Introduction
Colorectal cancer (CRC) is the third leading cause
of cancer-associated mortality in both men and women, and poses a
serious health problem (1).
Vermeulen et al (2)
previously revealed that by modulating Wnt signalling activity,
cancer cells can reversibly lose their tumorigenic capabilities,
which can be regained following stimulation by
myofibroblast-derived factors. This finding demonstrated the
plasticity of cancer stem cells, which can be modulated by
adjusting the activation status of Wnt signalling (3). The aberrant activation of the
Wnt/β-catenin signalling pathway is associated with a number of
human malignancies, including breast (4), liver (5), lung (6) and colorectal cancer (7).
Secreted frizzled-related proteins (SFRP1-5) belong
to a family of soluble proteins that are structurally related to
Frizzled proteins. They serve as extracellular inhibitors of Wnt
signalling by directly binding to Wnt ligands or Frizzled receptors
(8). Downregulated SFRP expression
by promoter hypermethylation has been previously demonstrated to
result in the constitutive activation of the Wnt pathway, which in
turn contributes to CRC malignancy (9). Promoter hypermethylation in the SFRP
family of genes have been previously revealed to occur early in the
colorectal and adenoma tumorigenesis process (10), where the adenocarcinoma and aberrant
crypt foci always maintain a high frequency of SFRP1 methylation,
with the rate reaching 94.4% (11).
SFRP gene promoter demethylation has been demonstrated to restore
SFRP expression and suppress Wnt signalling, leading to the
inhibition of tumour cell proliferation (10). This observation indicated that
promoter hypermethylation is associated with the silencing of SFRP
gene expression.
Epigenetic regulation of gene expression does not
lead to changes in the actual DNA sequence (12). In eukaryotic cells, gene expression
is predominantly regulated by epigenetic modifications the
chromatin, which can subsequently adopt active or repressive states
(13). For example, histone
acetylation, trimethylation and other regulatory factors can render
chromatin susceptible to transcriptional activation (13), whilst DNA hypermethylation,
chromatin repressors and specific codes of histone methylation can
compact the chromatin to repress transcription (12,13).
DNA methylation is a particularly important epigenetic modification
in the regulation of gene expression. Previous studies have
revealed that methylated CpG islands in the promoter region
protrude into the main groove, where transcription factors bind to
directly inhibit gene transcription (14).
The polycomb-group (PCG) proteins are a family of
epigenetic regulators that can repress transcription through
post-translational histone modifications or chromatin compaction
(15). Evidence suggests that the
overexpression of PCG proteins, and particularly that of BMI1 and
EZH2, results in the progression of a variety of cancer types
(16,17). The methyl-CpG-binding domain (MBD)
protein can specifically recognize and bind to methylated DNA,
recruit repressor complexes and histone deacetylases (HDACs) to
mediate epigenetic transcriptional repression (18,19).
However, to the best of our knowledge, the specific identities of
the PCG and MBD proteins that regulate SFRP1 promoter methylation
and subsequent SFRP1 expression remain to be determined. Therefore,
using CRC cell lines, the aim of the present study was to assess
the potential role of PCG and MBD proteins in the regulation of
SFRP1 gene expression in CRC.
Materials and methods
Cell culture
The colon adenocarcinoma cell lines used in the
present study were as follows: SW1116 (Dukes A), SW480 (Dukes B)
and HCT116 cells (Dukes D), were donated from the Department of
Gastroenterology, Zhongnan Hospital of Wuhan University (Wuhan,
China) (20). The human embryo
intestinal mucosa cell line CCC-HIE-2(21) (cat. no. 3111C0001CCC000178) was
purchased from the China Infrastructure of Cell Line Resources
(Beijing, China). All cells were cultured in DMEM (Gibco; Thermo
Fisher Scientific, Inc.) supplemented with 10% (v/v) FBS (Hangzhou
Sijiqing Biological Engineering Materials Co., Ltd.) and 1% (v/v)
penicillin/streptomycin (Thermo Fisher Scientific, Inc.) in a
humidified atmosphere at 37˚C and 5% CO2.
Methylation-specific PCR (MSP)
DNA was extracted from all cell lines using TIANamp
Genomic DNA kit (Tiangen Biotech Co., Ltd.) according to
manufacturer's protocol. Sodium bisulfite treatment was performed
using an EZ DNA Methylation kit (cat. no. D5020; Zymo Research
Corp.). The sequences of primers used for the present study were as
follows: SFRP1 unmethylated (U) forward,
5'-GTTTTGTAGTTTTTGGAGTTAGTGTTGTGT-3' and reverse,
5'-CCTACGATCGAAAACGACGCGAACG-3'; SFRP2 (U) forward,
5'-TTTTGGGTTGGAGTTTTTTGGAGTTGTGT-3' and reverse,
5'-AACCCACTCTCTTCACTAAATACAACTCA-3'; SFRP4 (U) forward,
5'-GGGGGTGATGTTATTGTTTTTGTATTGAT-3' and reverse,
5'-CACCTCCCCTAACATAAACTCAAAACA-3'; SFRP5 (U) forward,
5'-GTAAGATTTGGTGTTGGGTGGGATGTTT-3' and reverse,
5'-AAAACTCCAACCCAAACCTCACCATACA-3'; SFRP1 methylated (M) forward,
5'-TGTAGTTTTCGGAGTTAGTGTCGCGC-3' and reverse,
5'-CCTACGATCGAAAACGACGCGAACG-3', SFRP2 (M) forward,
5'-GGGTCGGAGTTTTTCGGAGTTGCGC-3' and reverse,
5'-CCGCTCTCTTCGCTAAATACGACTCG-3', SFRP4 (M) forward,
5'-GGGTGATGTTATCGTTTTTGTATCGAC-3' and reverse,
5'-CCTCCCCTAACGTAAACTCGAAACG-3'; SFRP5 (M) forward,
5'-AAGATTTGGCGTTGGGCGGGACGTTC-3' and reverse,
5'-ACTCCAACCCGAACCTCGCCGTACG-3'. The exact locations of the SFRP1,
SFRP2, SFRP4 and SFRP5 promoter regions analysed for methylation by
MSP are presented in Fig. S1A.
