Introduction
Renal cell carcinoma (RCC) accounts for 5% of all
adult malignancies with 300,000 new cases and 100,000 deaths per
year worldwide (1). Clear cell RCC
(ccRCC) is the most common subtype of renal cancer, which accounts
for ~75% of RCC worldwide (2).
ccRCC is resistant to chemotherapy and radiotherapy, therefore its
clinical management has become a difficult issue (3). Although the diagnosis, treatment and
overall survival rate of patients with ccRCC have been markedly
improved in recent years, long-term prognosis is still limited and
~30% of patients experience recurrence of renal cancer (4,5).
Therefore, further studies are required to clarify the underlying
mechanism of ccRCC progression and develop efficient therapeutic
targets for ccRCC.
Long non-coding RNAs (lncRNAs) are non-coding RNAs
with >200 nucleotides, and have been indicated to exert critical
roles in various cellular and biological processes (6). Accumulating evidence has suggested
that lncRNAs function as tumor suppressors or oncogenes in diverse
tumors, including ccRCC. Liu et al (7) demonstrated that knockdown of lncRNA
TP73-AS1 inhibited cell proliferation and invasion, and induced
cell apoptosis by binding to KISS1 in ccRCC. Hirata et al
(8) reported that lncRNA MALAT1
served as an oncogene by promoting proliferation and suppressing
apoptosis in ccRCC. Lung cancer-associated transcript 1 (LUCAT1)
has also been demonstrated to serve a vital role in ccRCC
progression. For instance, a previous study has reported that
LUCAT1 served as a poor prognostic factor in human ccRCC (9). Moreover, it has been demonstrated that
LUCAT1 overexpression induced cell proliferation and invasion via
the AKT/GSK-3β signaling pathway, indicating that LUCAT1 may be
used as a potential therapeutic target in ccRCC (10). Additionally, LUCAT1 has been
revealed to function as an oncogene via regulating the microRNA
(miRNA/miR)-495-3p/SATB1 axis, thereby promoting cell proliferation
and invasion in ccRCC (11).
However, the molecular mechanism underlying the biological function
of LUCAT1 in ccRCC remains to be fully elucidated.
miRNAs are non-coding RNAs with 19-22 nucleotides in
length, which serve as important regulators of gene expression via
binding to the 3'-untranslated region (3'-UTR) of target mRNAs
(12). Accumulating evidence has
indicated that abnormal expression of miRNAs, functioning as
oncogenes or tumor suppressors, was associated with the progression
and development of cancer, including ccRCC (13). Several studies have demonstrated
that miR-375 was downregulated in several types of cancer, such as
gastric cancer (14), glioma
(15) and cervical cancer (16). In renal cancer, miR-375 has been
reported to act as a tumor suppressor via inhibiting cell
proliferation, migration and invasion (17). Therefore, the aforementioned studies
have indicated the pivotal role of miR-375 in different types of
cancer, especially in ccRCC, while few studies have examined the
network of molecules upstream and downstream of miR-375 in
ccRCC.
Yes-associated protein 1 (YAP1), which is a
transcriptional co-activator, has also been associated with the
development of ccRCC. YAP1 has been demonstrated to serve an
oncogenic role in various types of cancer, including ccRCC
(18). Upregulation of YAP1 has
been revealed to accelerate cell growth, migration, invasion and
epithelial-mesenchymal transition via modulating arrestin
domain-containing protein 1/3(19),
suggesting that YAP1 may participate in ccRCC development.
The present study aimed to investigate the role and
underlying mechanism of LUCAT1 in ccRCC in vitro and in
vivo.
Materials and methods
Clinical specimens and cell
culture
ccRCC tissues (n=62) and paired adjacent healthy
tissues (n=62) were obtained from patients with ccRCC who were
hospitalized at Chenzhou No. 1 People's Hospital (Chenzhou, China)
between May 2016 and March 2018. The patient age range was 34-79
years, with a median age of 65 years, and 29 patients were females
and 33 were males. The inclusion criteria included a clinical and
imaging diagnosis of primary non-metastatic ccRCC. The exclusion
criteria included patients with active malignancy other than RCC,
cardiovascular disease, blood disease or other malignant tumor
types. The present study was approved by the Ethics Committee of
Chenzhou No. 1 People's Hospital and all participants provided
written informed consent.
A normal human proximal tubular epithelial cell line
(HK-2) and RCC cell lines (786-O, Caki-1, A498, 769-P and ACHN)
were obtained from American Type Culture Collection. All RCC cell
lines were cultured in RPMI-1640 (Invitrogen; Thermo Fisher
Scientific, Inc.) supplemented with 10% FBS (Gibco; Thermo Fisher
Scientific, Inc.) and 1% penicillin/streptomycin (5,000 U/ml,
Gibco; Thermo Fisher Scientific, Inc.). HK-2 cells were cultured in
DMEM supplemented with 10% FBS (both from Gibco; Thermo Fisher
Scientific, Inc.) and 1% penicillin/streptomycin. 293T cells were
purchased from The Cell Bank of Type Culture Collection of the
Chinese Academy of Sciences. 293T cells were cultured in DMEM
supplemented with 10% FBS and 1% penicillin/streptomycin. The cells
were maintained at 37˚C in a humidified incubator with 5%
CO2.
Cell transfection
LUCAT1 overexpression vector was constructed by
cloning full-length LUCAT1 into pcDNA3.1 (Invitrogen; Thermo Fisher
Scientific, Inc.), and the empty vector pcDNA3.1 was used as the
negative control. Small interfering (si)-RNAs targeting LUCAT1
(si-LUCAT1#1 and #2; final concentration, 20 nM) and its scrambled
negative control (NC) siRNA (si-NC; final concentration, 20 nM),
miR-375 inhibitor (final concentration, 30 nM) and control (miR-NC;
final concentration, 30 nM) and miR-375 mimics (final
concentration, 15 nM) and control (miR-NC mimics; final
concentration, 15 nM) were obtained from Shanghai GenePharma Co.,
Ltd. 1x106 Caki-1 and A498 cells were plated into
six-well plates for 12 h, then the constructs were transfected into
Caki-1 and A498 cells by mixing with Lipofectamine® 2000
reagent (Invitrogen; Thermo Fisher Scientific, Inc.) at 37˚C for 15
min following the manufacturer's instructions. A total of 24-72 h
after transfection, the transfected cells were collected for
subsequent experiments. The sequences of the transfected molecules
were as follows: si-LUCAT1#1, 5'-CCCAUCAGAAGAUGUCAGAAGAUAA-3';
si-LUCAT1#2, 5'-UGUCCCUCAGUGUUCUACUUCUUAA-3'; si-NC,
5'-AAUUCUCCGAACGUGUCACGU-3'; miR-375 inhibitor,
5'-UCACGCGAGCCGAACGAACAAA-3'; anti-miR-NC,
5'-CAGUACUUUUGUGUAGUACAAA-3'; miR-375 mimic,
5'-UUUGUUCGUUCGGCUCGCGUGA-3'; miR-NC,
5'-GGUUCGUACGUACACUGUUCA-3'.
