149-5p expression was significantly decreased in
KAT8-overexpression cells compared with vector control cells
(Fig. 3E). Collectively, the
results indicated that KAT8 may serve as a potential target of
miR-149-5p.
miR-149-5p negatively regulates
H4K16ac and promotes amyloid pathology by targeting KAT8 in
293/APPsw cells
To explore the effect of miR-149-5p on the
pathological alterations associated with KAT8 in patients with AD,
the levels of H416Kac, sAPPα, sAPPβ, BACE1 and BACE2 were assessed
following KAT8 knockdown and co-transfection with miR-149-5p
inhibitor in 293/APPsw cells. miR-149-5p inhibitor significantly
decreased miR-149-5p expression and increased KAT8 mRNA expression
in 293/APPsw cells compared with the inhibitor NC group (Fig. 4A and B). The results indicated that exogenous
miR-149-5p may inhibit KAT8 expression via mRNA destabilization.
Based on the finding that KAT8 promoted H4K16 acetylation, whether
KAT8 knockdown and co-transfection with miR-149-5p inhibitor
altered H4K16ac levels was investigated. The knockdown efficacy of
sh-KAT8 was assessed (Fig. 4C and
D). Compared with the inhibitor NC
group, the western blotting results indicated that miR-149-5p
inhibitor markedly increased H4K16ac levels, which were notably
suppressed by co-transfection with KAT8 shRNA. miR-149-5p inhibitor
markedly reduced sAPPβ and BACE1 expression levels, increased sAPPα
and BACE2 expression levels, and increased the
Aβ1-40/Aβ1-42 ratio compared with the
inhibitor NC group. Moreover, co-transfection of sh-KAT8 and
miR-149-5p inhibitor reversed miR-149-5p inhibitor-mediated
effects, resulting in increased levels of sAPPβ and BACE1, and a
decreased Aβ1-40/Aβ1-42 ratio, but reduced
APPα and BACE2 production (Fig. 4F
and G). In summary, the results
indicated that inhibiting miR-149-5p delivery could promote
amyloidogenic APP processing and decrease the
Aβ1-40/Aβ1-42 ratio by upregulating KAT8 and
H4K16ac expression.
 | Figure 4miR-149-5p inhibition exhibits
neuroprotective properties in 293/APPsw cells, which are blocked by
KAT8 knockdown. (A) Transfection efficiency of miR-149-5p
inhibitor. (B) Effect of miR-149-5p inhibitor on KAT8 expression.
Transfection efficiency of sh-KAT8 as determined by (C) reverse
transcription-quantitative PCR and (D) western blotting. (E) Effect
of miR-149-5p inhibitor and sh-KAT8 on H4K16ac, (F) APP, sAPPα,
sAPPβ, BACE1, BACE2 and (G) the ratio of Aβ1-40/Aβ1-42.
**P<0.01. miR, microRNA; KAT8, lysine
acetyltransferase 8; sh, short hairpin RNA; APP, amyloid precursor
protein; sAPP, soluble APP; BACE, β-secretase; Aβ, β-amyloid; NC,
negative control. |
Discussion
Several reports indicated that histone acetylation,
one of the main epigenetic modifications, could contribute to AD
onset and progression (17,19,42).
The present study compared acetylation of several core histones in
the plasma of patients with AD and cognitively healthy individuals.
H4K16ac was significantly downregulated in the plasma of patients
with AD compared with healthy subjects. In addition, KAT8
expression levels, the major lysine acetyltransferase responsible
for the acetylation of H4K16 in flies and mammals (29), were decreased in patients with AD
compared with healthy subjects, which implied that aberrant
expression patterns of H4K16ac and KAT8 could be related to the
progression of AD. AD is a multifactorial neurodegenerative
disease, and the abnormal deposition of Aβ is a critical feature of
the disease (43). The present
study indicated that KAT8 overexpression increased the level of
H4K16ac in 293/APPsw cells compared with the NC and vector groups.
Moreover, compared with the NC and vector groups, KAT8
overexpression also increased expression levels of neuroprotective
sAPPα and BACE2, and significantly decreased the levels of
neurotoxic sAPPβ and BACE1, which suggested that KAT8 may serve as
a critical player in APP processing via regulating acetylation of
core histones.
