Introduction
Glioblastoma multiforme (GBM) is one of the most
malignant tumor types of the central nervous system, with short
median survival and poor prognosis. The major treatment is combined
therapy, including surgery, radiotherapy and chemotherapy (1-3).
In spite of these combinations, the efficacy of GBM treatments
remains unsatisfactory, placing a serious burden on society and
affected families (4).
In recent years, the significant role of the immune
microenvironment in tumors has been increasingly elucidated. The
immune microenvironment has an important role in the prognosis of
patients with GBM (5-7).
In the immune microenvironment of tumors, immune cells and stromal
cells have an important role and affect the prognosis of patients
(8-13).
An algorithm called ESTIMATE was designed to predict immune
infiltration by estimating immune cells and stromal cells with
specific expression values (14).
In addition, transcription factors are able to bind to specific
sequences on the 5' end of the gene, thereby regulating gene
expression (15,16). Transcription factors have a key role
in the development of GBM (5,17-19).
The era of excessive data has provided a convenient
platform to explore the molecular mechanisms of various diseases.
Microarray analysis is of great research and application value and
has an important role in the discovery of molecular markers for
diagnosis and prognosis, as well as novel drug targets (20). In the present study, to further
investigate the association between transcription factors and
immune-regulatory genes, data from the Assay for
Transposase-Accessible Chromatin (ATAC) database from patients with
GBM were analyzed and peak coverage on chromosomes was determined.
Finally, an interaction network of the prognosis-associated
immune-regulatory genes and transcription factors was established,
providing a possible immune gene regulatory network for genes
associated with poor prognosis in GBM.
Materials and methods
Data sources
Gene Expression Omnibus (GEO) is a public functional
genomic database of integrated gene expression data, gene chips and
microarrays. The gene expression datasets used in the present study
were downloaded from the GEO online platform (https://www.ncbi.nlm.nih.gov/geo/) (21). The selected datasets were GSE2223,
including 4 normal glioma samples and 27 GBM samples, GSE50161,
including 13 normal samples and 34 GBM samples, and GSE4290,
including 23 normal glioma samples and 81 GBM samples. To further
verify the association between immune-regulatory genes and GBM, the
level-3 gene expression profile and clinical data of patients with
GBM were downloaded from The Cancer Genome Atlas (TCGA) database
(Data release/version 17.0; https://portal.gdc.cancer.gov/). The immune score and
stromal score were determined from data obtained from the database
using the ESTIMATE algorithm (14).
From the TCGA database, a small number of non-cancerous control
tissues from subjects without GBM were available. The most common
sites of GBM are the frontal lobe, temporal lobe and parietal lobe.
In order to reduce bias, the corresponding 102 normal samples were
downloaded from the University of California Santa Cruz (UCSC)
database (https://xena.ucsc.edu/) and the data
were then corrected using the ‘removevBatchEffect’ function of the
limma package (22). ATAC with
high-throughput sequencing (ATAC-seq) data for patients with GBM
were downloaded from the ATAC database (https://gdc.cancer.gov/about-data/publications/ATACseq-AWG).
Differentially expressed genes
(DEGs)
R software (version 3.5.1) was used to analyze the
three datasets GSE2223, GSE50161 and GSE4290, as well as the TCGA
and GTEX data. Screening criteria were set to P<0.05 and
log|fold change (FC)| >1, and finally, the common DEGs (co-DEGs)
of the datasets were obtained.
Retrieval of immune gene and
transcription factor data
The enrichment analysis of co-DEGS from the three
GEO datasets focused on results associated with immunization. For
further study, tumor-associated immune genes and transcription
factors were downloaded from the IMMPORT database (https://www.immport.org/) and the Cistrome database
(http://cistrome.org/). The DEGs among the
downloaded immune genes and transcription factors were obtained
using the screening conditions of P<0.05 and log|FC| >0.58
(since there were fewer eligible results when setting log|FC| >1
in the present study, log|FC| >0.58 was used).
Enrichment analysis of co-DEGs
The co-DEGs from the GEO datasets were uploaded to
the Funrich software (23) for Gene
Ontology and Kyoto Encyclopedia of Genes and Genomes enrichment
analysis and the corresponding functional terms and pathways were
obtained. P<0.05 was considered to indicate statistical
significance.
Immune gene and transcription factor
analysis
The intersection of the co-DEGs from GEO, the DEGs
from TCGA and the immune-associated genes was used to perform a
subsequent differential expression analysis. Pearson correlation
analysis between the differentially expressed immune genes and
transcription factors with the prognosis of GBM was then performed
with a correlation coefficient of the absolute value >0.4 and
P<0.05 considered to indicate statistical significance.
Construction of Cox risk model
First, the association of immune genes with the
prognosis of patients GBM was determined using univariate Cox
regression analysis and a total of 17 prognostic genes were
obtained. Subsequently, multivariate Cox regression analysis was
used to construct a best-fitting prognostic model and a risk score
formula was then established by including each of these selected
genes [in total, seven key genes were selected according to the
minimum criteria, these genes were calcium binding protein a10
(S100A10), mitogen-activated protein kinase 3 (MAPK3), serpin
family A member 3 (SERPINA3), Fc fragment of IgG receptor IIb
(FCGR2B), endothelin receptor type A (EDNRA) and p21 activated
kinase 1 (PAK1) and the patients were divided into high/low risk
score groups]. Patients were classified into high- or low
risk-groups with the median risk score as the cutoff.
Multivariate analysis
To determine whether the immune risk score
independently affects the survival of patients with GBM, univariate
and multivariate Cox proportional hazards model analyses of the
influence of risk scores and clinical features of patients with GBM
on survival were performed.
Interaction network
In the present study, the ‘corrplot’ package of R
software was used for interaction network correlation analysis and
a correlation coefficient >0.4 and P<0.001 were considered to
indicate a significant correlation. Subsequently, Cytoscape
software (24) was used to
construct an interaction network for the immune genes associated
with the prognosis of patients with GBM, differentially expressed
transcription factors and immune cells from the TIMER database
(https://cistrome.shinyapps.io/timer/).
ATAC-seq data analysis
ATAC standardized matrix data were downloaded from
the ATAC database and through ‘TxDb.Hsapiens’,
‘UCSC.hg38.knownGene, org.Hs.eg.db’, ‘clusterProfiler’,
‘karyoploteR’ and ‘ChIPseeker’ package analyses, the distribution
of ATAC-seq peaks on chromosomes was determined. To further assess
whether Snail family transcriptional repressor 2 (SNAI2)/MYC and
C-X-C motif chemokine receptor 4 (CXCR4)/Serpin family A member 3
(SERPINA3) may have binding sites, they were analyzed through the
UCSC Genome Browser tool (minimum score=400).