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA was extracted from cells (786-O, Caki-1,
A498, 769-P, ACHN and HK-2) and ccRCC tissues using
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc.) and cDNA synthesis was performed using Maxima Probe qPCR
Master mix (Fermentas; Thermo Fisher Scientific, Inc.) according to
the manufacturer's instructions. Quantitative analysis of LUCAT1
and YAP1 expression was performed using SYBR® Premix Ex
Taq™ reagent (Takara Biotechnology Co., Ltd.) and GAPDH was used as
the endogenous control. Thermocycling conditions were as follows:
94˚C for 3 min; 40 cycles of 94˚C for 30 sec, 55˚C for 30 sec and
72˚C for 1 min; and 72˚C for 10 min. The expression level of
miR-375 was analyzed using SYBR PrimeScript™ miRNA RT-PCR kit
(Takara Biotechnology Co., Ltd.) and U6 small nuclear RNA was used
as the internal reference. Thermocycling conditions were as
follows: Denaturation at 95˚C for 10 sec; 40 cycles of 95˚C for 5
sec and 60˚C for 20 sec; followed by dissociation curve analysis at
95˚C for 60 sec, 55˚C for 30 sec and 95˚C for 30 sec. Relative
expression levels were calculated using the 2-ΔΔCq
method (20). Primer sequences were
listed as follows: LUCAT1 forward, 5'-GTGTCAAGCTCGGATTGCCT-3' and
reverse, 5'-GAGCCCACACACTCAG-3'; GAPDH forward,
5'-AGAAGGCTGGGGCTCATTTG-3' and reverse, 5'-AGGGGCCATCCACAGTCTTC-3';
YAP1 forward, 5'-TGACCCTCGTTTTGCCATGA-3' and reverse,
5'-GTTGCTGCTGGTTGGAGTTG-3'; miR-375 forward, 5'-GTGCAGGGTCCGAGGT-3'
and reverse, 5'-AGCCGTTTGTTCGTTCGGCT-3'; U6 forward,
5'-CTCGCTTCGGCAGCACA-3' and reverse,
5'-AACGCTTCACGAATTTGCGT-3'.
Pathological analysis
In brief, the ccRCC tissues and paired adjacent
healthy tissues were processed according to standard procedures,
including 10% formalin fixation for 5 min at room temperature,
dehydration in graded alcohol series (30, 50, 70, 95 and twice 100%
alcohol) and xylene and embedding in paraffin. Subsequently, the
tissue biopsies (3 µm) were stained with hematoxylin for 5 min and
eosin for another 2 min at room temperature (Abcam). Finally, the
tissues were observed under a light microscope at x200
magnification and photographed.
Dual-luciferase reporter assay
The binding sites between miR-375 and LUCAT1, as
well as miR-375 and YAP were predicted by lncBase Predicted v.2
(http://www.microrna.gr/lncBase) or
TargetScan software (http://www.targetscan.org/vert_50/), respectively.
LUCAT1 and YAP1 3'-UTR containing the wild-type (WT) or mutant
(MUT) binding sites of miR-375 was synthesized and cloned into
pmirGLO luciferase reporter vector (Promega Corporation).
Subsequently, 1x106 293T cells were plated into six-well
plates for 12 h, then LUCAT1-WT, LUCAT1-MUT, YAP1-WT or YAP1-MUT
were co-transfected with miR-375 mimics or miR-NC into 293T cells
using Lipofectamine 2000 reagent (Invitrogen; Thermo Fisher
Scientific, Inc.). A total of 48 h following transfection, the
Renilla and firefly luciferase activity was measured using
Luc-Pair™ Duo-Luciferase Assay kit (Shanghai Yeasen Biotechnology
Co., Ltd.) according to the manufacturer's instructions.
Renilla luciferase activity was used for normalization.
Western blotting
Total protein was extracted from Caki-1 and A498
cells using a Cell lysis buffer for Western and IP (Beyotime
Institute of Biotechnology) following the manufacturer's protocol.
The protein samples were quantified using a BCA Protein Assay Kit
(Beyotime Institute of Biotechnology). A total of 30 µg
protein/sample were separated by 10% SDS-PAGE, and subsequently
transferred onto a nitrocellulose membrane (MilliporeSigma). The
membrane was blocked with 5% skimmed milk in TBS-Tween-20 (0.1%)
buffer for 1 h at room temperature. Subsequently, the membrane was
incubated with primary antibodies against YAP1 (1:600; cat. no.
ab52771) and GAPDH (1:4,000; cat. no. ab8245) (both from Abcam)
overnight at 4˚C. Subsequently, the membranes were incubated with
HRP-conjugated goat anti-rabbit IgG H&L secondary antibody
(1:20,000; cat. no. ab6721; Abcam) for 1 h at room temperature.
Finally, the protein bands were visualized with an ECL kit (GE
Healthcare) and analyzed using ImageJ v1.8.0 software (National
Institutes of Health).
Cell viability assay
A total of 2x103 Caki-1 and A498 cells
were seeded in a 96-well plate. After incubation for 24, 48 and 72
h, 10 µl CCK-8 solution (Beyotime Institute of Biotechnology) was
added to each well and incubated at 37˚C for 4 h according to the
manufacturer's protocol. Subsequently, the absorbance at 450 nm was
determined using an Elx800 Absorbance Microplate Reader (BioTek
Instruments, Inc.).
Cell migration and invasion assay
Transwell chambers (12-well; MilliporeSigma) were
used to assess the migratory and invasive abilities of Caki-1 and
A498 cells. In brief, 1x105 cells were seeded into the
upper chamber for the migration assay, while 2x105 cells
were plated into the upper chamber, which was pre-coated with 50 µl
Matrigel at 37˚C for 30 min (BD Biosciences), for the invasion
assay. The cells were seeded in the upper chambers in serum-free
RPMI-1640 medium, while RPMI-1640 supplemented with 10% FBS was
placed into the lower chamber. Following incubation for 24 h at
37˚C, the migrated/invaded cells were fixed with 100% methanol for
30 min and stained with 0.05% crystal violet for 20 min at room
temperature. Finally, the cells were photographed using an inverted
microscope at x400 magnification, and the average number of
migrated or invaded cells in five random microscopic fields per
Transwell was manually counted.
Tumor xenografts
The present study was approved by the Animal Ethics
Committee of Chenzhou No. 1 People's Hospital (Chenzhou, China).