Specific miRNAs have been reported to participate
during the initiation and progression of AD (9,44,45);
however, the effect of miRNAs on histone acetylation and
deacetylation during AD development is not completely understood.
To the best of our knowledge, for the first time, the present study
identified a novel inhibitory interaction between miR-149-5p and
KAT8 3'-UTR that participated in the pathological alterations of an
AD cell model via regulation of H416Kac levels. Numerous
dysregulated miRNAs such as miR-223(46), miR-455-3p (47), and miR-16(48) were reported among patients with AD,
animal AD models and AD model cell lines, which served an essential
role in amyloidogenic APP processing, synaptic dysfunction and
other pathophysiological processes of AD. Although previous studies
revealed that miR-149-5p is involved in cell migration,
proliferation, apoptosis and major signaling pathways in various
cell carcinomas (49,50), the expression profile of miR-149-5p
in AD has not been previously reported. The present study indicated
that miR-149-5p was significantly increased in the plasma of
patients with AD and the AD cell model compared with healthy
subjects, and NC and vector cells, respectively. Additionally, the
dual-luciferase reporter assay indicated a negative relationship
between miR-149-5p and KAT8 in the plasma of patients with AD,
which might promote AD pathogenesis via regulating KAT8 expression
levels. Furthermore, miR-149-5p displayed neurotoxic effects on
293/APPsw cells, as indicated by miR-149-5p inhibitor reducing APP
and Aβ levels in 293/APPsw cells via regulating KAT8 expression and
H4K16 acetylation.
The present study suggested that miR-149-5p could
negatively regulate KAT8 and H4K16ac in vitro. Since histone
acetylation serves a critical role in learning and memory
consolidation (37), the present
study revealed that KAT8 overexpression increased H4K16 acetylation
compared with the NC and vector groups. KAT8-induced increases in
H4K16 acetylation altered histone-associated heterochromatin
structures, which could regulate a variety of AD-related gene
transcriptions and suppress neurotoxic APP processing. The effect
of miR-149-5p knockdown on KAT8 expression and Aβ formation
requires further investigation. In conclusion, the present study
suggested a potential novel approach to attenuate the pathological
progression of AD by reducing miR-149-5p expression.
The present study had a key limitation; 293 cells
carrying the Swedish mutation of APP (293/APPsw) were used as an
established AD cell model. However, 293 cells lacking the Swedish
mutation of APP were not used as a control group to evaluate
whether the inhibitory effect of miR-149-5p on amyloid-β generation
was due to the APPsw mutation. Therefore, future studies should use
293 cells lacking the Swedish mutation of APP as a control group to
verify the results of the present study.
Supplementary Material
Expression profiles of H3K4ac, H3K27ac
and H4K12ac in the plasma of patients with Alzheimer's disease
(n=30) and healthy subjects (n=30).
Expression profiles of H3K4ac, H3K27ac
and H4K12ac in KAT8-overexpression 293/APPsw cells. KAT8, lysine
acetyltransferase 8; NC, negative control.
Acknowledgements
Not applicable.
Funding
The present study was supported by the National
Natural Science Foundation of China (grant nos. 81603684 and
81603686).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
FC contributed to designing the study, and
collecting, analyzing and interpreting the data. FC, HC, YJ, HL and
QT contributed to collecting the data, drafting the manuscript and
performing the literature search. XZ contributed to designing the
study and drafting the manuscript. All authors read and approved
the final version of the manuscript and agree to be accountable for
all aspects of the research in ensuring that the accuracy or
integrity of any part of the work are appropriately investigated
and resolved.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of First Teaching Hospital of Tianjin University of
Traditional Chinese Medicine. Written informed consent was obtained
from all patients and healthy subjects.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Nelson PT, Braak H and Markesbery WR:
Neuropathology and cognitive impairment in Alzheimer disease: A
complex but coherent relationship. J Neuropathol Exp Neurol.
68:1–14. 2009.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Gouras GK, Olsson TT and Hansson O:
β-Amyloid peptides and amyloid plaques in Alzheimer's disease.