Gold Star Taq DNA polymerase (CoWin Biosciences; cat. no. CW0938A)
was used to amplify the DNA treated with sodium bisufite. Each 50
µl reaction mixture consisted of 1 µl bisulfite-modified DNA
template, 10 µl 5X Gold Star Taq PCR Buffer, 4 µl dNTP Mix, 2 µl
forward and reverse primer (2.5 mM each), 0.5 µl Gold Star Taq DNA
Polymerase and 30.5 µl ddH2O. The thermocycling
conditions were as follows: Initial denaturation at 95˚C for 10
min, followed by 38 cycles of denaturation at 95˚C for 30 sec;
annealing at 60˚C for 30 sec, extension at 72˚C for 30 sec and full
extension at 72˚C for 5 min. The consequent PCR products were then
electrophoresed on a 2% agarose gel.
Chromatin immunoprecipitation
(ChIP)
ChIP assay was performed using a One-Day Chromatin
Immunoprecipitation kit (EMD Millipore) according to the
manufacturer's protocol. ChIP was performed using anti-MBD2 (1:50;
cat. no. AB38646; Abcam), BMI1 (1:50; cat. no. 5856; Cell
Signalling Technology, Inc.) and EZH2 antibodies (1:25; cat. no.
4905; Cell Signalling Technology, Inc.) for 12 h at 4˚C, with
normal rabbit IgG (cat. no. LK2001; 1:1,000; Sungene Biotech, Ltd.)
serving as a control. DNA fragments isolated from chromatin were
subjected to 40 cycles of RT-qPCR (cat. no. CW2627; CoWin
Biosciences) at 95˚C for 10 sec followed by annealing at 60˚C for
60 sec and extension at 95˚C for 15 sec. The enrichment of marks on
the target regions was quantified relative to the input amounts.
The region of the SFRP1 promoters analysed for methylation by ChIP
are presented in Fig. S1B.
RNA interference
All cells (5x105 cells/well) were seeded
into six-well plates in DMEM, then 100 nM MBD2, EZH2 or a negative
control small interfering (si)RNA was transfected into cells using
Lipofectamine JET (Invitrogen; Thermo Fisher Scientific, Inc; cat.
no. SL100468) according to the manufacturer's protocols. After 24 h
incubation at 37˚C in a CO2 incubator, the medium was
replaced with fresh DMEM containing 10% FBS and siRNA-Lipofectamine
JET complexes 24 h prior to further culture. The sequences of siRNA
used for the present study were as follows: MBD2-001 forward,
5'-GAGGCUACAAGGACUUAGUTT-3' and reverse,
5'-ACUAAGUCCUUGUAGCCUCTT-3'. MBD2-002, 5'-GAUGAUGCCUAGUAAAUUATT-3'
and reverse, 5'-UAAUUUACUAGGCAUCAUCTT-3'. MBD2-003,
5'-CCUGGGAAAUACUGUUGAUTT-3' and reverse,
5'-AUCAACAGUAUUUCCCAGGTT-3'. EZH2-2196 forward,
5'-GCAGCUUUCUGUUCAACUUTT-3' and reverse,
5'-AAGUUGAACAGAAAGCUGCTT-3'. EZH2-488 forward,
5'-GACUCUGAAUGCAGUUGCUTT-3' and reverse,
5'-AGCAACUGCAUUCAGAGUCTT-3'. EZH2-1952 forward,
5'-CCUGACCUCUGUCUUACUUTT-3' and reverse,
5'-AAGUAAGACAGAGGUCAGGTT-3'. A nonspecific control siRNA was used
as a control. NC forward, 5'-UUCUCCGAACGUGUCACGUTT-3' and reverse,
5'-ACGUGACACGUUCGGAGAATT-3'.
cDNA preparation and reverse
transcription-quantitative PCR (RT-qPCR)
Total RNA was isolated from cells using an
easy-spin™ Total RNA extraction kit (Beijing, Aidlab
Biotechnologies Co., Ltd.) according to the manufacturer's
protocols. The concentration of RNA was measured using a NanoDrop™
2000 spectrophotometer (Thermo Fisher Scientific, Inc.). A total of
1,000 ng total RNA was used for cDNA synthesis using a ReverTra
Ace® qPCR RT kit (Toyobo Life Science) and a temperature
protocol of 15 min at 37˚C, 5 min at 98˚C and 4˚C thereafter.