Lentiviral-based short hairpin (sh)-LUCAT1 vector and lentiviral
empty vector (sh-NC) were obtained from Shanghai GeneChem Co., Ltd.
The sequences of sh-LUCAT1 or sh-NC were cloned and inserted into
hU6-MCS-Ubiquitin-EGFP-IRES-puromycin lentiviral vector (Shanghai
GeneChem Co., Ltd.) to construct the recombinant lentiviral vector.
5x106 293T cells at the exponential growth phase were
plated into 100 mm cell culture plates for 12 h, then the
recombinant lentiviral vectors (16 µg) and 20 µl Lenti-Easy
Packaging Mix (Shanghai GeneChem Co., Ltd.) were co-transfected
into 293T cells using Lipofectamine 2000 for 48 h at 37˚C. A total
of 48 h after transfection, the cell supernatants were collected
and filtered using a 0.22 µM polyethersulfone membrane filter. The
supernatants were then centrifuged at 8,000 x g for 10 min at 4˚C
to collect the lentiviral particles. Subsequently, the titers were
determined by fluorescent microscopy. The titer was approximately
4-10x108 infectious units/ml. To establish the stable
LUCAT1 knockdown cell line, 1x106 Caki-1 cells were
cultured in a six-well plate with culture media containing
lentivirus at 50 MOI for 72 h. A total of 3 days after infection,
the cells were incubated with 2 µg/ml puromycin for 2 weeks to
select the stably transfected cells.
Five-week-old male BALB/c nude mice with an average
body weight of ~20 g were provided by the Animal Center of Nanjing
University (Nanjing, China), and were divided into two groups (6
mice in total; n=3 per group). All introduced mice were
bio-purified and were entered into the animal facility after
veterinary authorization. The mice were housed in a pathogen-free
environment at 25˚C, 45-50% humidity and a programmed 12 h light/12
h dark cycle. All mice were allowed free access to drinking water
and a sterilized standard diet. Any effort was made to avoid
unnecessary pain of the animals. Stably transfected Caki-1 cells
(3x106 cells in 100 µl PBS) were subcutaneously injected
into the left flank of the nude mice. The volume of the tumors was
measured every 6 days after 6 days of injection. A total of 30 days
after injection, the mice were sacrificed by cervical dislocation
after deep anesthesia with 2% isoflurane (Baxter International
Inc.), and the tumors were excised and weighed. The level of LUCAT1
in the excised tumors was detected by RT-qPCR. Additionally, the
humane endpoint criteria used in the present study were as follows:
During the post-operative period, the mice were inspected daily to
ensure their ability to eat, drink and ambulate normally; when they
exhibited apparent physical defects, such as dry or dull eyes,
sticky mucous membranes, ambulation difficulties, dyspnea or
cachexia (loss of 15% of animal original body weight), the mice
were deemed to meet the criteria of euthanasia and were sacrificed
to minimize suffering and distress.
Statistical analysis
Statistical analysis was performed using SPSS v20.0
software (IBM Corp.). All experiments were analyzed in triplicate
and are presented as the mean ± SD. Pearson's correlation was used
to assess the correlation between LUCAT1 and miR-375, as well as
miR-375 and YAP1. Comparisons between two groups were performed
using paired or unpaired Student's t-test, while comparisons among
≥3 groups were performed using one-way ANOVA followed by Tukey's
post hoc test. Pearson's χ2 test or Fisher's exact test
were used to assess categorical variables. P<0.05 was considered
to indicate a statistically significant difference.
Results
LUCAT1 and YAP1 expression levels are
increased in ccRCC tissues and cells
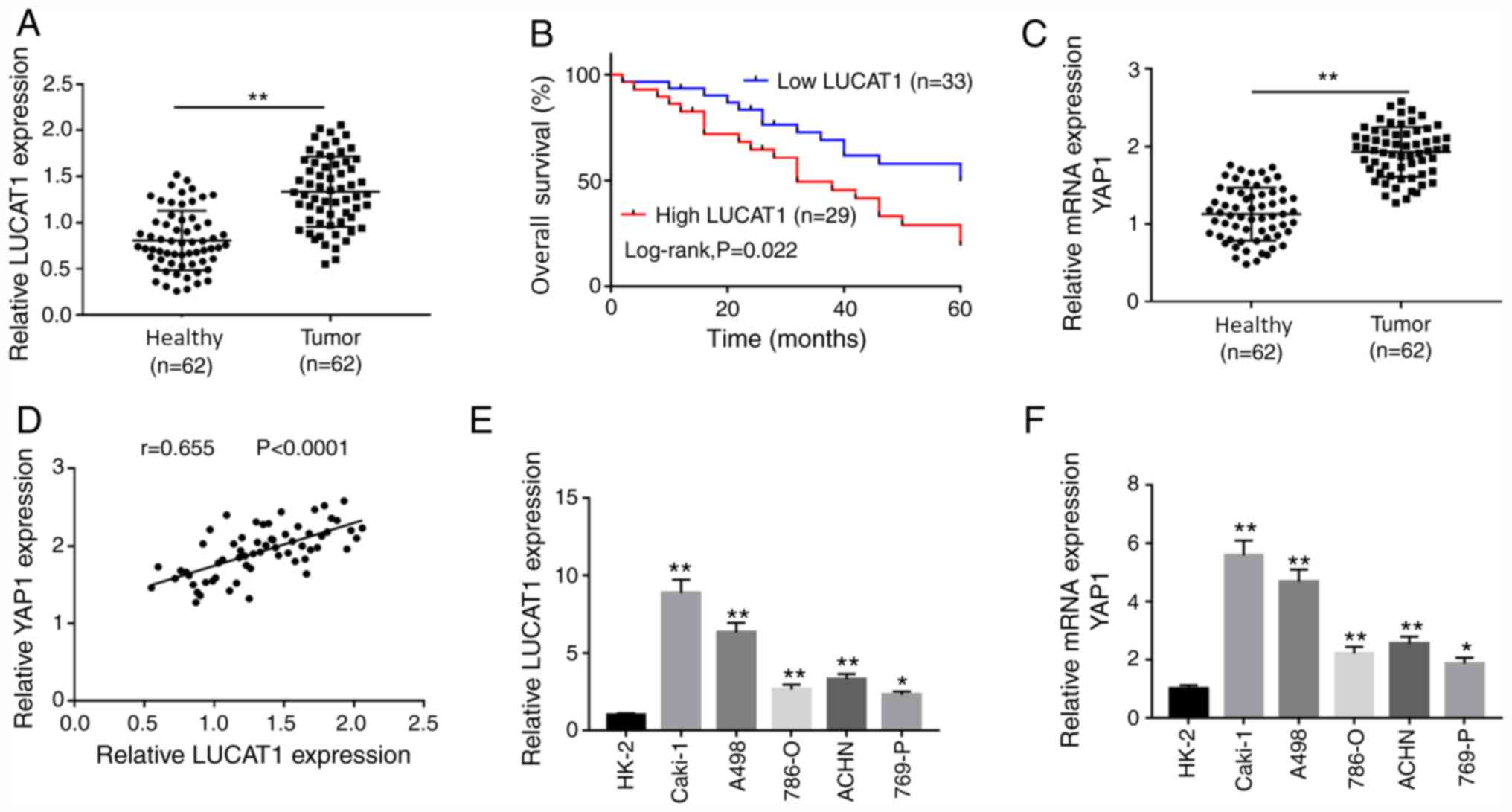
LUCAT1 and YAP1 mRNA expression levels were
significantly increased in ccRCC tissues, compared with the
adjacent healthy tissues (Fig. 1A
and C). Similarly, LUCAT1 and YAP1
mRNA expression levels were upregulated in ccRCC cell lines (786-O,
Caki-1, A498, 769-P and ACHN) compared with the human normal
proximal tubular epithelial cell line HK-2 (Fig. 1E and F). Pearson's correlation analysis
demonstrated that the expression of LUCAT1 was positively
correlated to that of YAP1 in ccRCC tissues (Fig. 1D). To explore whether the expression