Correlation analysis between risk
score and immune cells
The TCGA expression data of immune cells of patients
with GBM were downloaded from the TIMER database. The ‘corrplot’
package of R software was used to perform a Pearson correlation
analysis of risk scores with the immune cells infiltration
level.
Statistical analysis
All statistical analyses were performed using R
software (v. 3.5.1). An independent-samples t-test was used for
comparison between groups. The Cox proportional hazards regression
model was used for logistic regression. Missing values from
downloaded datasets were filled using multiple imputation methods
(25).
Results
Identification of DEGs
The datasets from the GSE2223, GSE50161 and GSE4290
gene chips were processed using R software. The screening
conditions were set to log|FC|>1 and P<0.05, and 1,420
co-DEGs were obtained (Fig. 1A).
Furthermore, 48 immune-associated genes were obtained after the
DEGs and the immune-associated genes were taken together (Fig. 1B). The differentially expressed
immune-associated genes and transcription factors (27
differentially expressed transcription factors) are presented in
Fig. 1C and D.
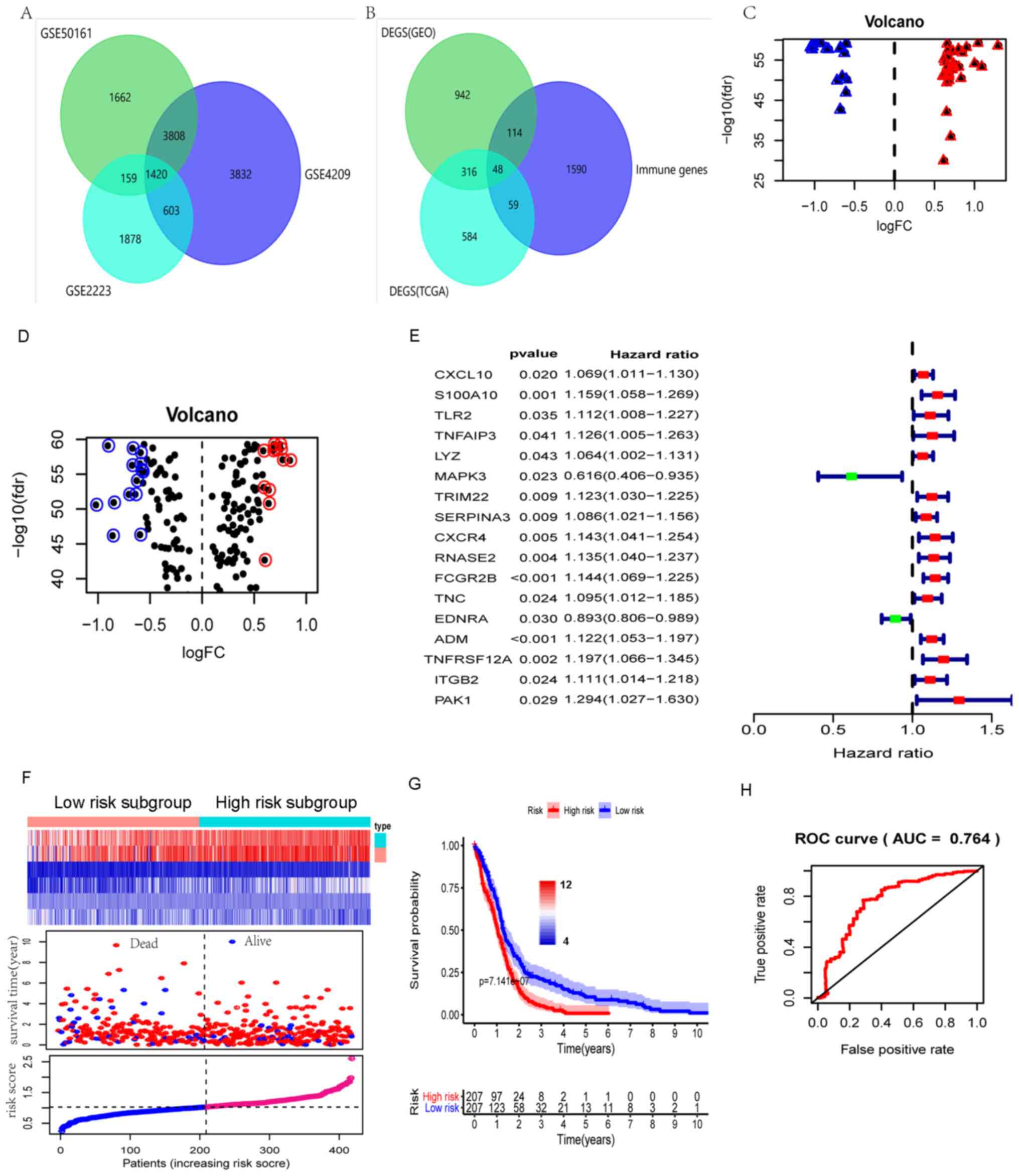 | Figure 1Screening of prognosis-related immune
genes and differentially expressed transcription factors and
construction of risk model. (A) Venn diagram of common DEGs among
the GSE4290, GSE50161 and GSE2223 gene expression datasets. (B)
Venn diagram of the intersection of DEGs and known
immune-associated genes. (C) Volcano plots displaying differential
expression analysis of immune-associated genes and (D)
transcription factors. Red indicates high expression and blue
indicates low expression. Screening conditions were set to
P<0.05 and log|FC| >0.58. (E) Univariate Cox regression
analysis identified 17 immune-associated genes affecting the
prognosis of patients with GBM and the HR of most genes was >1.
(F) Risk scoring model for immune gene construction. Red indicates
death and blue denotes survival. (G) Impact of high and low risk
scores on the prognosis of patients with GBM. A higher risk score
indicates a worse prognosis. (H) Receiver operating characteristic
curve of the risk scoring model (area under the curve=0.764). DEG,
differentially expressed gene; GEO, gene expression omnibus; GBM,
glioblastoma multiforme; TCGA, The Cancer Genome Atlas; FC, fold
change; FDR, false discovery rate; HR, hazard ratio; ROC, receiver
operating characteristic; AUC, area under the ROC curve; CXCL10,
C-X-C Motif Chemokine 10; S100A10, S100 Calcium Binding Protein
A10; TLR2, Toll like receptor 2; TNFAIP3, TNF Alpha Induced Protein
3; LYZ, Lysozyme; MAPK3, Mitogen-Activated Protein Kinase 3;
TRIM22, Tripartite Motif Containing 22; SERPINA3, Serpin family A
member 3; CXCR4, C-X-C motif chemokine receptor 4; RNASE2,
Ribonuclease A Family Member 2; FCGR2B, Fc fragment of IgG receptor
IIb; TNC, Tenascin C; EDNRA, Endothelin Receptor Type A; ADM,
Adrenomedullin; TNFRSF12A, TNF Receptor Superfamily Member 12A;
ITGB2, Integrin Subunit Beta 2; PAK1, P21 (RAC1) Activated Kinase
1. |
Enrichment analysis
Enrichment analysis of co-DEGs indicated that the
major functional terms and pathways were cell communication,
regulation of enzyme activity, immune response, nervous system, p38
signaling mediated by mitogen-activated protein kinase-activated
protein kinase (MAPKAP) kinases, cytokine signaling in immune
system and PI3K signaling events mediated by AKT (Fig. 2). Gene set variation (GSVA)
enrichment analysis of CXCR4 and SERPINA3 indicated a significant
positive correlation with AKT signaling pathways, which were
mutually verified in the TCGA and Chinese Glioma Genome Atlas
(CGGA) databases (http://www.cgga.org.cn/; Fig. S1).