RT-qPCR was subsequently performed using SYBR® Green PCR
Master Mix (cat. no. CW2627; ComWin Biosciences) in a Bio-Rad 7500
Real-time PCR System (Bio-Rad Laboratories, Inc.). Primers used for
qPCR contained the following sequences: GAPDH forward,
5'-AGAAGGCTGGGGCTCATTTG-3' and reverse,
5'-GCAGGAGGCATTGCTGATGAT-3'; EZH2 forward,
5'-TCCTACATCCTTTTCATGCAACAC-3' and reverse,
5'-GCTCCCTCCAAATGCTGGTA-3'; MBD2 forward, 5'-GCAAGCCTCAGTTGGCAAG-3'
and reverse, 5'-ATCGTTTCGCAGTCTCTGTTT-3'; SFRP1 forward,
5'-TGGCCCGAGATGCTTAAGTG-3', and reverse, 5'-GACACACCGTTGTGCCTTG-3';
SFRP2 forward, 5'-CGACATGCTTGAGTGCGAC-3' and reverse,
5'-CTTTGGAGTTCCTCGGTGG-3'; SFRP4 forward,
5'-TCACCCATCCCTCGAACTCA-3' and reverse, 5'-CATCATCCTTGAGCGCCACT-3';
SFRP5 forward, 5'-CCTCCAGTGACCAAGATCTGC-3' and reverse,
5'-TCCTTGATGCGCATTTTGACC-3'. GAPDH was used as an internal
reference. The thermocycling conditions were as follows: Initial
denaturation at 95˚C for 10 min, followed by 39 cycles of
denaturation at 95˚C for 10 sec, annealing at 60˚C for 20 sec,
extension at 72˚C for 10 sec and final extension at 72˚C for 3 min.
All reactions were performed in triplicate, and the results were
analysed and expressed relative to threshold cycle (Cq) values and
then converted to fold change values (2-ΔΔCq) (22).
Statistical analysis
SPSS 20.0 software (IBM Corp.) was used for data
analysis. Data presented were analysed using one-way ANOVA followed
by a Bonferroni's correction and presented as mean ± standard
deviation in the text and figures. P<0.05 was considered to
indicate a statistically significant difference.
Results
DNA promoter hypermethylation and
reduced SFRP1 mRNA expression in CRC cell lines
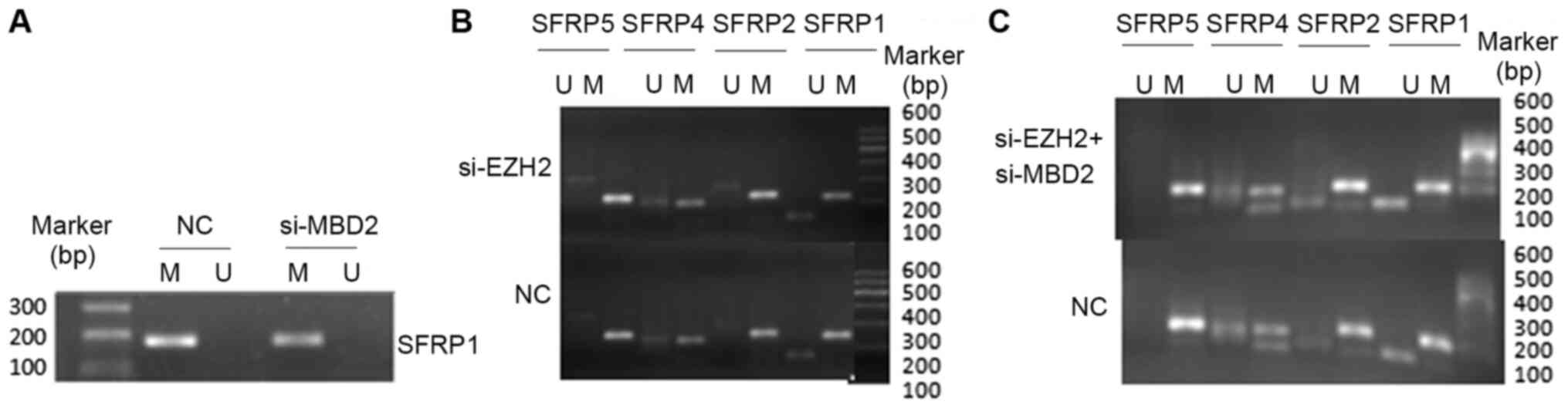
The SFRP1 gene promoter was demonstrated to be
completely methylated in SW1116, SW480 and HCT116 cells but only
partially methylated in CCC-HIE-2 cells according to MSP analysis
(Fig. 1). The mRNA levels of SFRP1
was significantly lower in CRC cell lines compared with that of
CCC-HIE-2 (Fig. 2).