of LUCAT1 was associated with prognosis, patients with ccRCC were
divided in low (n=33) and high LUCAT1 group (n=29) according to the
median expression of LUCAT1. The results revealed that the low
LUCAT1 group exhibited a higher overall survival rate compared with
the high LUCAT1 group (Fig. 1B),
suggesting a close association between LUCAT1 expression level and
prognosis. To further assess the role of LUCAT1 in the prognosis of
ccRCC, patients were divided into two groups (high and low
expression) based on the median expression levels of LUCAT1, and
the association between LUCAT1 expression and clinicopathological
factors of patients with ccRCC was analyzed. The
clinicopathological characteristics of all 62 patients are
presented in Table I. Statistical
analysis indicated that LUCAT1 expression was associated with the
tumor stage, tumor size and lymph node metastasis of ccRCC. In
addition, a significant association was observed between YAP1
expression level (low and high YAP1 expression level was divided
based on the mean value of YAP1 expression level) and tumor stage,
tumor size or lymph node metastasis in patients with ccRCC
(Table SI). Furthermore, H&E
staining of the adjacent healthy kidney group and ccRCC tissues is
presented in Fig. S1. Compared
with the adjacent healthy kidney group, the tumors displayed dense
cellularity with brightly eosinophilic cytoplasm, and were arranged
in nests. These results indicated that LUCAT1 and YAP1 were
associated with ccRCC progression.
 | Table IAssociation of clinicopathological
features with LUCAT1 expression in patients with clear cell renal
cell carcinoma. |
Table I
Association of clinicopathological
features with LUCAT1 expression in patients with clear cell renal
cell carcinoma.
| | LUCAT1
expression | |
|---|
| Parameter | Number of
cases | Low (n=33) | High (n=29) |
P-valuea |
|---|
| Sex | | | | |
|
Female | 30 | 16 | 14 | 0.124 |
|
Male | 32 | 17 | 15 | |
| Age, years | | | | |
|
≤55 | 27 | 15 | 12 | 0.074 |
|
>55 | 35 | 18 | 17 | |
| Tumor stage | | | | |
|
T1-2 | 40 | 27 | 13 | 0.003b |
|
T3-4 | 22 | 6 | 16 | |
| Tumor size | | | | |
|
≤4 cm | 28 | 20 | 8 | 0.005b |
|
>4
cm | 34 | 13 | 21 | |
| Lymph node
metastasis | | | | |
|
No | 36 | 22 | 14 | 0.011b |
|
Yes | 26 | 11 | 15 | |
| Distant
metastasis | | | | |
|
No | 39 | 21 | 18 | 0.086 |
|
Yes | 23 | 12 | 11 | |
LUCAT1 directly targets miR-375 in RCC
cells
Increasing evidence has suggested that lncRNAs act
as molecular sponges or competing endogenous RNAs (ceRNAs),
regulating miRNA expression, thereby affecting their biological
function (21). Using the
bioinformatics tool lncBase Predicted v.2, LUCAT1 was predicted to
contain complementary bases to miR-375 (Fig. 2A). Transfection of miR-375 mimics
decreased the luciferase activity of the LUCAT1-WT group compared
with the miR-NC mimics transfection (Fig. 2B), while there was little change in
the luciferase activity of the LUCAT1-MUT group, indicating that
LUCAT1 directly targeted miR-375. A negative correlation was also
observed between miR-375 and LUCAT1 expression level in ccRCC
tissues (Fig. 2D). In addition, the
results indicated that the expression level of miR-375 was
decreased in ccRCC tissues and cell lines compared with healthy
tissues and normal cells, respectively (Fig. 2C and E). Moreover, based on the higher or lower
than the median expression levels of miR-375, the patients were
divided into two groups, and the association between miR-375
expression and clinicopathological factors of patients with ccRCC
was analyzed. Our results indicated that the expression level of
miR-375 was associated with tumor stage, tumor size, lymph node
metastasis and distant metastasis in patients with ccRCC (Table SII). Furthermore, LUCAT1 was
upregulated, while miR-375 was downregulated in
LUCAT1-overexpressing Caki-1 and A498 cells compared with the
vector-NC group (Fig. 2F and
H). In Caki-1 and A498 cells
transfected with si-LUCAT1#1 or si-LUCAT1#2, LUCAT1 expression
level was decreased, while miR-375 expression level was increased
compared with the si-NC group (Fig.
2G and I). These results
indicated that LUCAT1 interacted with miR-375.