Evaluation of risk models
In total, 14 immune genes associated with GBM
prognosis were obtained by univariate COX regression analysis
(Fig. 1E), A risk-score formula was
created based on the expression values of these six key genes
(Fig. 1F), as follows: Risk score =
(0.08 x expression value of S100A10) + (-0.466 x expression value
of MAPK3) + (0.095 x expression value of SERPINA3) + (0.071 x
expression value of FCGR2B) + (-0.154 x expression value of EDNRA)
+ (0.253 x expression value of PAK1). Finally, the patients were
divided into high/low risk groups. The prognosis of the two groups
was significantly different, where the high risk group was worse
than the low risk group (Fig. 1G).
The risk model constructed from prognosis-associated
immune-regulatory genes had high accuracy in predicting the
prognosis of patients with GBM (AUC=0.764) (Fig. 1H), which was higher than that of a
previously reported model constructed from prognosis-associated
biomarkers (AUC=0.667; Fig. S2)
(10).
Interaction network
In the present study, Cytoscape software was used to
construct an interaction network for the immune-regulatory genes
(associated with the prognosis of GBM), transcription factors and
immune cells (Fig. 3A). The circles
indicate the immune-regulatory genes, the diamonds indicate the
transcription factors and the squares indicate the immune cells.
Red nodes indicate high expression and blue indicates low
expression. Red lines indicate positive regulation and blue lines
indicate negative regulation. There were 41 nodes in the network,
where CXCR4 and SERPINA3 were found to be the key core node genes.
Survival analysis identified four transcription factors linked to
immune-regulatory genes that affected the prognosis of GBM, namely
BRF1, MYC, SNAI2 and SOX4 (Fig.
3B-E, respectively).
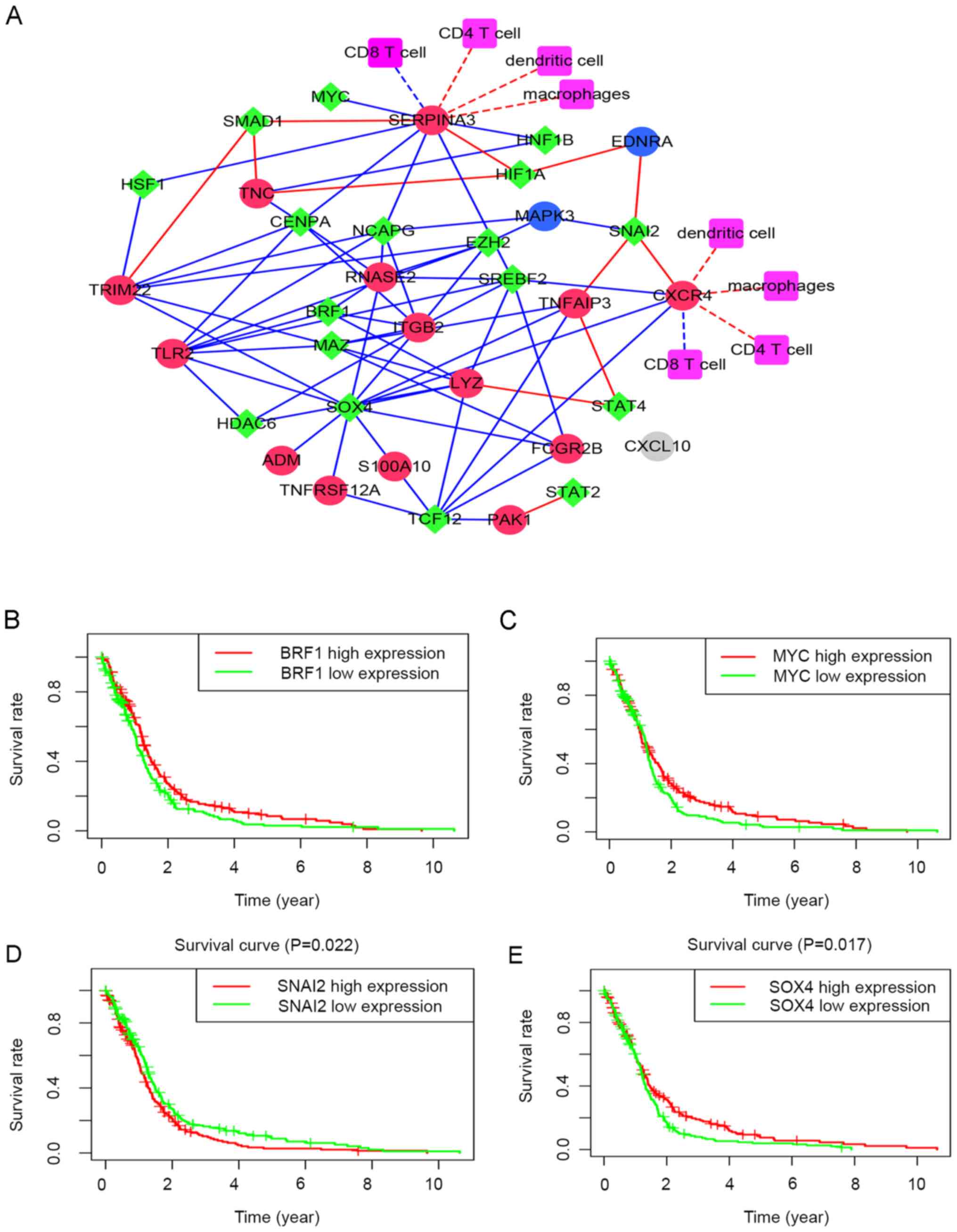 | Figure 3Search for transcription factors
related to immune genes and prognosis, and construction of
interaction network. (A) Interaction network of
prognosis-associated immune genes, transcription factors and immune
cells. The circles indicate the immune genes, the diamonds indicate
transcription factors, red nodes indicate high expression and blue
nodes indicate low expression, red lines indicate positive
regulation, blue lines indicate negative regulation and gray
indicates no obvious correlation; red/blue dashed lines indicate
possible positive/negative effects. (B-E) Survival analysis
identified four transcription factors linked to immune genes
affecting the prognosis of glioblastoma multiforme. (B) There were
BRF1, (C) MYC, (D) SNAI2 and (E) SOX4. BRF1, BRF1 RNA polymerase
III transcription initiation factor subunit; MYC, MYC
proto-oncogene, BHLH transcription factor; SNAI2, Snail family
transcriptional repressor 2, SOX4, SRY-Box Transcription Factor
4. |
Clinical data of patients with
GBM
From the GBM patient data that were downloaded
(Table I), patients with no
survival information were removed and the 417 remaining patients
were analyzed. The cohort comprised 248 male and 169 female
patients with an age range of 10-89 years, an immune score of
-1,448 to 3,210 and a stromal score of -3,055 to 2,016. The
classification was divided into four subtypes as follows: Classical
(30.7%), mesenchymal (29.3%), neural (15.3%) and proneural (24.7%).
