Introduction
Breast cancer is one of the most common
malignancies, accounting for 29% of all cancer cases in females
worldwide (1). In the Middle East,
the prevalence of breast cancer remains lower than that in North
America and Europe, where it accounts for 14-42% of all female
cancer cases (2). In Syria, there
are no official statistics on the prevalence of breast cancer or
other malignancies (3). According
to the International Agency for Research on Cancer in 2012, the
prevalence ratio of breast cancer in females in Syria is 36.4% and
the mortality rate is 24.8% (4). In
recent years, numerous methods have been used for screening,
diagnosing and monitoring breast cancer, including physical
examination, mammography and biopsy (5). However, each of these techniques has
numerous drawbacks, making it necessary to identify a novel
diagnostic biomarker or method (6).
In addition, despite the significant progress in the development of
cancer therapies, cancer-associated mortality remains high
(7). Previous research has
increasingly focused on the role of the quantitative and
qualitative changes of circulating cell-free nuclear DNA (ccfnDNA)
and circulating cell-free mitochondrial DNA (ccfmtDNA) in numerous
different types of disease, but particularly in cancer (8,9).
Cancer cells release their nucleic acids (both n/mDNA and RNAs)
into the systemic circulation through cellular necrosis and
apoptosis (10,11); this DNA is mixed with DNA released
from normal cells (12). In
previous studies, changes in the ccfnDNA and ccfmtDNA levels were
identified not only in tissue samples but also in body fluids such
as plasma, which makes them attractive noninvasive potential
biomarkers (13,14). Several studies have reported
elevated levels of ccfnDNA or ccfmtDNA in different types of
cancer, while others reported decreased levels, thus the results
remain conflicting (15-17).
Due to these results, the present study aimed to determine their
role in breast cancer by comparing the absolute quantities of
ccfnDNA and ccfmtDNA, as well as the ccfmtDNA/ccfnDNA ratio, among
malignant and benign tumor groups and healthy controls.
Materials and methods
Patients
In total, 84 Syrian females were included in the
present study. Informed consent forms were signed by all
participants and the study was approved by the Syrian Higher
Commission for Scientific Research, Ministry of Higher Education,
Damascus, Syria. The study cohort (n=84) was divided into 3 groups:
i) Malignant disease group (n=33); ii) benign disease group (n=26);
and iii) healthy control group (n=25). Groups 1 and 2 contained
patients encountered at the general surgery department of Al-Assad
University Hospital-Damascus (Damascus, Syria) for breast
mastectomy; the diagnosis was confirmed by biopsy for all cases.
The healthy control group used in this study had neither a history
of cancer nor suffered from any other severe or autoimmune disease.
All blood samples were taken prior to any invasive procedures or
therapeutic treatments between February 2013 and June 2014. The
clinical data of each patient [age, smoking status, menopausal
status, cancer stage, grade, tumor size, lymph node involvement,
estrogen receptor (ER), progesterone receptor (PR) and human
epidermal growth factor receptor (Her2-neu) status] were obtained
from the pathological reports (Table
I). The blood samples (2 ml) were centrifuged twice, first at
1,600 x g for 10 min at 4˚C and then at 16,000 x g for 10 min at
4˚C (each time with the Centrifuge 5417R; Eppendorf). The
supernatant was stored at -80˚C until the next step. The DNA was
extracted using the GeneJETGenomic DNA purification kit (Thermo
Fisher Scientific, Inc.) according to the manufacturer's protocol,
except for a change of the elution buffer volume from 200 to 100
µl. DNA extracts were stored at -80˚C until further use. CcfnDNA
and ccfmtDNA levels were measured via real-time quantitative PCR
using a LightCycler instrument (Roche Diagnostics GmbH). One primer
pair specific for the nDNA (β2M) and another primer pair for the
mDNA (D-loop) were used for quantification (TIB Molbiol). The
primers pairs were selected after reviewing a large number of
scientific articles (15) and their
specificity was analyzed using BLAST (https://blast.ncbi.nlm.nih.gov/; date of accession
April 25th, 2019) and Primer BLAST (https://www.ncbi.nlm.nih.gov/tools/primer-blast/;
date of accession April 25th, 2019). The primer sequences are
listed in Table II. Standard
curves for nDNA and mtDNA were generated using homemade synthetic
target amplicons. The standards were synthesized through DNA
isolation, conventional PCR, PCR product purification, DNA
sequencing, measuring the concentration and finally confirming that
the sample and the standard were amplified with identical
efficiencies. Each sample was assayed in duplicate using FastStart
DNA Master SYBR Green 1 (Roche Diagnostics GmbH) using a different
primer pair each time. qPCR was performed in a total reaction
volume of 20 µl, containing 3 mM MgCl2, 0.5 µM forward
primer and 0.5 µM reverse primer. The following thermocycling
conditions were used: Initial denaturation at 95˚C for 10 min,
followed by 55 cycles of denaturation at 95˚C for 10 sec, annealing
at 60˚C for 10 sec and elongation at 72˚C for 7 sec (18). All of the reactions were followed by
melting curve analysis. The copy numbers per ml of plasma for
ccfnDNA and ccfmtDNA in samples were quantified using the following
equation:
 | Table IParameters of the study cohort. |
Table I
Parameters of the study cohort.