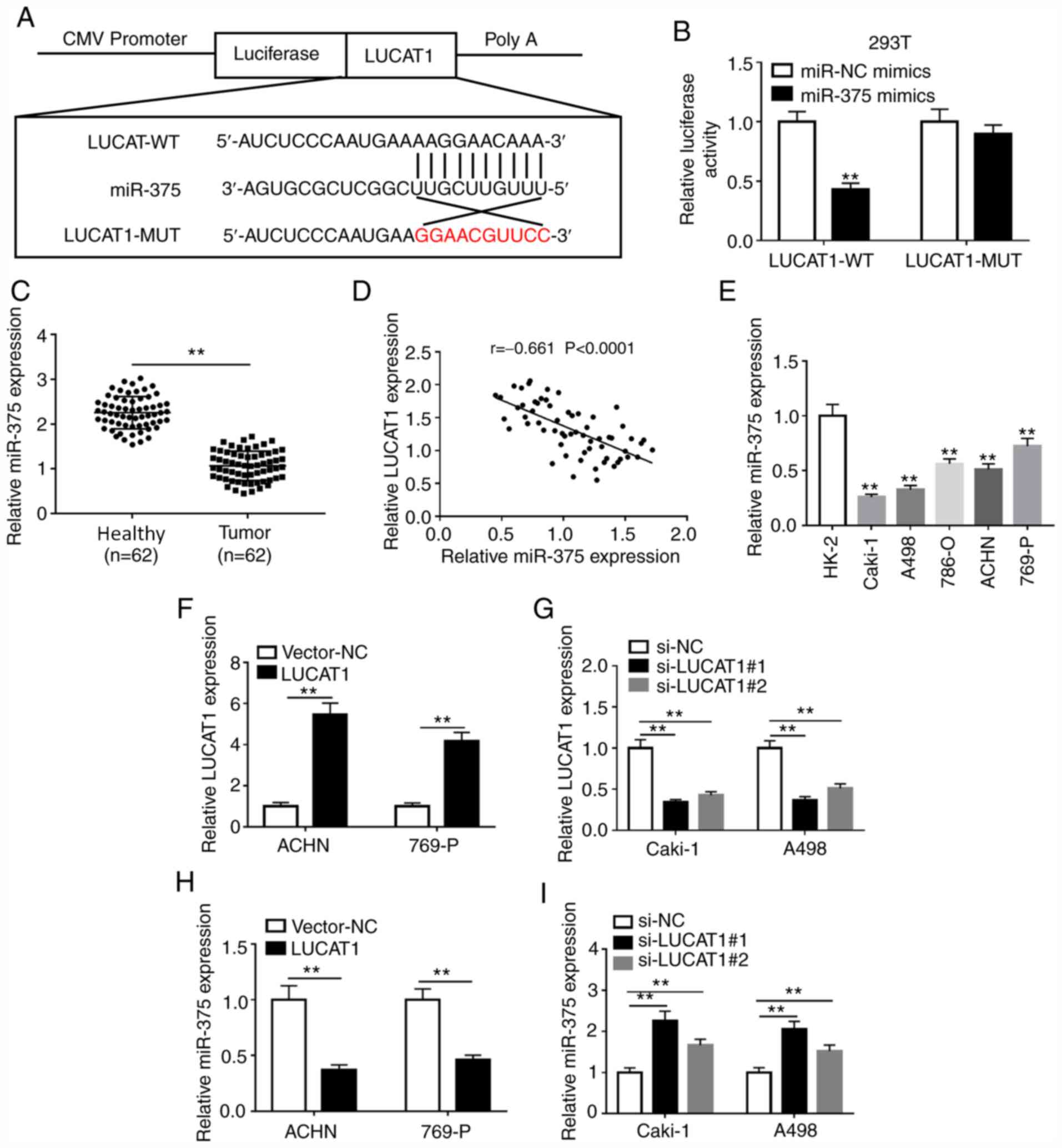 | Figure 2LUCAT1 directly binds to miR-375. (A)
The binding sites between LUCAT1 and miR-375 were predicted using
the lncBase Predicted v.2 software. (B) The effects of miR-375
overexpression on the luciferase activity of LUCAT1-WT or
LUCAT1-MUT reporter were detected using a dual-luciferase reporter
assay. (C) miR-375 expression in 62 pairs of ccRCC tissues and
adjacent healthy tissues was measured by RT-qPCR. (D) The
correlation between LUCAT1 and miR-375 expression levels in ccRCC
tissues was analyzed by Pearson's correlation analysis. (E) miR-375
expression in the normal human proximal tubular epithelial cell
line HK-2 and RCC cell lines (786-O, Caki-1, A498, 769-P and ACHN)
was measured by RT-qPCR. (F) Overexpression or (G) knockdown
efficiency of LUCAT1 was determined by RT-qPCR in ACHN and 769-P
cells or Caki-1 and A498 cells, respectively. The effects of LUCAT1
(H) overexpression or (I) knockdown on miR-375 level in ACHN and
769-P cells or Caki-1 and A498 cells, respectively, were measured
by RT-qPCR. **P<0.01 vs. the indicated groups or HK-2
cells. RT-qPCR, reverse transcription-quantitative PCR; LUCAT1,
lung cancer-associated transcript 1; RCC, renal cell carcinoma;
ccRCC, clear cell RCC; miR, microRNA; WT, wild-type; MUT, mutant;
si, small interfering; NC, negative control. |
LUCAT1 knockdown inhibits cell
proliferation, migration and invasion via regulating miR-375 in RCC
cells
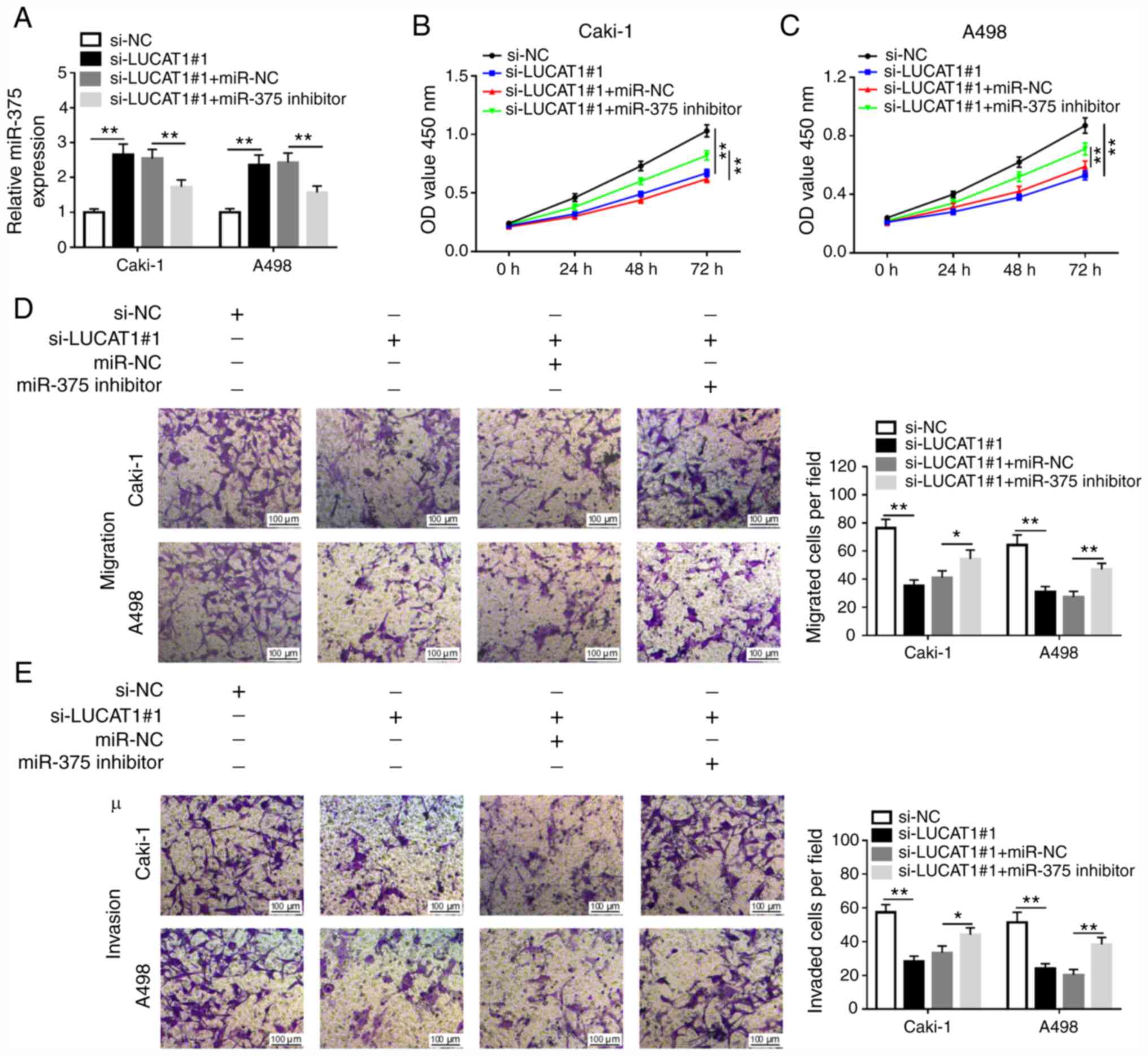
To explore the effects of LUCAT1 and miR-375 in