Among the patients, 272 received chemotherapy and 140 did not;
furthermore, 365 received radiotherapy and 52 did not. The median
follow-up duration was 12.8 months (range, 0-129.4 months).
 | Table IThe Cancer Genome Atlas Glioblastoma
patient characteristics. |
Table I
The Cancer Genome Atlas Glioblastoma
patient characteristics.
| Clinical
characteristics | Total (n=417) | % |
|---|
| Age (10-89
years) | | |
|
<75
years | 366 | 87.8 |
|
≥75
years | 51 | 12.2 |
| Sex | | |
|
Male | 248 | 59.5 |
|
Female | 169 | 40.5 |
| Immune score | | |
|
-1,448-3,210 | 417 | 100 |
| Stromal score | | |
|
-3,055-2,016 | 417 | 100 |
| Radiotherapy | | |
|
Yes | 365 | 87.5 |
|
No | 52 | 12.5 |
| Chemotherapy | | |
|
Yes | 272 | 66 |
|
Yes | 140 | 34 |
| Subtype | | |
|
Classical | 128 | 30.7 |
|
Mesenchymal | 122 | 29.3 |
|
Neural | 64 | 15.3 |
|
Proneural | 103 | 24.7 |
Analysis of the association between
risk score and characteristics of patients with GBM
The association between the risk score and the
characteristics of patients with GBM is presented in Fig. 4A-F. The risk score was significantly
associated with the prognosis, stromal score and immune score of
patients with GBM. Multivariate analysis indicated that the risk
score was an independent predictive factor of poor prognosis in
patients with GBM (Fig. 4B). High
risk scores predicted adverse outcomes in patients with GBM. The
immune score and stromal score were significantly associated with
the prognosis of patients with GBM (Fig. 4E and F).
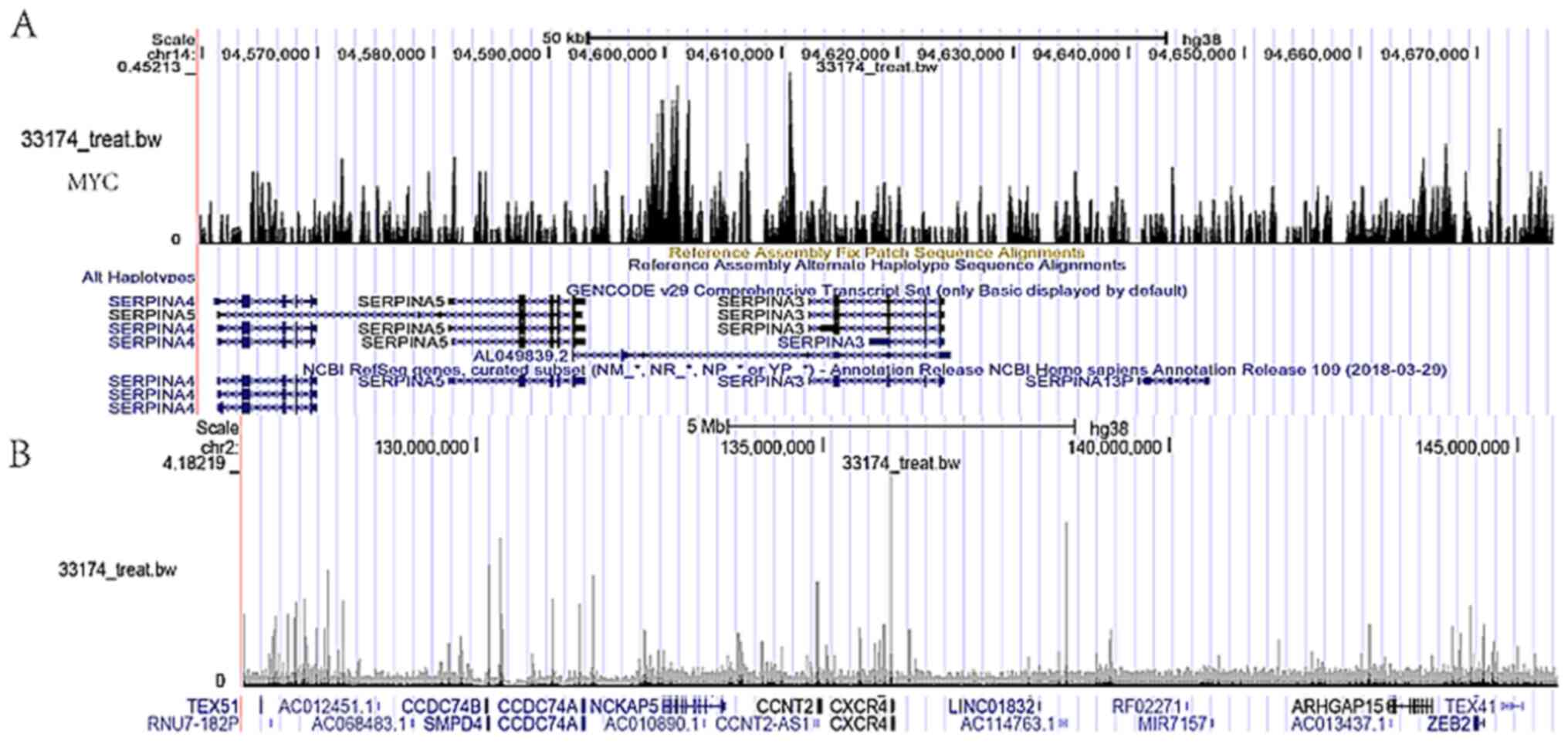
ATAC data analysis and search for
upstream transcription factors of SERPINA3 and CXCR4
Analysis of the ATAC data indicated that, in
addition to the Y chromosome, there was a large number of peaks in
other chromosomes. Further analysis suggested that the most binding
sites were situated near the promoter (≤1 kb), and in the heatmap,
a large number of peaks were enriched near the transcription start
site (TSS), while the peak enrichment gradually decreased with the
distance from the TSS. At the same time, most peaks existed in
multiple regions, consistent with previous studies (Fig. 4G-I). In a further search for
possible upstream transcription factors of SERPINA3 and CXCR4,
possible transcription factors were predicted using the UCSC
database, and MYC and SNAI2 were identified, which were also
associated with the prognosis of patients with GBM. Further
analysis indicated that the promoter regions of SERPINA3 had
prospective binding peak sites for MYC and CXCR4 had prospective
binding peak sites for SNAI2 (Fig.