| | Smoking status
(%) | Menopausal status
(%) |
|---|
| Group | N | Age (years) | Smoking | Non-smoker | Premenopause | Postmenopause |
|---|
| Malignant
disease | 33 | 46±12.8 | 30 | 70 | 70 | 30 |
| Benign disease | 26 | 37±13.6 | 19 | 81 | 89 | 11 |
| Healthy control | 25 | 43±13.6 | 28 | 72 | 86 | 32 |
 | Table IIPrimer sequences used for PCR. |
Table II
Primer sequences used for PCR.
| Gene ID | Primer name | Sequence (5'-3') | Amplicon length |
|---|
| β2M | hβ2MF2 |
GCTGGGTAGCTCTAAACAATGTATTCA | 94 |
| | hβ2MR2 |
CCATGTACTAACAAATGTCTAAAATGGT | |
| D-loop | hmitoF5 |
CTTCTGGCCACAGCACTTAAAC | 64 |
| | hmitoR5 |
GCTGGTGTTAGGGTTCTTTGTTTT | |
Qnuclear is the ccfnDNA copy number per milliliter,
C is the ccfnDNA concentration (pg/µl) determined by qPCR targeting
the β2M gene sequence and 3.3 pg is the human haploid genome mass.
Velution is the volume of ccfDNA extract (µl) and
Vplasma is the volume of plasma used for the extraction
(ml).
Qmitochondrial is the ccfmtDNA copy
number per milliliter and C is the ccfmtDNA concentration (pg/µl)
determined by a qPCR targeting the mitochondrial D-loop. Na is
Avogadro's number (6.02x1023 molecules per mole), Ln is
the nucleotide length and MW is the molecular weight of one
nucleotide (g/mol). Velution is the elution volume of
ccfDNA extract (µl) and Vplasma is the volume of plasma
used for the extraction (ml).
Statistical analysis
All statistical analyses were performed using SPSS
statistics 13.0 software (SPSS, Inc.). Statistical differences
between ccfnDNA or ccfmtDNA concentrations and the ccfmtDNA/ccfnDNA
ratio were compared using ANOVA followed by Bonferroni's post-hoc
test. The Kolmogorov-Smirnov test confirmed that the data were
normally distributed. The correlation of the mean values with other
parameters was assessed by calculating Pearson's and Spearman's
correlation coefficients. Student's t-test was used to determine
the association of ccfnDNA, ccfmtDNA or the ccfnDNA/ccfmtDNA ratio
with menopause, smoking and lymph node status, while Pearson's
correlation was used to study the correlation between ccfnDNA,
ccfmtDNA and ccfnDNA/ccfmtDNA levels with age and tumor size.
Furthermore, Spearman's correlation analysis was used to study the
correlation between ccfnDNA, ccfmtDNA and ccfnDNA/ccfmtDNA levels
with the tumor stage or grade, or with the ER, PR and Her2/neu
receptor status. P<0.05 was considered to indicate statistical
significance.