ccRCC cells, in vitro assays were performed. The
transfection efficiency of miR-375 mimics and inhibitor compared
with their respective NCs in Caki-1 and A498 cells was examined via
RT-qPCR (Fig. S2A and B). Caki-1 and A498 cells were transfected
with si-NC, si-LUCAT1#1, si-LUCAT1#1 + miR-NC or si-LUCAT1#1 +
miR-375 inhibitor. RT-qPCR analysis indicated that miR-375
expression was increased in Caki-1 and A498 cells transfected with
si-LUCAT1#1, but was inhibited by miR-375 inhibitor (Fig. 3A). Subsequently, CCK-8 and Transwell
assays were performed to examine the role of LUCAT1 and miR-375 in
ccRCC progression. As demonstrated in Fig. 3B and C, LUCAT1 knockdown inhibited the
proliferation of Caki-1 and A498 cells, which was reversed by
miR-375 inhibition. Transwell assay revealed that LUCAT1 silencing
significantly inhibited the migratory and invasive abilities of
Caki-1 and A498 cells, while miR-375 inhibitor inverted the
si-LUCAT1#1-induced inhibitory effects on cell migration and
invasion of Caki-1 and A498 cells (Fig.
3D and E). Furthermore, the
results of the present study also suggested that the overexpression
of LUCAT1 promoted the proliferation, migration and invasion of
Caki-1 and A498 cells (Fig. S3).
Together, these results demonstrated that LUCAT1 regulated cell
proliferation, migration and invasion via targeting miR-375 in RCC
cells.
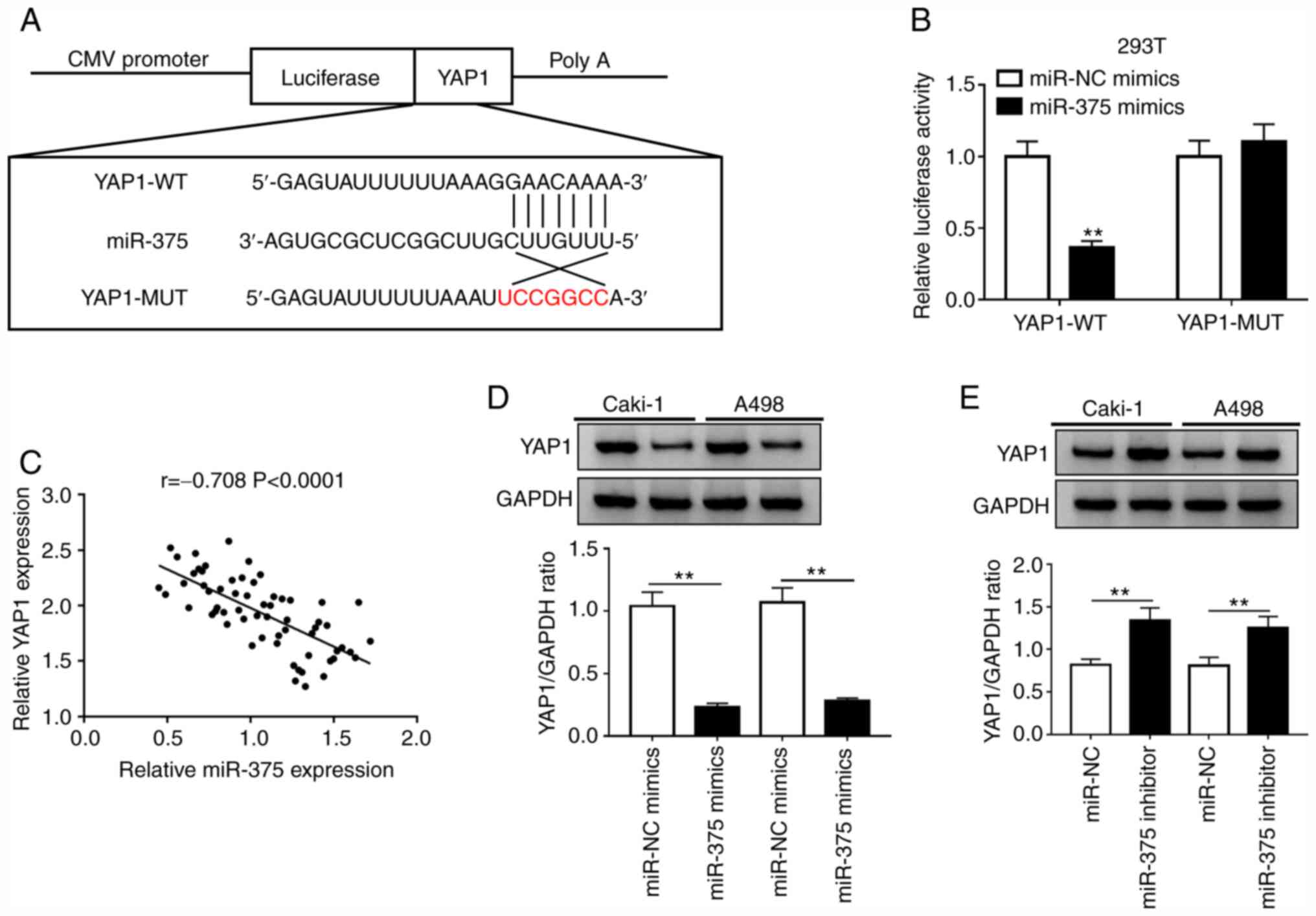
YAP1 is a target of miR-375 in RCC
cells
Previous studies have reported that miRNAs exert
their function by targeting the 3'-UTR of mRNAs (11,12).