Neurotherapeutics. 12:3–11. 2015.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Hussain I, Powell D, Howlett DR, Tew DG,
Meek TD, Chapman C, Gloger IS, Murphy KE, Southan CD, Ryan DM, et
al: Identification of a novel aspartic protease (Asp 2) as
beta-secretase. Mol Cell Neurosci. 14:419–427. 1999.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Yan R, Bienkowski MJ, Shuck ME, Miao H,
Tory MC, Pauley AM, Brashier JR, Stratman NC, Mathews WR, Buhl AE,
et al: Membrane-anchored aspartyl protease with Alzheimer's disease
beta-secretase activity. Nature. 402:533–537. 1999.PubMed/NCBI View
Article : Google Scholar
|
|
5
|
Ourdev D, Foroutanpay BV, Wang Y and Kar
S: The effect of Aβ1-42 oligomers on APP processing and Aβ1-40
generation in cultured U-373 astrocytes. Neurodegener Dis.
15:361–368. 2015.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Zhang YW, Thompson R, Zhang H and Xu H:
APP processing in Alzheimer's disease. Mol Brain.
4(3)2011.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Kapsimali M, Kloosterman WP, de Bruijn E,
Rosa F, Plasterk RH and Wilson SW: MicroRNAs show a wide diversity
of expression profiles in the developing and mature central nervous
system. Genome Biol. 8(R173)2007.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Konopka W, Kiryk A, Novak M, Herwerth M,
Parkitna JR, Wawrzyniak M, Kowarsch A, Michaluk P, Dzwonek J,
Arnsperger T, et al: MicroRNA loss enhances learning and memory in
mice. J Neurosci. 30:14835–14842. 2010.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Li J and Wang H: miR-15b reduces amyloid-β
accumulation in SH-SY5Y cell line through targetting NF-κB
signaling and BACE1. Biosci Rep. 38(BSR20180051)2018.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Santa-Maria I, Alaniz ME, Renwick N, Cela
C, Fulga TA, Van Vactor D, Tuschl T, Clark LN, Shelanski ML, McCabe
BD and Crary JF: Dysregulation of microRNA-219 promotes
neurodegeneration through post-transcriptional regulation of tau. J
Clin Invest. 125:681–686. 2015.PubMed/NCBI View
Article : Google Scholar
|
|
11
|
Ruan D, Liu Y, Wang X, Yang D and Sun Y:
miR-149-5p protects against high glucose-induced pancreatic beta
cell apoptosis via targeting the BH3-only protein BIM. Exp Mol
Pathol. 110(104279)2019.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Xiang F, Fan Y, Ni Z, Liu Q, Zhu Z, Chen
Z, Hao W, Yue H, Wu R and Kang X: Ursolic acid reverses the
chemoresistance of breast cancer cells to paclitaxel by targeting
miRNA-149-5p/MyD88. Front Oncol. 9(501)2019.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Lu H, Liu X, Deng Y and Qing H: DNA
methylation, a hand behind neurodegenerative diseases. Front Aging
Neurosci. 5(85)2013.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Mastroeni D, Grover A, Delvaux E,
Whiteside C, Coleman PD and Rogers J: Epigenetic mechanisms in
Alzheimer's disease. Neurobiol Aging. 32:1161–1180. 2011.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Aubry S, Shin W, Crary JF, Lefort R,
Qureshi YH, Lefebvre C, Califano A and Shelanski ML: Assembly and
interrogation of Alzheimer's disease genetic networks reveal novel
regulators of progression. PLoS One. 10(e0120352)2015.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Min SW, Cho SH, Zhou Y, Schroeder S,
Haroutunian V, Seeley WW, Huang EJ, Shen Y, Masliah E, Mukherjee C,
et al: Acetylation of tau inhibits its degradation and contributes
to tauopathy. Neuron. 67:953–966. 2010.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Lu X, Deng Y, Yu D, Cao H, Wang L, Liu L,
Yu C, Zhang Y, Guo X and Yu G: Histone acetyltransferase p300
mediates histone acetylation of PS1 and BACE1 in a cellular model
of Alzheimer's disease. PLoS One. 9(e103067)2014.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Marques SC, Lemos R, Ferreiro E, Martins
M, de Mendonça A, Santana I, Outeiro TF and Pereira CM: Epigenetic
regulation of BACE1 in Alzheimer's disease patients and in
transgenic mice. Neuroscience. 220:256–266. 2012.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Peleg S, Sananbenesi F, Zovoilis A,
Burkhardt S, Bahari-Javan S, Agis-Balboa RC, Cota P, Wittnam JL,
Gogol-Doering A, Opitz L, et al: Altered histone acetylation is
associated with age-dependent memory impairment in mice. Science.