5).
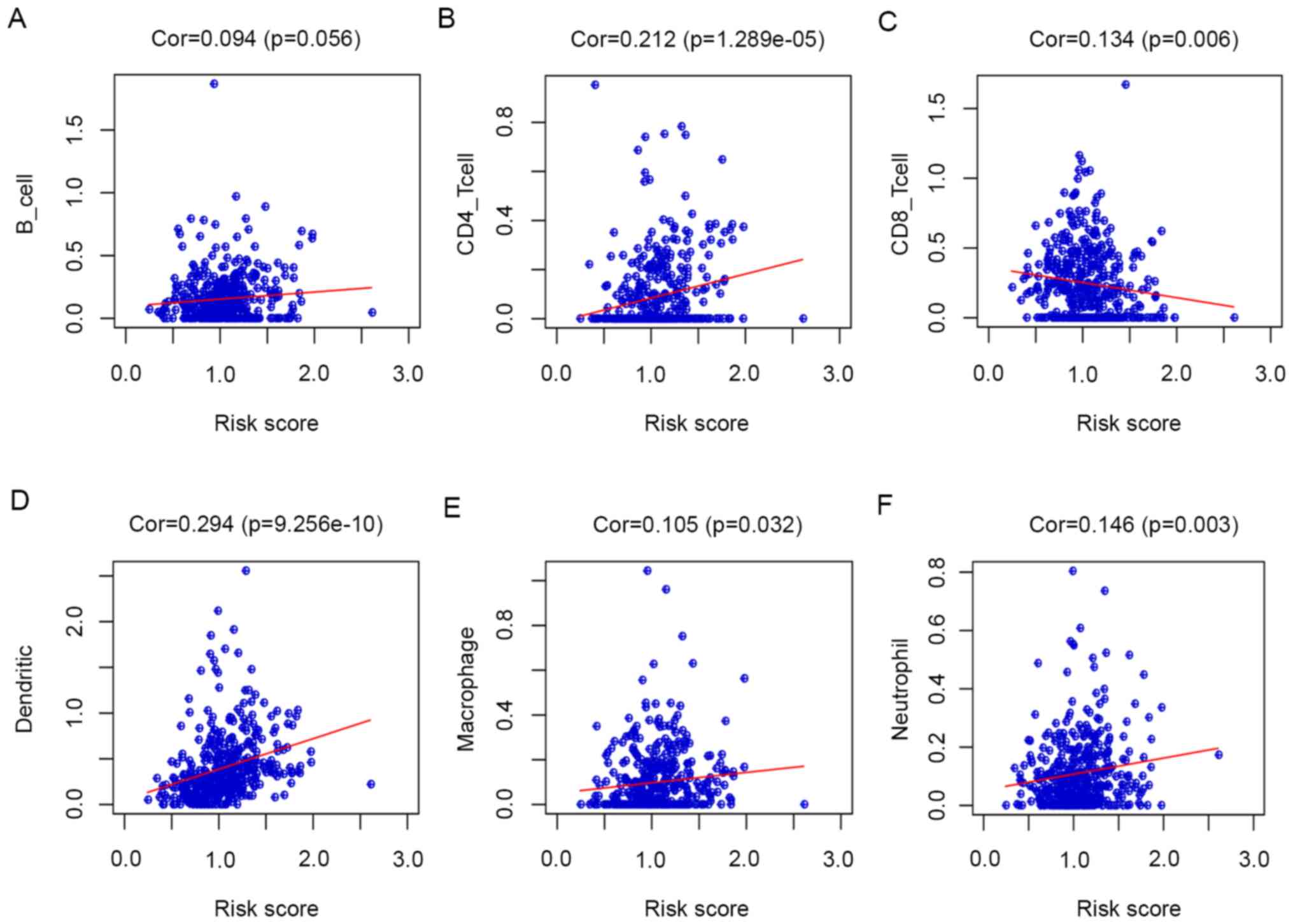
Correlation analysis between risk
score and immune cells
The ‘corrplot’ package of R software was used to
determine the correlation of the risk scores with immune cells. The
results indicated that the risk score was positively correlated
with CD4 T cells, dendritic cells, neutrophil cells and macrophages
and negatively correlated with CD8 T cells, but the correlation
with B-cells did not reach statistical significance (Fig. 6).
Discussion
In the present study, data of patients with GBM were
downloaded from the GEO, TCGA, TIMER, IMMPORT and CISTROME
databases. Co-DEGs were mainly enriched in cell communication,
regulation of enzyme activity, immune response, nervous system, p38
signaling mediated by MAPKAP kinases, cytokine signaling in the
immune system and PI3K signaling events mediated by AKT. The
further GSVA enrichment analysis of CXCR4 and SERPINA3 indicated a
positive association with macrophage activation, differentiation
and regulation of the AKT signaling pathway.