Results
Plasma levels of ccfnDNA, ccfmtDNA and
ccfnDNA/ccfmtDNA between the study groups
The mean ccfnDNA concentration in the breast cancer,
benign tumor and the control group was 6,448, 6,197 and 6,071
copies/ml respectively, while the mean value of the ccfmtDNA
concentration was 20,770,515, 6,572,829 and 5,188,000 copies/ml,
respectively. The mean value of the ccfmtDNA/ccfnDNA ratio was
154,017, 2,155 and 24,192 in the breast cancer, benign tumor and
control group, respectively. When comparing the plasma levels of
ccfnDNA and ccfmtDNA and the ccfmtDNA/ccfnDNA ratio between the
three study groups using ANOVA, no significant differences were
identified between the groups (P=0.979, 0.542 and 0.447,
respectively; P>0.05) data not shown.
Correlation between ccfnDNA, ccfmtDNA
or ccfmtDNA/ccfnDNA levels and demographic features (age, menopause
and smoking status)
The mean age for females with breast cancer, benign
tumor and control subjects was 46 (range, 28-90), 37 (range, 21-75)
and 43 (range, 24-76) years, respectively. When calculating
Pearson's correlation coefficients for ccfnDNA or ccfmtDNA levels
or the ccfmtDNA/ccfnDNA ratio with age, menopause or smoking status
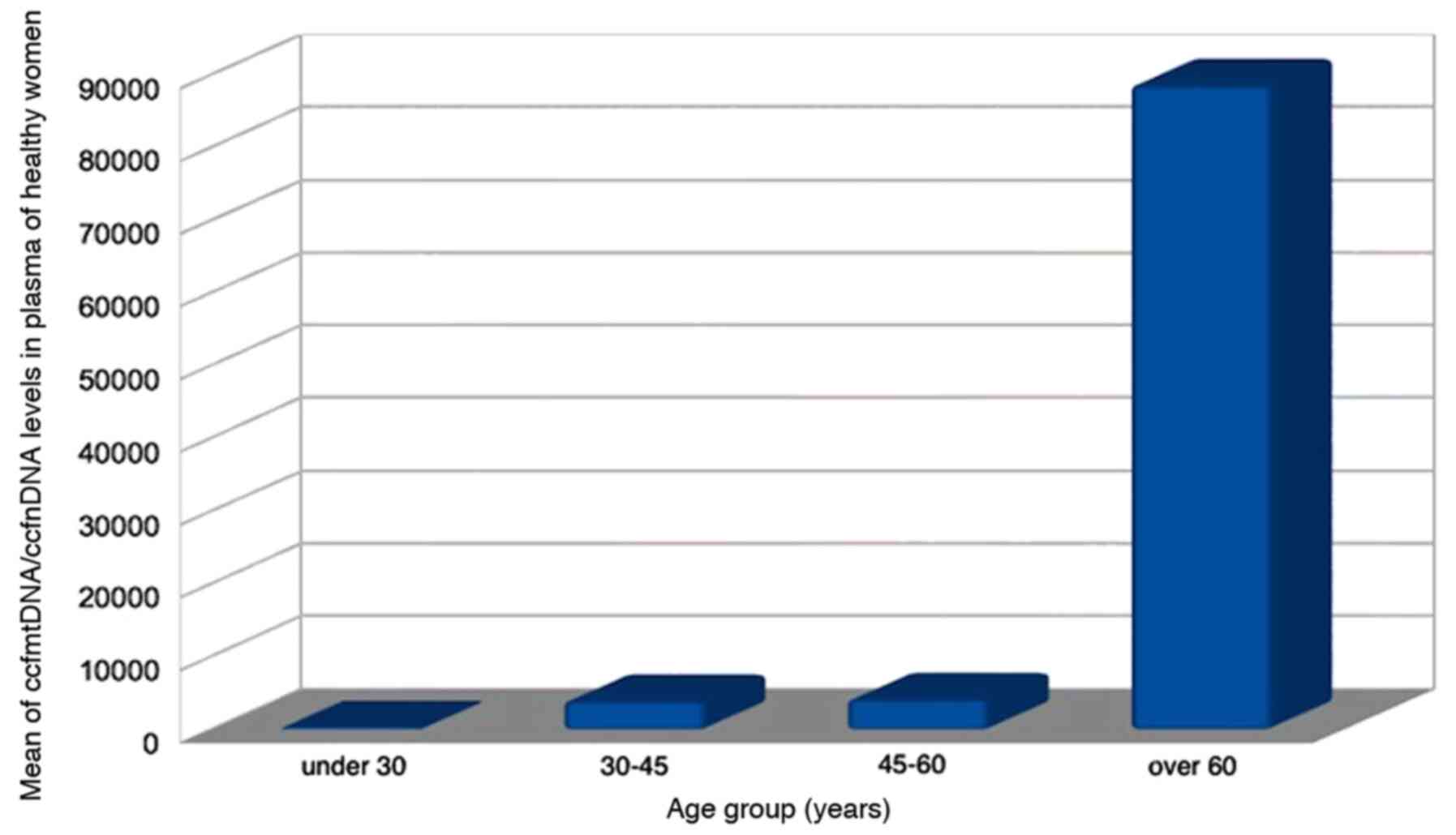
in the three study groups, a positive correlation was only
identified between the ccfmtDNA/ccfnDNA ratio and age in the
control group (P=0.012, r=0.505; Fig.