328:753–756. 2010.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Walker MP, LaFerla FM, Oddo SS and Brewer
GJ: Reversible epigenetic histone modifications and Bdnf expression
in neurons with aging and from a mouse model of Alzheimer's
disease. Age (Dordr). 35:519–531. 2013.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Lau P, Frigerio CS and De Strooper B:
Variance in the identification of microRNAs deregulated in
Alzheimer's disease and possible role of lincRNAs in the pathology:
The need of larger datasets. Ageing Res Rev. 17:43–53.
2014.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Goldberg AD, Allis CD and Bernstein E:
Epigenetics: A landscape takes shape. Cell. 128:635–638.
2007.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Kouzarides T: Chromatin modifications and
their function. Cell. 128:693–705. 2007.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Yamakawa H, Cheng J, Penney J, Gao F,
Rueda R, Wang J, Yamakawa S, Kritskiy O, Gjoneska E and Tsai LH:
The transcription factor Sp3 cooperates with HDAC2 to regulate
synaptic function and plasticity in neurons. Cell Rep.
20:1319–1334. 2017.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Chuang DM, Leng Y, Marinova Z, Kim HJ and
Chiu CT: Multiple roles of HDAC inhibition in neurodegenerative
conditions. Trends Neurosci. 32:591–601. 2009.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Gräff J and Mansuy IM: Epigenetic
dysregulation in cognitive disorders. Eur J Neurosci. 30:1–8.
2009.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Sharma GG, So S, Gupta A, Kumar R, Cayrou
C, Avvakumov N, Bhadra U, Pandita RK, Porteus MH, Chen DJ, et al:
MOF and histone H4 acetylation at lysine 16 are critical for DNA
damage response and double-strand break repair. Mol Cell Biol.
30:3582–3595. 2010.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Marioni RE, Harris SE, Zhang Q, McRae AF,
Hagenaars SP, Hill WD, Davies G, Ritchie CW, Gale CR, Starr JM, et
al: GWAS on family history of Alzheimer's disease. Transl
Psychiatry. 8(99)2018.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Chatterjee A, Seyfferth J, Lucci J,
Gilsbach R, Preissl S, Böttinger L, Mårtensson CU, Panhale A,
Stehle T, Kretz O, et al: MOF acetyl transferase regulates
transcription and respiration in mitochondria. Cell. 167:722–738
e723. 2016.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Guo R, Fan G, Zhang J, Wu C, Du Y, Ye H,
Li Z, Wang L, Zhang Z, Zhang L, et al: A 9-microRNA signature in
serum serves as a noninvasive biomarker in early diagnosis of
Alzheimer's disease. J Alzheimers Dis. 60:1365–1377.
2017.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Tamaoka A: Alzheimer's disease: Definition
and national institute of neurological and communicative disorders
and stroke and the Alzheimer's disease and related disorders
association (NINCDS-ADRDA). Nihon Rinsho. 2 (Suppl 69):S240–S245.
2011.PubMed/NCBI(In Japanese).
|
|
32
|
Haris M, Singh A, Cai K, McArdle E, Fenty
M, Davatzikos C, Trojanowski JQ, Melhem ER, Clark CM and Borthakur
A: T(1p) MRI in Alzheimer's disease: Detection of pathological
changes in medial temporal lobe. J Neuroimaging. 21:e86–e90.
2011.
|
|
33
|
Reichenberg A, Harvey PD, Bowie CR,
Mojtabai R, Rabinowitz J, Heaton RK and Bromet E:
Neuropsychological function and dysfunction in schizophrenia and
psychotic affective disorders. Schizophr Bull. 35:1022–1029.
2009.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Mews P, Donahue G, Drake AM, Luczak V,
Abel T and Berger SL: Acetyl-CoA synthetase regulates histone
acetylation and hippocampal memory. Nature. 546:381–386.
2017.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Gräff J and Tsai LH: The potential of HDAC
inhibitors as cognitive enhancers. Annu Rev Pharmacol Toxicol.