The present study focused on the tumor immune
microenvironment. In the immune microenvironment, immune cells and
stromal cells have a key role and affect the prognosis of patients
with cancer (14). In the present
study, the risk scores were different between subgroups of patients
with high and low immune scores and stromal scores, and the risk
score was able to be used as an independent risk factor for the
prognosis of patients with GBM with statistical significance, and
immune scores and stromal scores also affected the prognosis of
GBM, which was consistent with previous studies (8,9). In
the present study, a total of 48 immune-associated genes were used
to perform a prognostic analysis of differentially expressed
immune-regulatory genes, yielding 17 genes significantly associated
with the prognosis of patients with GBM. The risk model constructed
from prognosis-associated immune-regulatory genes had high accuracy
in predicting the prognosis of patients with GBM (AUC=0.764), which
was higher than that of a previously reported model constructed
from prognosis-associated biomarkers (AUC=0.667) (10). This result suggested that the immune
genes screened in the present study were able to better reflect the
prognosis of patients with GBM and patients with a high-risk score
had a worse prognosis than those with a lower risk score. For
further investigation, an interaction network was constructed from
prognosis-associated immune genes, differentially expressed
transcription factors and immune cells. At the same time,
differentially expressed transcription factors were used to predict
the prognosis of patients with GBM, indicating that BRF1, MYC,
SNAI2 and SOX4 affected the survival of patients with GBM. Combined
with the results of the analysis of the UCSC database, SNAI2 and
MYC appeared promising, and it was predicted that SNAI2 is able to
positively regulate the immune genes CXCR4, and that MYC is able to
positively regulate the immune genes SERPINA3. CXCR4 is a CXC
chemokine receptor specific for stromal cell-derived factor-1; the
protein has 7 transmembrane regions and is located on the cell
surface (26). SNAI2 encodes a
member of the Snail family of C2H2-type zinc finger transcription
factors (27). Previous studies
indicated that CXCR4 affects the proliferation, invasion and
angiogenesis of glioma cells by regulating the AKT signaling
pathway (28-30),
which was consistent with the results of the present enrichment
analysis. SNAI2 may be used as a GBM marker, which participates in
the epithelial-to-mesenchymal transition and thus affects drug
resistance (31,32); it may also enhance the development
of tumors by affecting the AKT signaling pathway (32-34).
However, whether SNAI2 is able to regulate CXCR4 expression, affect
the ATK signaling pathway and thus affect the phenotype of glioma
remains to be elucidated. The protein encoded by SERPINA3 is a
plasma protease inhibitor and member of the serine protease
inhibitor class. Serine protease has an important role in the
development of glioma, which may promote the migration and invasion
of glioma, but serine protease inhibitor has the opposite role
(35). The present study indicated
that circulating SERPINA3 may be a marker in GBM and promote the
invasion of glioblastic stem cells (36,37),
and a positive correlation with the AKT signaling pathway was
identified in the GSVA enrichment analysis. However, the specific
mechanisms remain elusive. MYC is a proto-oncogene and encodes a
nuclear phosphoprotein that has a role in cell cycle progression,
apoptosis and cellular transformation of glioma (38,39). A
negative regulatory association between MYC and SERPINA3 was
indicated in the regulatory network generated in the present study.
In GBM, MYC is mainly highly expressed, and it may therefore be
speculated that high expression of MYC promotes the development of
GBM through inhibition of SERPINA3, which requires further
verification. The present study aimed to explore whether binding
sites exist between SNAI2 and CXCR4, and between MYC and SERPINA3.
The results indicated that there was a large number of peaks on
chromosomes of patients with GBM, suggesting that there were
numerous transcription factor binding sites on the chromosome and
most binding sites were located near the promoter (≤1 kb). The UCSC
database was used for analysis, suggesting that the CXCR4 and
SERPINA3 gene sequences had peaks near the promoter of the
transcription factor SNAI2 and MYC, respectively, indicating that
there were binding sites between them.
Finally, the association between the risk score and
immune cells was analyzed, revealing that the risk score was linked
to the immune microenvironment and that the risk score was
positively correlated with CD4 T cells and negatively correlated
with CD8 T cells. This suggests that a high-risk score may be
associated with a secondary elevation of CD4 T cells, dendritic
cells and macrophages, as well as a secondary reduction of CD8 T
cells. Previous studies have indicated that CD8 T cells and
dendritic cells have a positive role in the development of a normal
body (40-44).
This may indicate that an increase in the risk score may cause a
secondary decrease in CD8 T cells, thereby promoting tumor
development. It may be hypothesized that high expression of
immune-regulatory genes, including SERPINA3 and CXCR4, increased
the risk score of patients with GBM, which led to poor prognosis,
as well as secondary changes in immune cells in the immune
microenvironment, including a secondary decline of CD8 T cells.
However, this requires to be confirmed by future studies.
In conclusion, the immune gene interaction network
constructed in the present study helps to understand the mechanisms
associated with poor prognosis of patients with GBM. A risk scoring
system was established in the present study, and high-risk scores
indicated poor prognosis of patients with GBM and may be used as an
independent risk factor for assessing the prognosis of patients
with GBM.
Supplementary Material
GSVA enrichment Analysis of CXCR4 and
SERPINA3. (A) Gene set variation enrichment analysis of CXCR4 in
TCGA, (B) Gene set variation enrichment analysis of SERPINA3 in
TCGA, (C) Gene set variation enrichment analysis of CXCR4 in CGGA,
(D) Gene set variation enrichment analysis of SERPINA3 in CGGA. The
greater the clockwise angle of the pie chart, the higher the degree
of positive correlation, and the greater the counterclockwise
angle, the higher the degree of negative correlation. The red
sector of the diagram indicates a positive association and the
green sector of the diagram indicates a negative association. A
darker color indicates a higher association. The results suggested
that CXCR4 and SERPINA3 were positively associated with the
activation and regulation of AKT signaling pathway. P<0.05 was
considered to indicate statistical significance. GO terms:
GO:0032148, activation of protein kinase B activity; GO:0043491,
protein kinase B signaling; GO:0051896, regulation of protein
kinase B signaling; GO:0051897, positive regulation of protein
kinase B signaling. GO, gene ontology; GSVA, gene set variation;
TCGA, The Cancer Genome Atlas; CGGA, Chinese Glioma Genome Atlas;
CXCR4, C-X-C motif chemokine receptor 4; SERPINA3, Serpin family A
member 3.
Construction of risk model based on
other biomarkers. A risk model based on previously reported
glioblastoma multiforme biomarker genes is able to better predict
the prognosis of patients, but the accuracy is poorer than that of
the model constructed in the present study. (A) Risk scoring model
for biomarker genes construction. Red indicates death and blue
denotes survival. (B) Impact of high and low risk scores on the
prognosis of patients with GBM, a higher risk score indicates a
worse prognosis. (C) Receiver operating characteristic curve of the
risk scoring model (area under the curve=0.667). ROC, receiver
operating characteristic; AUC, area under the ROC curve.
Acknowledgements
Not applicable.
Funding
The present study was funded by the Natural Science
Foundation of China (grant no. 81660421), The First People's
Hospital of Zunyi Joint Research and Development of Science and
Technology projects [grant nos. (2010)66 and (2011)24].
Availability of data and materials
The data that support the findings of this study are
openly available in TCGA at http://www.tcga.org/, CGGA at http://www.cgga.org.cn/, TIMER at https://cistrome.shinyapps.io/timer/,
UCSC at https://xenabrowser.net/datapages/ and GEO at
https://www.ncbi.nlm.nih.gov/geo/.