Expression of EZH2, BMI1 and MBD2 are
upregulated with SFRP1 methylation in HCT116 and SW480 cells
EZH2 mRNA expression was revealed to be
significantly increased in CRC cell lines compared with CCC-HIE-2
cells. MBD2 and BMI1 expression were significantly higher in SW480
and HCT116 cells compared with CCC-HIE-2 cells, but was markedly
lower in SW1116 cells compared with CCC-HIE-2 cells (Fig. 3).
EZH2 and MBD2 can bind to the promoter
region of SFRP1
To examine if PCG and MBD proteins can interact with
the SFRP1 promoter directly, ChIP assay was performed in HCT116,
SW480 and CCC-HIE-2 cell samples. Enrichment of EZH2 (% input
=2.899) on the SFRP1 promoter region was observed in CCC-HIE-2 cell
samples. In SW480 cells, increased binding of MBD2 (% input =3.244)
and reduced binding of EZH2 (% input =1.147) to the SFRP1 promoter
were observed compared with samples from CCC-HIE-2 cells. However,
no enrichment was observed in samples from HCT116 cells (Fig. 4).
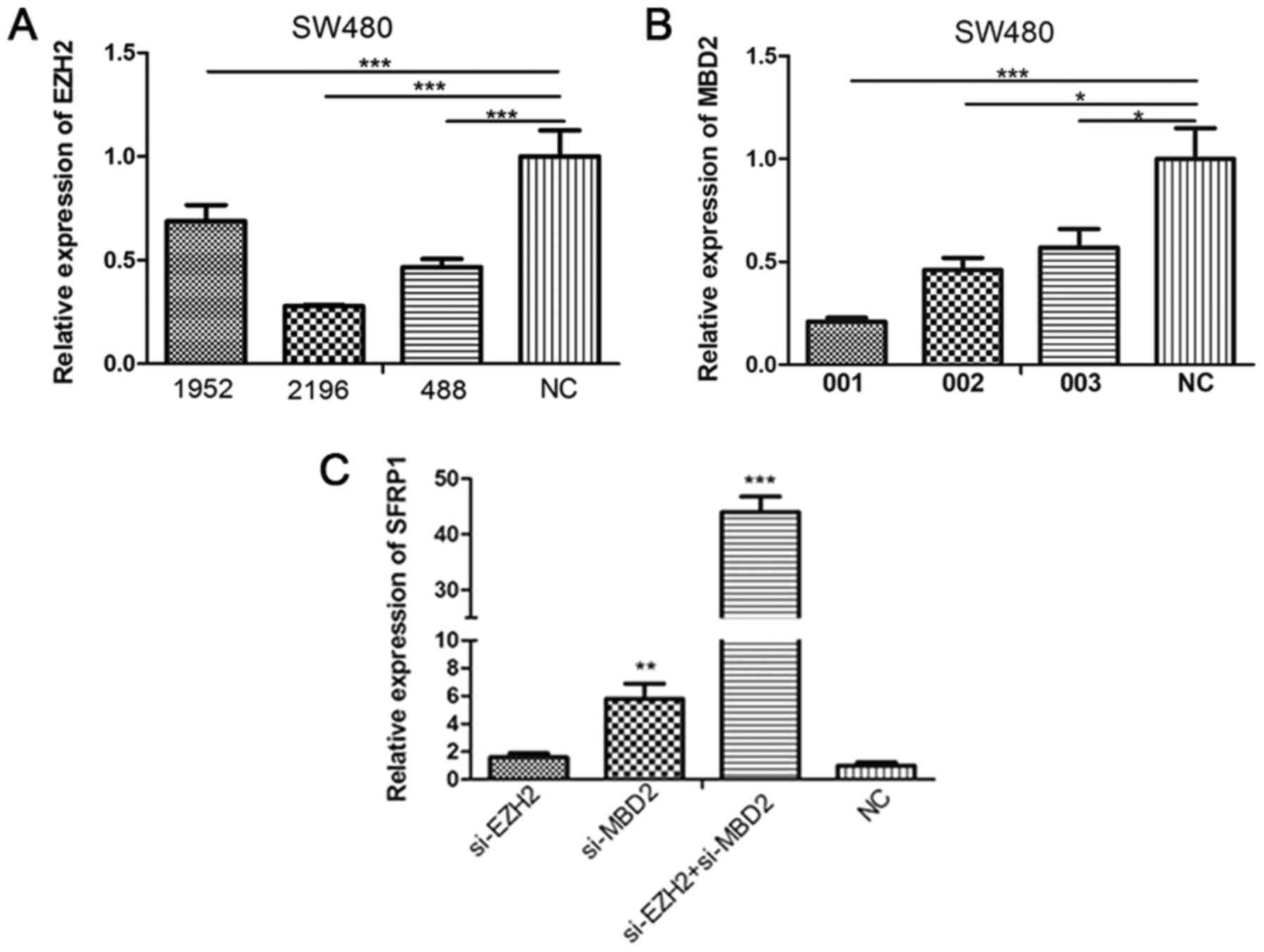
Knockdown of MBD2 and EZH2 expression
increases SFRP1 expression in SW480 cells
To investigate the effect of MBD2 and EZH2 on SFRP1
gene expression and promoter methylation, MBD2 and EZH2 knockdown
experiments were performed on SW480 cells following confirmation of
siRNA transfection efficiency (Fig.