1). However, in the three study groups, no statistical
significance was obtained for the correlation between the levels of
ccfnDNA or ccfmtDNA or the ccmtDNA/ccfnDNA ratio and demographic
features (data not shown; P>0.05).
Correlation between ccfnDNA, ccfmtDNA
or ccfmtDNA/ccfnDNA levels in breast cancer and the
clinicopathological parameters (stage, grade, lymph node
involvement, tumor size, ER, PR and Her2/neu receptor status)
In the breast cancer group, no statistically
significant correlations between the levels of ccfnDNA or ccfmtDNA
or the ccfnDNA/ccfmtDNA ratio and the stage, grade, lymph node
status or tumor size were obtained (P>0.05). When calculating
the Spearman's correlation coefficient for the correlation between
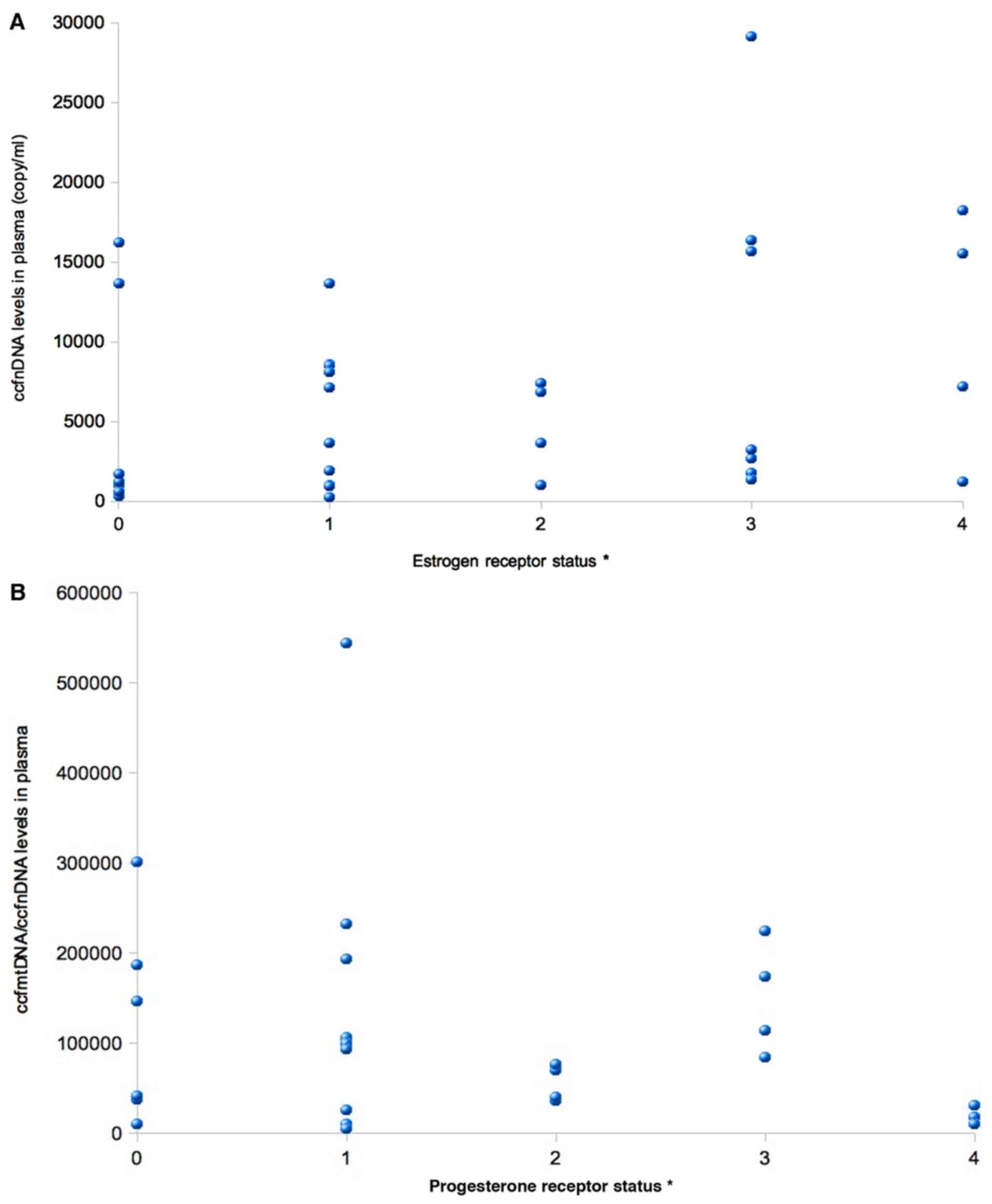
ccfnDNA or ccfmtDNA or the ccfmtDNA/ccfnDNA ratio with the ER, PR
or Her2/neu receptors status, a positive correlation was identified
between ccfnDNA levels and ER status (P=0.045, r=0.416; Fig. 2A), while a negative correlation was
obtained between the ccfmtDNA/ccfnDNA ratio and PR status (P=0.045;
r=-0.478; Fig. 2B and Table III).
 | Table IIICorrelations between ccfnDNA or
ccfmtDNA levels, or the rcfnDNA/ccfmtDNA ratio and
clinicopathological parameters in the malignant disease group. |
Table III
Correlations between ccfnDNA or
ccfmtDNA levels, or the rcfnDNA/ccfmtDNA ratio and
clinicopathological parameters in the malignant disease group.
| Parameter | Patients, n (%) | ccfmtDNA
(copies/ml) | P-value, r-value | ccfnDNA
(copies/ml) | P-value, r-value | ccfmtDNA/ccfnDNA | P-value, r-value |
|---|
| Stagea | | | 0.886, 0.032 | | 0.552, -0.139 | | 0.641, 0.112 |
|
I | 8 (24.2) | 6,772,500
(1,360,000-19,500,000) | | 5,790.3
(2.51-16,200) | | 5,704,798
(9,690-39,840,637) | |
|
II | 13 (39.4) | 2,647,076.9
(652,000-6,240,000) | | 8,767.3
(17-29,400) | | 8,427
(4,170-23,141) | |
|
III | 12 (36.4) | 49,736,250
(945,000-549,000,000) | | 4,374.25
(38.6-13,600) | | 343,351
(3,610-300,000) | |
| Gradea | | | 0.254, 0.246 | | 0.282, 0.198 | | 0.626, -0.139 |
|
I | 5 (15.2) | 2,914,000
(1,360,000-5,590,000) | | 5,950
(2.51-16,200) | | 8,501
(3,489-2,237.2) | |