Authors' contributions
XH, JC and SY conceived the current study,
participated in design and coordination, performed the
bioinformatic analysis and drafted the manuscript. XH, QZ, and FC
participated in the design of the study and performed statistical
analysis. YF, CX and GZ participated in the acquisition of data and
analysis. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Louis DN, Ohgaki H, Wiestler OD, Cavenee
WK, Burger PC, Jouvet A, Scheithauer BW and Kleihues P: The 2007
WHO classification of tumours of the central nervous system. Acta
Neuropathol. 114:97–109. 2007.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Dubrow R, Darefsky AS, Jacobs DI, Park LS,
Rose MG, Laurans MS and King JT Jr: Time trends in glioblastoma
multiforme survival: The role of temozolomide. Neuro-oncol.
15:1750–1761. 2013.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Stupp R, Hegi ME, Mason WP, van den Bent
MJ, Taphoorn MJ, Janzer RC, Ludwin SK, Allgeier A, Fisher B,
Belanger K, et al: European Organisation for Research and Treatment
of Cancer Brain Tumour and Radiation Oncology Groups; National
Cancer Institute of Canada Clinical Trials Group: Effects of
radiotherapy with concomitant and adjuvant temozolomide versus
radiotherapy alone on survival in glioblastoma in a randomised
phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet
Oncol. 10:459–466. 2009.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Alexiou GA, Tsiouris S, Kyritsis AP,
Voulgaris S, Argyropoulou MI and Fotopoulos AD: Glioma recurrence
versus radiation necrosis: Accuracy of current imaging modalities.
J Neurooncol. 95:1–11. 2009.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Cooper LA, Gutman DA, Chisolm C, Appin C,
Kong J, Rong Y, Kurc T, Van Meir EG, Saltz JH, Moreno CS, et al:
The tumor microenvironment strongly impacts master transcriptional
regulators and gene expression class of glioblastoma. Am J Pathol.
180:2108–2119. 2012.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Galon J, Pagès F, Marincola FM, Thurin M,
Trinchieri G, Fox BA, Gajewski TF and Ascierto PA: The immune score
as a new possible approach for the classification of cancer. J
Transl Med. 10(1)2012.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Şenbabaoğlu Y, Gejman RS, Winer AG, Liu M,
Van Allen EM, de Velasco G, Miao D, Ostrovnaya I, Drill E, Luna A,
et al: Tumor immune microenvironment characterization in clear cell
renal cell carcinoma identifies prognostic and
immunotherapeutically relevant messenger RNA signatures. Genome
Biol. 17(231)2016.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Chang N, Ahn SH, Kong DS, Lee HW and Nam
DH: The role of STAT3 in glioblastoma progression through dual
influences on tumor cells and the immune microenvironment. Mol Cell
Endocrinol. 451:53–65. 2017.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Zhang B, Shen R, Cheng S and Feng L:
Immune microenvironments differ in immune characteristics and
outcome of glioblastoma multiforme. Cancer Med. 8:2897–2907.
2019.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Jia D, Li S, Li D, Xue H, Yang D and Liu
Y: Mining TCGA database for genes of prognostic value in
glioblastoma microenvironment. Aging (Albany NY). 10:592–605.
2018.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Akimoto K, Kimura K, Nagano M, Takano S,
To'a Salazar G, Yamashita T and Ohneda O: Umbilical cord
blood-derived mesenchymal stem cells inhibit, but adipose
tissue-derived mesenchymal stem cells promote, glioblastoma
multiforme proliferation. Stem Cells Dev. 22:1370–1386.
2013.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Boonstra MC, Verbeek FP, Mazar AP, Prevoo
HA, Kuppen PJ, van de Velde CJ, Vahrmeijer AL and Sier CF:
Expression of uPAR in tumor-associated stromal cells is associated
with colorectal cancer patient prognosis: A TMA study. BMC Cancer.
14(269)2014.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Miki Y, Yashiro M, Okuno T, Kuroda K,
Togano S, Hirakawa K and Ohira M: Clinico-pathological significance
of exosome marker CD63 expression on cancer cells and stromal cells
in gastric cancer. PLoS One. 13(e0202956)2018.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Yoshihara K, Shahmoradgoli M, Martínez E,
Vegesna R, Kim H, Torres-Garcia W, Treviño V, Shen H, Laird PW,
Levine DA, et al: Inferring tumour purity and stromal and immune
cell admixture from expression data. Nat Commun.
4(2612)2013.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Lambert SA, Jolma A, Campitelli LF, Das
PK, Yin Y, Albu M, Chen X, Taipale J, Hughes TR and Weirauch MT:
The Human Transcription Factors. Cell. 172:650–665. 2018.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Malysheva V, Mendoza-Parra MA, Saleem MA
and Gronemeyer H: Reconstruction of gene regulatory networks
reveals chromatin remodelers and key transcription factors in
tumorigenesis. Genome Med. 8(57)2016.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Wang Q, Hu B, Hu X, Kim H, Squatrito M,
Scarpace L, deCarvalho AC, Lyu S, Li P, Li Y, et al: Tumor
Evolution of Glioma-Intrinsic Gene Expression Subtypes Associates
with Immunological Changes in the Microenvironment. Cancer Cell.
32:42–56.e6. 2017.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Chong YK, Sandanaraj E, Koh LW,
Thangaveloo M, Tan MS, Koh GR, Toh TB, Lim GG, Holbrook JD, Kon OL,
et al: ST3GAL1-Associated Transcriptomic Program in Glioblastoma
Tumor Growth, Invasion, and Prognosis. J Natl Cancer Inst.
108(djv326)2015.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Yan ZH, Bao ZS, Yan W, Liu YW, Zhang CB,
Wang HJ, Feng Y, Wang YZ, Zhang W, You G, et al: Upregulation of
DLX2 confers a poor prognosis in glioblastoma patients by inducing
a proliferative phenotype. Curr Mol Med. 13:438–445.
2013.PubMed/NCBI
|
|
20
|
Wang S, Liu F, Wang Y, Fan W, Zhao H, Liu
L, Cen C, Jiang X, Sun M and Han P: Integrated analysis of 34
microarray datasets reveals CBX3 as a diagnostic and prognostic
biomarker in glioblastoma. J Transl Med. 17(179)2019.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Barrett T, Wilhite SE, Ledoux P,
Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH,
Sherman PM, Holko M, et al: NCBI GEO: Archive for functional
genomics data sets - update. Nucleic Acids Res. 41D:D991–D995.