5A and B). SFRP1 expression was
demonstrated to be significantly higher following transfection with
MBD2 siRNA but not EZH2 siRNA (Fig.
5C). However, co-transfection of SW480 cells with EZH2 siRNA
alongside MBD2 siRNA significantly potentiated the increase in
SFRP1 gene expression compared with MBD2 siRNA alone (Fig. 5C). The methylation status of the
SFRP promoters was therefore indicated to not be altered by MBD2 or
EZH2 knockdown, either alone or in combination (Fig. 6A-C).
Discussion
SFRP proteins have been demonstrated to directly and
indirectly inhibit Wnt signalling (23). Accumulating evidence has suggested
that SFRP expression is tightly regulated by promoter methylation
(24). In the present study, SFRP1
mRNA expression was significantly downregulated in CRC cell lines
compared with CCC-HIE-2 cells, a non-cancerous intestinal mucosal
cell line, with concomitant hypermethylation in the SFRP1 promoter
region. Therefore, it was hypothesized that the silencing of SFRP1
expression was due to methylation of the SFRP1 gene promoter.
DNA methyltransferase (DNMT), which catalyses the
transfer of methyl groups to the carbon-5 position of cytosine in
the dinucleotide sequence CpG, is associated with establishing and
maintaining the profile of DNA methylation (25). A previous study has demonstrated
that treatment of SW480 and HCT116 cells with the DNA methylation
inhibitor decitabine (DAC) and HDAC inhibitor trichostatin A (TSA)
resulted in the restoration of SFRP1 expression, which was markedly
higher compared with that observed with DAC treatment only; though
TSA treatment alone did not restore the expression of
SFRP1(11). DAC is a DNMT inhibitor
that specifically promotes the degradation of DNMT1, whereas TSA is
a histone deacetylase inhibitor resulting in hyperacetylation on
histones H3 and H4(26). This
result suggests that promoter hypermethylation of SFRP1 was partly
mediated by DNMT1 and downregulation of SFRP1 in SW480 and HCT116
cells was mediated by both DNA methylation and histone
deacetylation.
A previous study reported that BMI1 interacts
directly with DNMT1-associated protein 1, which in turn interacts
with DNMT1 to mediate transcriptional repression (27). EZH2 can recruit DNMT1, DNMT3A and
DNMT3B to promote DNA methylation at CpG islands and establish
stable repressive chromatin structures (19). MBD2, which is a member of the MBD
protein family, can specifically recognize and bind to methylated
CpG sites, recruit co-repressors and modify the chromatin structure
to inhibit gene transcription, with HDAC as an example (28). MBD2 provides a potential link
between DNA methylation, chromatin remodelling and histone
modification. Enhanced binding of DNMT1, HDAC1 and MBD proteins to
the transcriptional start site of SFRP1 in hepatocarcinoma cells
has been previously reported to result in the epigenetic silencing
of SFRP1 expression (29).
According to results in the present study, the
transcription levels of MBD2 and BMI1 were only observed to be
upregulated in SW480 and HCT116 cells, whilst EZH2 expression was
increased from the early stages of CRC onwards, suggesting that
EZH2 may be involved in the occurrence of early CRC. The
involvement of BMI1, MBD2 and EZH2 in the regulation of DNMT and
HDAC migration mainly occurred during the middle to late stages of
CRC development.
To clarify the mechanism involved in the silencing
of SFRP1 by PCG and MBD proteins, potential PCG-SFRP1 promoter and
MBD-SFRP1 promoter complexes were detected using ChIP in CCC-HIE-2,
SW480 and HCT116 cells. BMI1 did not combine with the SFRP1
promoter region in the three cell lines, suggesting that it did not
participate in SFRP1 promoter methylation. The enrichment of EZH2
on the SFRP1 promoter region of SW480 cells was considerably lower
compared with that in CCC-HIE-2 cells, suggesting that it may be
associated with SFRP1 promoter methylation in CCC-HIE-2 cells but
not in SW480 cells. MBD2 was only indicated on the SFRP1 promoter
in SW480 cells, indicative of a role in SFRP1 promoter methylation
during the middle stages of CRC. In addition, it was revealed that
MBD2 and EZH2 bound to the SFRP1 gene promoter in SW480 cells but
not in HCT116 cells, suggesting that these two proteins may serve
distinct regulatory functions during CRC tumorigenesis. In SW480
cells, SFRP1 gene expression was restored following MBD2 knockdown
but not after EZH2 knockdown. In contrast, when MBD2 and EZH2 were
knocked down simultaneously, the expression of SFRP1 was found to
be significantly increased. However, neither MBD2 or EZH2 knockdown
could reverse SFRP1 gene promoter hypermethylation. These
observations suggest that SFRP1 gene inactivation in CRC is
associated with an additional mechanism that is distinct from
promoter hypermethylation.
MBD2 has been previously revealed to bind to
methylated DNA in the promoter region and recruit the
Mi-2/nucleosome remodelling and deacetylase nucleosome remodelling
complex, resulting in histone deacetylation and chromatin
remodelling followed by inhibition of gene transcription (18,30).