|
II | 19 (57.6) | 33,083,000
(652,000-549,000,000) | | 5,265.7
(17-15,600) | | 2,689,931
(4,170-39,840,637) | |
|
III | 9 (27.3) | 4,697,777.7
(1,790,000-9,040,000) | | 9,221.1
(38.6-29,400) | | 6,496
(3,610-1,134.8) | |
| Tumor size
(cm)b | | | 0.659, 0.148 | | 0.839, -0.051 | | 0.296, -0.176 |
|
<2 | 6 (18.2) | 10,800,000
(1,570,000-19,500,000) | | 1,800
(2.51-16,200) | | 52,521
(1,478-39,840,637) | |
|
≥2-
<5 | 16 (48.5) | 2,550,000
(652,000-9,040,000) | | 4,510
(17-29,400) | | 4,105
(4,170-300,000) | |
|
≥5 | 11 (33.3) | 2,640,000
(652,000-549,000,000) | | 2,690
(38.6-13,600) | | 956
(3,610-300,000) | |
| Lymph node
involvement | | | 0.512
t(31)=-0.748 | | 0.785
t(31)=0.490 | | 0.365
t(26)=1.332 |
|
No | 12 (36.4) | 5,354,333.3
(652,000-19,500,000) | | 6,527.7
(2.51-16,200) | | 39,952
(4,170-39,840,637) | |
|
Yes | 21 (63.6) | 29,579,761.9
(945,000-549,000,000) | | 6,402.6
(17-29,400) | | 17,623
(3610-300,000) | |
| Estrogen receptor
statusa | | | 0.702, 0.136 | | 0.045, 0.416 | | 0.239, -0.311 |
|
Negative | 8 (24.2) | 2,390,000
(652,000-10,000,000) | | 7,410
(2.51-15,600) | | 39,276
(4,170-398,406,375) | |
|
Positive | | | | | | | |
|
+ | 11 (33.3) | 3,070,000
(945,000-549,000,000) | | 1,930
(38.6-13,100) | | 18,661
(1,478-30,000,000) | |
|
++ | 5 (15.2) | 3,010,000
(1,410,000-19,500,000) | | 3,590
(27.3-29,400) | | 543,175
(1,023-16,400,000) | |
|
+++ | 9 (21.3) | 3,030,000
(1,490,000-8,360,000) | | 4,900
(1,770-15,600) | | 69,197
(9,550-223,729) | |
| Progesterone
receptor statusa | | | 0.297, -0.179 | | 0.119, 0.334 | | 0.045, -0.78 |
|
Negative | 10 (30.3) | 2,550,000
(1,500,000-549,000,000) | | 1,210
(2.51-15,500) | | 39,276
(4,170-398,406,375) | |
|
Positive | | | | | | | |
|
+ | 10 (30.3) | 3,720,000
(652,000-19,500,000) | | 6,030
(38.6-29,400) | | 91,335
(955-543,175) | |
|
++ | 7 (21.1) | 1,870,000
(1,490,000-3,190,000) | | 5,180
(27.3-16,200) | | 36,100
(1,478-816,061) | |
|
+++ | 6 (18.2) | 4,740,000
(1,360,000-7,440,000) | | 3,800
(1,770-8,040) | | 103,010
(30,909-223,729) | |
| Her2-neu receptor
statusa | | | 0.594, -0.122 | | 0.289, -0.228 | | 0.985, 0.005 |
|
Negative | 24 (72.7) | 3,050,000