2013.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Gentleman R, Carey VJ, Huber W, Irizarry
RA and Dudoit S: limma: Linear models for microarray data In:
Bioinformatics and Computational Biology Solutions Using R and
Bioconductor. Gentleman R, Carey VJ, Huber W, Irizarry RA and
Dudoit S (eds). Springer, New York, NY, pp397-420, 2005.
|
|
23
|
Pathan M, Keerthikumar S, Ang CS, Gangoda
L, Quek CY, Williamson NA, Mouradov D, Sieber OM, Simpson RJ, Salim
A, et al: FunRich: An open access standalone functional enrichment
and interaction network analysis tool. Proteomics. 15:2597–2601.
2015.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Lee KJ and Simpson JA: Introduction to
multiple imputation for dealing with missing data. Respirology.
19:162–167. 2014.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Rummel PC, Thiele S, Hansen LS, Petersen
TP, Sparre-Ulrich AH, Ulven T and Rosenkilde MM: Extracellular
disulfide bridges serve different purposes in two homologous
chemokine receptors, CCR1 and CCR5. Mol Pharmacol. 84:335–345.
2013.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Cobaleda C, Pérez-Caro M, Vicente-Dueñas C
and Sánchez-García I: Function of the zinc-finger transcription
factor SNAI2 in cancer and development. Annu Rev Genet. 41:41–61.
2007.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Yi L, Zhou X, Li T, Liu P, Hai L, Tong L,
Ma H, Tao Z, Xie Y, Zhang C, et al: Notch1 signaling pathway
promotes invasion, self-renewal and growth of glioma initiating
cells via modulating chemokine system CXCL12/CXCR4. J Exp Clin
Cancer Res. 38(339)2019.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Ping YF, Yao XH, Jiang JY, Zhao LT, Yu SC,
Jiang T, Lin MC, Chen JH, Wang B, Zhang R, et al: The chemokine
CXCL12 and its receptor CXCR4 promote glioma stem cell-mediated
VEGF production and tumour angiogenesis via PI3K/AKT signalling. J
Pathol. 224:344–354. 2011.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Liu Y, Wei S, Zou Q and Luo Y: Stachydrine
suppresses viability & migration of astrocytoma cells via
CXCR4/ERK & CXCR4/Akt pathway activity. Future Oncol.
14:1443–1459. 2018.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Cheng Q, Huang C, Cao H, Lin J, Gong X, Li
J, Chen Y, Tian Z, Fang Z and Huang J: A Novel Prognostic Signature
of Transcription Factors for the Prediction in Patients With GBM.
Front Genet. 10(906)2019.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Liao H, Bai Y, Qiu S, Zheng L, Huang L,
Liu T, Wang X, Liu Y, Xu N, Yan X, et al: miR-203 downregulation is
responsible for chemoresistance in human glioblastoma by promoting
epithelial-mesenchymal transition via SNAI2. Oncotarget.
6:8914–8928. 2015.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Fenouille N, Tichet M, Dufies M, Pottier
A, Mogha A, Soo JK, Rocchi S, Mallavialle A, Galibert MD, Khammari
A, et al: The epithelial-mesenchymal transition (EMT) regulatory
factor SLUG (SNAI2) is a downstream target of SPARC and AKT in
promoting melanoma cell invasion. PLoS One.
7(e40378)2012.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Laakkonen JP, Lappalainen JP, Theelen TL,
Toivanen PI, Nieminen T, Jauhiainen S, Kaikkonen MU, Sluimer JC and
Ylä-Herttuala S: Differential regulation of angiogenic cellular
processes and claudin-5 by histamine and VEGF via PI3K-signaling,
transcription factor SNAI2 and interleukin-8. Angiogenesis.
20:109–124. 2017.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Huasong G, Zongmei D, Jianfeng H, Xiaojun
Q, Jun G, Sun G, Donglin W and Jianhong Z: Serine protease
inhibitor (SERPIN) B1 suppresses cell migration and invasion in
glioma cells. Brain Res. 1600:59–69. 2015.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Miyauchi E, Furuta T, Ohtsuki S, Tachikawa
M, Uchida Y, Sabit H, Obuchi W, Baba T, Watanabe M, Terasaki T, et
al: Identification of blood biomarkers in glioblastoma by SWATH
mass spectrometry and quantitative targeted absolute proteomics.
PLoS One. 13(e0193799)2018.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Li Y, Dong X, Cai J, Yin S, Sun Y, Yang D
and Jiang C: SERPINA3 induced by astroglia/microglia co-culture
facilitates glioblastoma stem-like cell invasion. Oncol Lett.
15:285–291. 2018.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Ding Z, Liu X, Liu Y, Zhang J, Huang X,
Yang X, Yao L, Cui G and Wang D: Expression of far upstream element
(FUSE) binding protein 1 in human glioma is correlated with c-Myc
and cell proliferation. Mol Carcinog. 54:405–415. 2015.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Chen H, Guo Y, Sun J, Dong J, Bao Q, Zhang
X and Fu F: Preferential Expression of B7-H6 in Glioma Stem-Like
Cells Enhances Tumor Cell Proliferation via the c-Myc/RNMT Axis. J
Immunol Res. 2020(2328675)2020.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Eiraku Y, Terunuma H, Yagi M, Deng X,
Nicol AJ and Nieda M: Dendritic cells cross-talk with tumour
antigen-specific CD8+ T cells, Vγ9γδT cells and Vα24NKT
cells in patients with glioblastoma multiforme and in healthy
donors. Clin Exp Immunol. 194:54–66. 2018.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Prasad S, Gaedicke S, Machein M, Mittler
G, Braun F, Hettich M, Firat E, Klingner K, Schüler J, Wider D, et
al: Effective Eradication of Glioblastoma Stem Cells by Local
Application of an AC133/CD133-Specific T-cell-Engaging Antibody and
CD8 T Cells. Cancer Res. 75:2166–2176. 2015.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Xu X, Stockhammer F, Schmitt A,
Casalegno-Garduno R, Enders A, Mani J, Classen CF, Linnebacher M,
Freund M and Schmitt M: Therapeutical doses of temozolomide do not
impair the function of dendritic cells and CD8+ T cells.
Int J Oncol. 40:764–772. 2012.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Pellegatta S, Poliani PL, Stucchi E, Corno
D, Colombo CA, Orzan F, Ravanini M and Finocchiaro G: Intra-tumoral
dendritic cells increase efficacy of peripheral vaccination by
modulation of glioma microenvironment. Neuro-oncol. 12:377–388.
2010.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Candolfi M, King GD, Yagiz K, Curtin JF,
Mineharu Y, Muhammad AK, Foulad D, Kroeger KM, Barnett N, Josien R,
et al: Plasmacytoid dendritic cells in the tumor microenvironment:
Immune targets for glioma therapeutics. Neoplasia. 14:757–770.
2012.PubMed/NCBI View Article : Google Scholar
|