This effect can be potentiated by EZH2, which also functions in
chromatin remodelling (18,30).
Inhibition of MBD2 and EZH2 can reduce the activity
of histone deacetylase and alleviate the repressive state of
chromatin, restoring gene expression (31). This phenomenon is an indirect
mechanism of inhibiting gene expression without changing DNA
methylation status. However, a previous study demonstrated that
histone deacetylation was not associated with the suppression of
SFRP1 gene expression, as inhibition of histone deacetylase alone
did not restore SFRP1 expression (11). This mechanism is not sufficient to
explain the role of MBD2 knockdown in the restoration of
expression. A possible reason could be that SFRP1 transcription
persisted even with DNA methylation.
Studies have indicated that certain transcription
factors can also bind methylated DNA (32,33).
CG dinucleotides in the promoter region of SFRP1 can associate with
different types of transcription factors depending on the
methylation status of the sequence. In CRC cells, the
hypermethylated promoter of SFRP1 blocks transcription factor
binding to unmethylated CG, which may explain why the
hypermethylation of the SFRP1 promoter in colorectal tumours
effectively restores gene expression, whilst the other
transcription factors that preferentially bind to methylated CG of
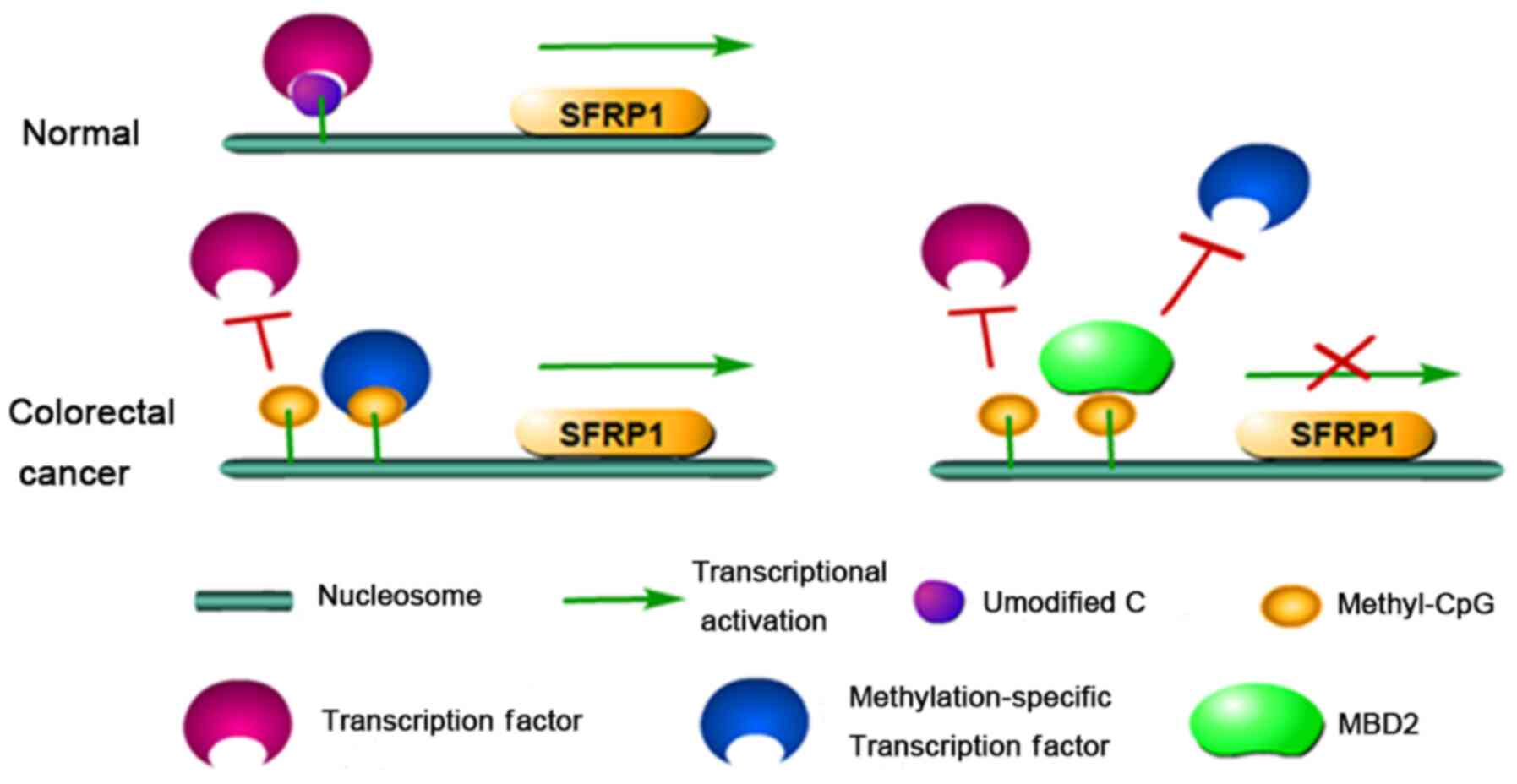
SFRP1 can still maintain gene transcription (34). Therefore, it can be speculated that
MBD2 may recognize and bind to the methylated promoter of SFRP1,
outcompeting other transcription factors that would normally bind
to the methylated CG, resulting in the complete inactivation of the
SFRP1 gene in CR tumours (Fig. 7).