(945,000-549,000,000) | | 4,000
(2.51-16,200) | | 84,141
(4,170-398,406,375) | |
|
Positive | | | | | | | |
|
+ | 2(6) | 3,450,000
(1,790,000-5,100,000) | | 10,400
(7,100-13,600) | | 31,356
(25,211-37,500) | |
|
++ | 2(6) | 2,540,000
(652,000-4,420,000) | | 8,760
(1,910-15,600) | | 117,797
(4,179-231,414) | |
|
+++ | 5 (15.3) | 2,090,000
(1,500,000-1,1000,000) | | 1,210
(17-29,400) | | 83,969
(1,032-146,281) | |
Discussion
For breast cancer in general, but particularly in
the Middle East, there remains a lack of research in the field of
ccfDNA, and it is required to further investigate its role. In the
present study, PCR with SYBR-Green I was used for the
quantification and numerous previous studies were reviewed to
ensure that the most specific primers for both nDNA and mDNA were
used, and their specificity was tested prior to using them. Each
PCR was also followed up by melting curve analysis to ensure the
accuracy of the results. Neither the concentrations of ccfnDNA and
ccfmtDNA, nor the ccfmtDNA/ccfnDNA ratio were significantly
different between the three study groups, which differs from the
results of other previous studies (15-17).
This difference may be explained through ethnic differences,
limited sample sizes and differences in the methods used for
quantification. Furthermore, the results of the present study
indicated no correlations between ccfnDNA, ccfmtDNA or
ccfmtDNA/ccfnDNA levels and demographic features or
clinicopathological parameters, which is consistent with the
results of other previous studies, except that the present study
reported a positive correlation between the ccfmtDNA/ccfnDNA ratio
and age in the healthy control group. This result may be due to
increased levels of ccfmDNA, decreased levels of ccfnDNA or the
impaired interaction between mtDNA and nDNA. In addition, it may be
explained through the increased levels of reactive oxygen species
with aging, which is directly responsible for damaging cells and
releasing their nDNA and mtDNA into the blood stream (19) or increasing the mutation rate for
both nDNA and mtDNA, but with much higher rates for mtDNA (20,21).