The TargetScan software indicated the existence of binding sites
between miR-375 and YAP1 (Fig. 4A).
To further validate this prediction, YAP1-WT or YAP1-MUT reporter
plasmids and miR-NC or miR-375 mimics were co-transfected into 293T
cells. The luciferase activity of the YAP1-WT reporter was
suppressed in cells transfected with miR-375 mimics, while a small
alteration was observed in the luciferase activity of the YAP1-MUT
group compared with that of the miR-NC group (Fig. 4B). Moreover, the expression level of
miR-375 was negatively correlated with that of YAP1 in ccRCC
tissues (Fig. 4C). Subsequent
western blot assays confirmed that overexpression of miR-375
reduced the protein level of YAP1, while the protein level of YAP1
was increased by miR-375 knockdown in Caki-1 and A498 cells
(Fig. 4D and E). These results indicated that miR-375
directly targeted YAP1 and inhibited the transcription and
translation of YAP1 in RCC cells.
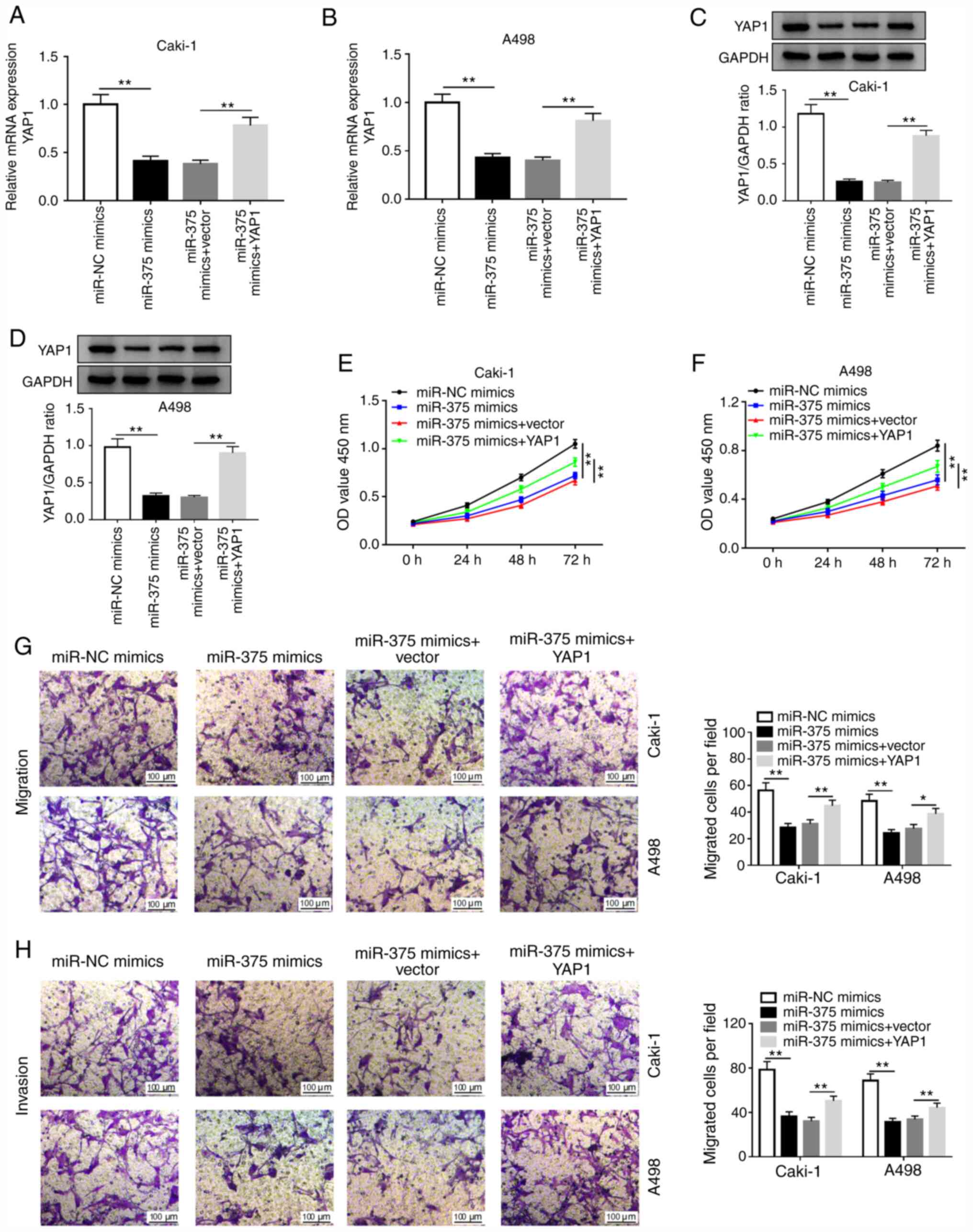
miR-375 suppresses cell proliferation,
migration and invasion by targeting YAP1 in RCC cells
To further investigate whether miR-375 exerted its
function by targeting YAP1 in RCC, Caki-1 and A498 cells were
transfected with miR-NC, miR-375 mimics, miR-375 mimics+vector or
miR-375 mimics+YAP1. The transfection efficiency of the YAP1
overexpressing vector in Caki-1 and A498 cells compared with its
respective NC is presented in Fig.
S2C and D. As depicted in
Fig. 5A and B, the level of YAP1 mRNA was significantly
decreased in miR-375-overexpressing Caki-1 and A498 cells, which
was partially reversed by the overexpression of YAP1.
Overexpression of miR-375 decreased the protein level of YAP1,
while this decrease was abolished by YAP1 overexpression (Fig. 5C and D). Furthermore, miR-375 overexpression
promoted cell proliferation, migration and invasion in Caki-1 and
A498 cells, while these effects were reversed by YAP1
overexpression (Fig. 5E-H). These
data suggested that miR-375 suppressed RCC phenotypes via targeting
YAP1.