This could explain the restoration of SFRP1 gene expression without
changes in the methylation status of the SFRP1 promoter following
EZH2 and MBD2 knockdown.
The present study demonstrated that MBD2 and EZH2
regulated SFRP1 expression without affecting SFRP1 methylation in
CRC cell lines. Further studies should be undertaken by profiling
transcription factors that can bind to the SFRP1 gene promoters. In
particular, the role of the typical transcription factors on the
restoration of SFRP1 expression, the precise mechanistic function
of MBD2 and EZH2 in addition to the regulatory mechanism of DNA
methylation on SFRP1 expression in CR tumours require further
study.
Supplementary Material
The regions and corresponding
sequences of SFRP promoters analyzed for methylation. (a) Regions
and corresponding sequences of SFRP1, SFRP2, SFRP4 and SFRP5
analyzed for methylation-specific PCR. (b) Regions and
corresponding sequences of the SFRP1 promoter analyzed for
chromatin immunoprecipitation. SFRP, secreted frizzled-related
protein. SFRP, secreted frizzled-related protein.
Acknowledgements
The abstract was presented at the 26th United
European Gastroenterology (UEG) Week, October 20-24, 2018 in
Vienna, Austria.
Funding
This project was supported by the Chinese National
Science Foundation Projects (grant no. 81101868) and the Applied
Basic Research Programs of Wuhan Science and Technology Department
(grant no. 2015061701011642).
Authors' contributions
All authors were responsible for the conception and
design of the present study. JY, YX and YL were responsible for the
provision of the study materials. JY and YX were responsible for
the collection and assembly of the data. JY, YX, YL, FW, ML and JQ
performed the data analysis and interpretation. JY, YX and JQ
contributed in writing the manuscript. JY, YX, YL, FW, ML and JQ
read and gave the final approval of the manuscript. All authors
read and approved the final manuscript.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel R, Desantis C and Jemal A:
Colorectal cancer statistics, 2014. CA Cancer J Clin. 64:104–117.
2014.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Vermeulen L, De Sousa E Melo F, van der
Heijden M, Cameron K, de Jong JH, Borovski T, Tuynman JB, Todaro M,
Merz C, Rodermond H, et al: Wnt activity defines colon cancer stem
cells and is regulated by the microenvironment. Nat Cell Biol.
12:468–476. 2010.PubMed/NCBI View
Article : Google Scholar
|
|
3
|
Jensen M, Hoerndli FJ, Brockie PJ, Wang R,
Johnson E, Maxfield D, Francis MM, Madsen DM and Maricq AV: Wnt
signaling regulates acetylcholine receptor translocation and
synaptic plasticity in the adult nervous system. Cell. 149:173–187.
2012.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Lv C, Li F, Li X, Tian Y, Zhang Y, Sheng
X, Song Y, Meng Q, Yuan S, Luan L, et al: MiR-31 promotes mammary
stem cell expansion and breast tumorigenesis by suppressing Wnt
signaling antagonists. Nat Commun. 8(1036)2017.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Russell JO and Monga SS: Wnt/β-catenin
signaling in liver development, homeostasis, and pathobiology. Annu
Rev Pathol. 13:351–378. 2018.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Tammela T, Sanchez-Rivera FJ, Cetinbas NM,
Wu K, Joshi NS, Helenius K, Park Y, Azimi R, Kerper NR, Wesselhoeft
RA, et al: A Wnt-producing niche drives proliferative potential and
progression in lung adenocarcinoma. Nature. 545:355–359.
2017.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Klaus A and Birchmeier W: Wnt signalling
and its impact on development and cancer. Nat Rev Cancer.
8:387–398. 2008.PubMed/NCBI View
Article : Google Scholar
|
|
8
|
Rattner A, Hsieh JC, Smallwood PM, Gilbert
DJ, Copeland NG, Jenkins NA and Nathans J: A family of secreted
proteins contains homology to the cysteine-rich ligand-binding
domain of frizzled receptors. Natl Acad Sci USA. 94:2859–2863.
1997.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Suzuki H, Watkins DN, Jair KW, Schuebel
KE, Markowitz SD, Chen WD, Pretlow TP, Yang B, Akiyama Y, Van
Engeland M, et al: Epigenetic inactivation of SFRP genes allows
constitutive WNT signaling in colorectal cancer. Nat Genet.
36:417–422. 2004.PubMed/NCBI View
Article : Google Scholar
|
|
10
|
Elzi DJ, Song MH, Hakala K, Weintraub ST
and Shiio Y: Wnt antagonist SFRP1 functions as a secreted mediator
of senescence. Mol Cell Biol. 32:4388–4399. 2012.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Qi J, Zhu YQ, Luo J and Tao WH:
Hypermethylation and expression regulation of secreted
frizzled-related protein genes in colorectal tumor. World J
Gastroenterol. 12:7113–7117. 2006.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Margueron R and Reinberg D: Chromatin
structure and the inheritance of epigenetic information. Nat Rev
Genet. 11:285–296. 2010.PubMed/NCBI View
Article : Google Scholar
|
|
13
|
Allis CD and Jenuwein T: The molecular
hallmarks of epigenetic control. Nat Rev Genet. 17:487–500.