On the other hand, due to the complex relationship between mtDNA
and nDNA, mutations in mtDNA may affect the way mtDNA and nDNA
communicate with each other (22).
The present study also revealed a positive correlation between
ccfnDNA levels and ER status, which may be linked to the role of ER
signaling in downregulating the DNA damage and repair pathway in
mammary tissues (23). In healthy
tissues, ER activity was discovered to be governed by highly
regulated genetics that keep ER activity under control (24). Furthermore, dysregulated ER signals
were indicated to lead to increased proliferation, the accumulation
of DNA damage and eventually tumorigenesis (23,25).
This increase in the rate of proliferation was reported to lead to
increased levels of ccfnDNA in the plasma of patients with breast
cancer (17). On the other hand, a
negative correlation was identified between the ccfmtDNA/ccfnDNA
ratio and the PR status. Similar to ER-positive cells, PR-positive
cells demonstrate loss of or alterations in the DNA damage and
repair pathway or checkpoint controlling system, and an increased
proliferation rate, eventually leading to cancer formation
(26). Of note, all previous
research in this field has reported positive trends in these
particular parameters, which is promising (15-17);
however, the conflicting results should be followed up with
large-scale studies and a mutation analysis to clarify the
relationship between ccfmtDNA, ccfnDNA and breast cancer.
In conclusion, a positive correlation was identified
between ccfmtDNA/ccfnDNA levels and age in healthy individuals. In
addition, a positive correlation was observed between ccfnDNA and
the ER status, while a negative correlation was observed between
ccfmtDNA/ccfnDNA levels and the PR status. Despite the small sample
size, these results suggested that the plasma levels of ccfmtDNA
and ccfnDNA may serve as a non-invasive biomarkers; however,
large-scale studies are required to validate the results and
clarify the conflicting results.
Acknowledgements
The authors would like to thank Professor Dr Fawza
Monem and Dr Wafa Habbal from the Laboratories Department for
facilitating the work in the laboratories of Al-Asaad Hospital
(Damascus, Syria).
Funding
The present study was funded by the Scientific
Research Office, Damascus University.
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
MS designed the study and performed the molecular
biology studies. MS and ARN analyzed the data; MS drafted the
manuscript and ARN revised it. All authors read and approved the
final manuscript.
Ethics approval and consent to
participate
Informed consent forms were signed by all
participants and the study was approved by the Syrian Higher
Commission for Scientific Research, Ministry of Higher Education,
Damascus, Syria.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Luffarelli P, Manna E and Fortunato L:
Epidemiology and risk factors. In: Ductal carcinoma in situ of the
breast. Mariotti C (ed). Springer, Cham, pp23-28, 2017.
|
|
2
|
Chouchane L, Boussen H and Sastry KS:
Breast cancer in Arab populations: Molecular characteristics and
disease management implications. Lancet Oncol. 14:e417–e424.
2013.PubMed/NCBI View Article : Google Scholar
|
|
3
|
El Saghir NS and Abulkhair O: Epidemiology
prevention and management guidelines for breast cancer in Arab
countries. Pan Arab J Oncol. 3:12–18. 2010.
|
|
4
|
World Health Organization: Estimated
Cancer Incidence, Mortality and Prevelence Worldwide in 2012.
Available from urihttp://globocan.iarc.fr/Pages/fact_sheets_population.aspx?country=818simplehttp://globocan.iarc.fr/Pages/fact_sheets_population.aspx?country=818.
Accessed 30 October, 2017.
|
|
5
|
Marmot MG, Altman DG, Cameron DA, Dewar
JA, Thompson SG and Wilcox M: The benefits and harms of breast
cancer screening: An independent review. Br J Cancer.
108:2205–2240. 2013.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Freimanis RI and Yacobozzi M: Breast
cancer screening. NC Med J. 75:117–120. 2014.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Alix-Panabières C, Schwarzenbach H and
Pantel K: Circulating tumor cells and circulating tumor DNA. Annu
Rev Med. 63:199–215. 2012.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Elshimali YI, Khaddour H, Sarkissyan M, Wu
Y and Vadgama JV: The clinical utilization of circulating cell free
DNA (CCFDNA) in blood of cancer patients. Int J Mol Sci.