53:311–330. 2013.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Levenson JM, O'Riordan KJ, Brown KD, Trinh
MA, Molfese DL and Sweatt JD: Regulation of histone acetylation
during memory formation in the hippocampus. J Biol Chem.
279:40545–40559. 2004.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Basi G, Frigon N, Barbour R, Doan T,
Gordon G, McConlogue L, Sinha S and Zeller M: Antagonistic effects
of beta-site amyloid precursor protein-cleaving enzymes 1 and 2 on
beta-amyloid peptide production in cells. J Biol Chem.
278:31512–31520. 2003.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Sun X, He G and Song W: BACE2, as a novel
APP theta-secretase, is not responsible for the pathogenesis of
Alzheimer's disease in down syndrome. FASEB J. 20:1369–1376.
2006.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Lu TX and Rothenberg ME: MicroRNA. J
Allergy Clin Immunol. 141:1202–1207. 2018.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Zetterberg H and Burnham SC: Blood-based
molecular biomarkers for Alzheimer's disease. Mol Brain.
12(26)2019.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Gräff J, Rei D, Guan JS, Wang WY, Seo J,
Hennig KM, Nieland TJF, Fass DM, Kao PF, Kahn M, et al: An
epigenetic blockade of cognitive functions in the neurodegenerating
brain. Nature. 483:222–226. 2012.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Dorszewska J, Prendecki M, Oczkowska A,
Dezor M and Kozubski W: Molecular basis of familial and sporadic
Alzheimer's disease. Curr Alzheimer Res. 13:952–963.
2016.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Wang L, Liu J, Wang Q, Jiang H, Zeng L, Li
Z and Liu R: MicroRNA-200a-3p mediates neuroprotection in
Alzheimer-related deficits and attenuates amyloid-beta
overproduction and tau hyperphosphorylation via coregulating BACE1
and PRKACB. Front Pharmacol. 10(806)2019.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Long JM, Ray B and Lahiri DK:
MicroRNA-339-5p down-regulates protein expression of β-site amyloid
precursor protein-cleaving enzyme 1 (BACE1) in human primary brain
cultures and is reduced in brain tissue specimens of Alzheimer
disease subjects. J Biol Chem. 289:5184–5198. 2014.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Jia LH and Liu YN: Downregulated serum
miR-223 servers as biomarker in Alzheimer's disease. Cell Biochem
Funct. 34:233–237. 2016.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Kumar S, Vijayan M and Reddy PH:
MicroRNA-455-3p as a potential peripheral biomarker for Alzheimer's
disease. Hum Mol Genet. 26:3808–3822. 2017.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Zhang B, Chen CF, Wang AH and Lin QF:
miR-16 regulates cell death in Alzheimer's disease by targeting
amyloid precursor protein. Eur Rev Med Pharmacol Sci. 19:4020–4027.
2015.PubMed/NCBI
|
|
49
|
Chen W, Zhang J, Xu H, Dai J and Zhang X:
The negative regulation of miR-149-5p in melanoma cell survival and
apoptosis by targeting LRIG2. Am J Transl Res. 9:4331–4340.
2017.PubMed/NCBI
|
|
50
|
Xu RD, Feng F, Yu XS, Liu ZD and Lao LF:
miR-149-5p inhibits cell growth by regulating TWEAK/Fn14/PI3K/AKT
pathway and predicts favorable survival in human osteosarcoma. Int
J Immunopathol Pharmacol. 32(2058738418786656)2018.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Sundermann EE, Heaton RK, Pasipanodya E,
Moore RC, Paolillo EW, Rubin LH, Ellis R, Moore DJ and Group HNRP:
Sex differences in HIV-associated cognitive impairment. AIDS.
32:2719–2726. 2018.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Norman MA, Moore DJ, Taylor M, Franklin D
Jr, Cysique L, Ake C, Lazarretto D, Vaida F, Heaton RK and Group
HNRC: Demographically corrected norms for African Americans and
caucasians on the hopkins verbal learning test-revised, brief
visuospatial memory test-revised, stroop color and word test, and
wisconsin card sorting test 64-card version. J Clin Exp
Neuropsychol. 33:793–804. 2011.PubMed/NCBI View Article : Google Scholar
|