2016.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Hughes TR and Lambert SA: Transcription
factors read epigenetics. Science. 356:489–490. 2017.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Riising EM, Comet I, Leblanc B, Wu X,
Johansen JV and Helin K: Gene silencing triggers polycomb
repressive complex 2 recruitment to CpG islands genome wide. Mol
Cell. 55:347–360. 2014.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Pan H, Bilinovich SM, Kaur P, Riehn R,
Wang H and Williams DC Jr: CpG and methylation-dependent DNA
binding and dynamics of the methylcytosine binding domain 2 protein
at the single-molecule level. Nucleic Acids Res. 45:9164–9177.
2017.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Negishi M, Saraya A, Miyagi S, Nagao K,
Inagaki Y, Nishikawa M, Tajima S, Koseki H, Tsuda H, Takasaki Y, et
al: Bmi1 cooperates with Dnmt1-associated protein 1 in gene
silencing. Biochem Biophys Res Commun. 353:992–998. 2007.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Wood KH and Zhou Z: Emerging molecular and
biological functions of MBD2, a reader of DNA methylation. Fron
Genet. 7(93)2016.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Vire E, Brenner C, Deplus R, Blanchon L,
Fraga M, Didelot C, Morey L, Van Eynde A, Bernard D, Vanderwinden
JM, et al: The Polycomb group protein EZH2 directly controls DNA
methylation. Nature. 439:871–874. 2006.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Ahmed D, Eide PW, Eilertsen IA, Danielsen
SA, Eknæs M, Hektoen M, Lind GE and Lothe RA: Epigenetic and
genetic features of 24 colon cancer cell lines. Oncogenesis.
2(e71)2013.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Bai J, Xu J, Zhao J and Zhang R: lncRNA
SNHG1 cooperated with miR-497/miR-195-5p to modify
epithelial-mesenchymal transition underlying colorectal cancer
exacerbation. J Cell Physiol. 235:1453–1468. 2020.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Cruciat CM and Niehrs C: Secreted and
transmembrane wnt inhibitors and activators. Cold Spring Harb
Perspect Biol. 5(a015081)2013.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Yu J, Xie Y, Li M, Zhou F, Zhong Z, Liu Y,
Wang F and Qi J: Association between SFRP promoter hypermethylation
and different types of cancer: A systematic review and
meta-analysis. Oncol Lett. 18:3481–3492. 2019.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Tatematsu KI, Yamazaki T and Ishikawa F:
MBD2-MBD3 complex binds to hemi-methylated DNA and forms a complex
containing DNMT1 at the replication foci in late S phase. Genes
Cells. 5:677–688. 2000.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Jones PA and Taylor SM: Cellular
differentiation, cytidine analogues and DNA methylation. Cell.
20:85–93. 1980.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Cramer JM, Scarsdale JN, Walavalkar NM,
Buchwald WA, Ginder GD and Williams DC Jr: Probing the dynamic
distribution of bound states for methylcytosine-binding domains on
DNA. J Biol Chem. 289:1294–1302. 2014.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Wood KH and Zhou Z: Emerging molecular and
biological functions of MBD2, a reader of DNA methylation. Front
Genet. 7(93)2016.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Quan H, Zhou F, Nie D, Chen Q, Cai X, Shan
X, Zhou Z, Chen K, Huang A, Li S and Tang N: Hepatitis C virus core
protein epigenetically silences SFRP1 and enhances HCC
aggressiveness by inducing epithelial-mesenchymal transition.
Oncogene. 33:2826–2835. 2014.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Desai MA, Webb HD, Sinanan LM, Scarsdale
JN, Walavalkar NM, Ginder GD and Williams DC Jr: An intrinsically
disordered region of methyl-CpG binding domain protein 2 (MBD2)
recruits the histone deacetylase core of the NuRD complex. Nucleic
Acids Res. 43:3100–3113. 2015.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Berger J and Bird A: Role of MBD2 in gene
regulation and tumorigenesis. Biochem Soc Trans. 33:1537–1540.
2005.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Tuğrul M, Paixão T, Barton NH and Tkačik
G: Dynamics of transcription factor binding site evolution. PLoS
Genet. 11(e1005639)2015.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Liu Y, Toh H, Sasaki H, Zhang X and Cheng
X: An atomic model of Zfp57 recognition of CpG methylation within a
specific DNA sequence. Genes Dev. 26:2374–2379. 2012.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Yin Y, Morgunova E, Jolma A, Kaasinen E,
Sahu B, Khund-Sayeed S, Das PK, Kivioja T, Dave K, Zhong F, et al:
Impact of cytosine methylation on DNA binding specificities of
human transcription factors. Science. 356(eaaj2239)2017.PubMed/NCBI View Article : Google Scholar
|