14:18925–18958. 2013.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Vymetalkova V, Cervena K, Bartu L and
Vodicka P: Circulating cell-free DNA and colorectal cancer: A
systematic review. Int J Mol Sci. 19(3356)2018.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Yu M, Stott S, Toner M, Maheswaran S and
Haber DA: Circulating tumor cells: Approaches to isolation and
characterization. J Cell Biol. 192:373–382. 2011.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Zhou Q, Li W, Leng B, Zheng W, He Z, Zuo M
and Chen A: Circulating cell free DNA as the diagnostic marker for
ovarian cancer: A systematic review and meta-analysis. PLoS One.
11(e0155495)2016.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Rykova EY, Morozkin ES, Ponomaryova AA,
Loseva EM, Zaporozhchenko IA, Cherdyntseva NV, Vlassov VV and
Laktionov PP: Cell-free and cell-bound circulating nucleic acid
complexes: Mechanisms of generation, concentration and content.
Expert Opin Biol Ther. 12 (Suppl 1):S141–S153. 2012.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Ignatiadis M, Lee M and Jeffrey SS:
Circulating tumor cells and circulating tumor DNA: Challenges and
opportunities on the path to clinical utility. Clin Cancer Res.
21:4786–4800. 2015.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Schwarzenbach H, Hoon DS and Pantel K:
Cell-free nucleic acids as biomarkers in cancer patients. Nat Rev
Cancer. 11:426–437. 2011.PubMed/NCBI View
Article : Google Scholar
|
|
15
|
Malik AN, Shahni R, Rodriguez-de-Ledesma
A, Laftah A and Cunningham P: Mitochondrial DNA as a non-invasive
biomarker: Accurate quantification using real time quantitative PCR
without co-amplification of pseudogenes and dilution bias. Biochem
Biophys Res Commun. 412:1–7. 2011.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Kohler C, Radpour R, Barekati Z,
Asadollahi R, Bitzer J, Wight E, Bürki N, Diesch C, Holzgreve W and
Zhong XY: Levels of plasma circulating cell free nuclear and
mitochondrial DNA as potential biomarkers for breast tumors. Mol
Cancer. 8(105)2009.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Meddeb R, Dache ZAA, Thezenas S, Otandault
A, Tanos R, Pastor B, Sanchez C, Azzi J, Tousch G, Azan S, et al:
Quantifying circulating cell-free DNA in humans. Sci Rep.
9(5220)2019.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Springer J, Loeffler J, Heinz W,
Schlossnagel H, Lehmann M, Morton O, Rogers TR, Schmitt C, Frosch
M, Einsele H and Kurzai O: Pathogen-specific DNA enrichment does
not increase sensitivity of PCR for diagnosis of invasive
aspergillosis in neutropenic patients. J Clin Microbiol.
49:1267–1273. 2011.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Sena LA and Chandel NS: Physiological
roles of mitochondrial reactive oxygen species. Mol Cell.
48:158–167. 2012.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Desler C, Marcker ML, Singh KK and
Rasmussen LJ: The importance of mitochondrial DNA in aging and
cancer. J Aging Res. 2011(407536)2011.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Veitia R, Govindaraju D, Bottani S and
Birchler J: Aging: Somatic mutations, epigenetic drift and gene
dosage imbalance. Trends Cell Biol. 27:299–310. 2017.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Doynova MD, Berretta A, Jones MB, Jasoni
CL, Vickers MH and O'Sullivan JM: Interactions between
mitochondrial and nuclear DNA in mammalian cells are non-random.
Mitochondrion. 30:187–196. 2016.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Matta J, Morales L, Ortiz C, Adams D,
Vargas W, Casbas P, Dutil J, Echenique M and Suárez E: Estrogen
receptor expression is associated with DNA repair capacity in
breast cancer. PLoS One. 11(e0152422)2016.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Caldon CE: Estrogen signaling and the DNA
damage response in hormone dependent breast cancers. Front Oncol.
4(106)2014.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Finlay-Schultz J, Gillen AE, Brechbuhl HM,
Ivie JJ, Matthews SB, Jacobsen BM, Bentley DL, Kabos P and
Sartorius CA: Breast cancer suppression by progesterone receptors
is mediated by their modulation of estrogen receptors and RNA
polymerase III. Cancer Res. 77:4934–4946. 2017.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Liou GY and Storz P: Reactive oxygen
species in cancer. Free Radic Res. 44:479–496. 2010.PubMed/NCBI View Article : Google Scholar
|