Knockdown of LUCAT1 suppresses cell
proliferation, migration and invasion by regulating the
miR-375/YAP1 axis in RCC cells
As aforementioned, the present study hypothesized
that LUCAT1 exerted its carcinogenic role via the
LUCAT1/miR-375/YAP1 regulatory axis. RT-qPCR results demonstrated
that knockdown of LUCAT1 decreased the mRNA level of YAP1, which
was abolished by miR-375 inhibition in RCC cells (Fig. 6A). Western blotting results
confirmed that miR-375 inhibitor reversed the si-LUCAT1-mediated
inhibitory effect on YAP1 protein expression level in Caki-1 and
A498 cells (Fig. 6B and C). Furthermore, LUCAT1 knockdown decreased
the level of YAP1 mRNA and YAP1 overexpression abolished this
effect (Fig. 6D). LUCAT1 silencing
also suppressed YAP1 protein level, which was abolished by YAP1
overexpression (Fig. 6E and
F). Therefore, the
LUCAT1/miR-375/YAP1 regulatory axis may participate in the
development of RCC. To further verify this hypothesis, CCK-8 and
Transwell experiments were performed. LUCAT1 knockdown suppressed
the proliferation, migration and invasion of Caki-1 and A498 cells,
which were reversed by the restoration of YAP1 expression (Fig. 6G-J). These results indicated that
LUCAT1 knockdown suppressed RCC phenotypes via the miR-375/YAP1
regulatory axis.
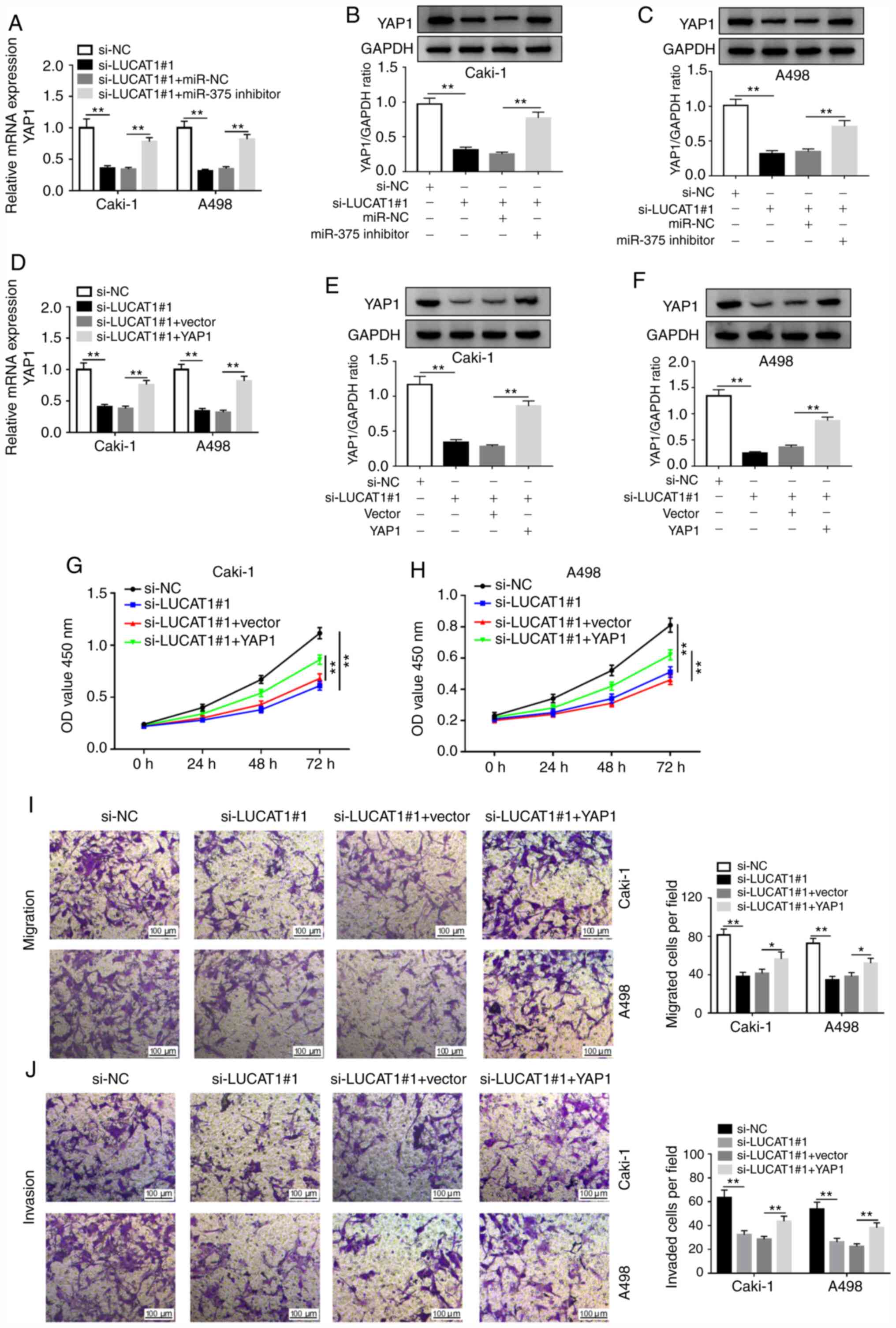 | Figure 6LUCAT1 enhances the proliferation,
migration and invasion of renal cell carcinoma cells by targeting
miR-375 to regulate YAP1 in vitro. (A) YAP1 mRNA expression
was assessed by RT-qPCR in Caki-1 and A498 cells transfected with
si-NC, si-LUCAT1#1, si-LUCAT1#1+miR-NC or si-LUCAT1#1+miR-375
inhibitor. YAP1 protein expression was examined by western blotting
in (B) Caki-1 and (C) A498 cells transfected si-NC, si-LUCAT1#1,
si-LUCAT1#1+miR-NC or si-LUCAT1#1+miR-375 inhibitor. (D) YAP1 mRNA
level was assessed by RT-qPCR in Caki-1 and A498 cells transfected
with si-NC, si-LUCAT1#1, si-LUCAT1#1+vector or si-LUCAT1#1+YAP1.
YAP1 protein level was examined by western blotting in (E) Caki-1
and (F) A498 cells transfected si-NC, si-LUCAT1#1,
si-LUCAT1#1+vector or si-LUCAT1#1+YAP1. Cell proliferation was
measured using Cell Counting Kit-8 assay in transfected (G) Caki-1
and (H) A498 cells. (I) Cell migratory and (J) invasive
capabilities were assessed by Transwell assay in transfected Caki-1
and A498 cells. *P<0.05; **P<0.01.
LUCAT1, lung cancer-associated transcript 1; YAP1, yes-associated
protein 1; miR, microRNA; NC, negative control; si, small
interfering; RT-qPCR, reverse transcription-quantitative PCR; OD,
optical density. |
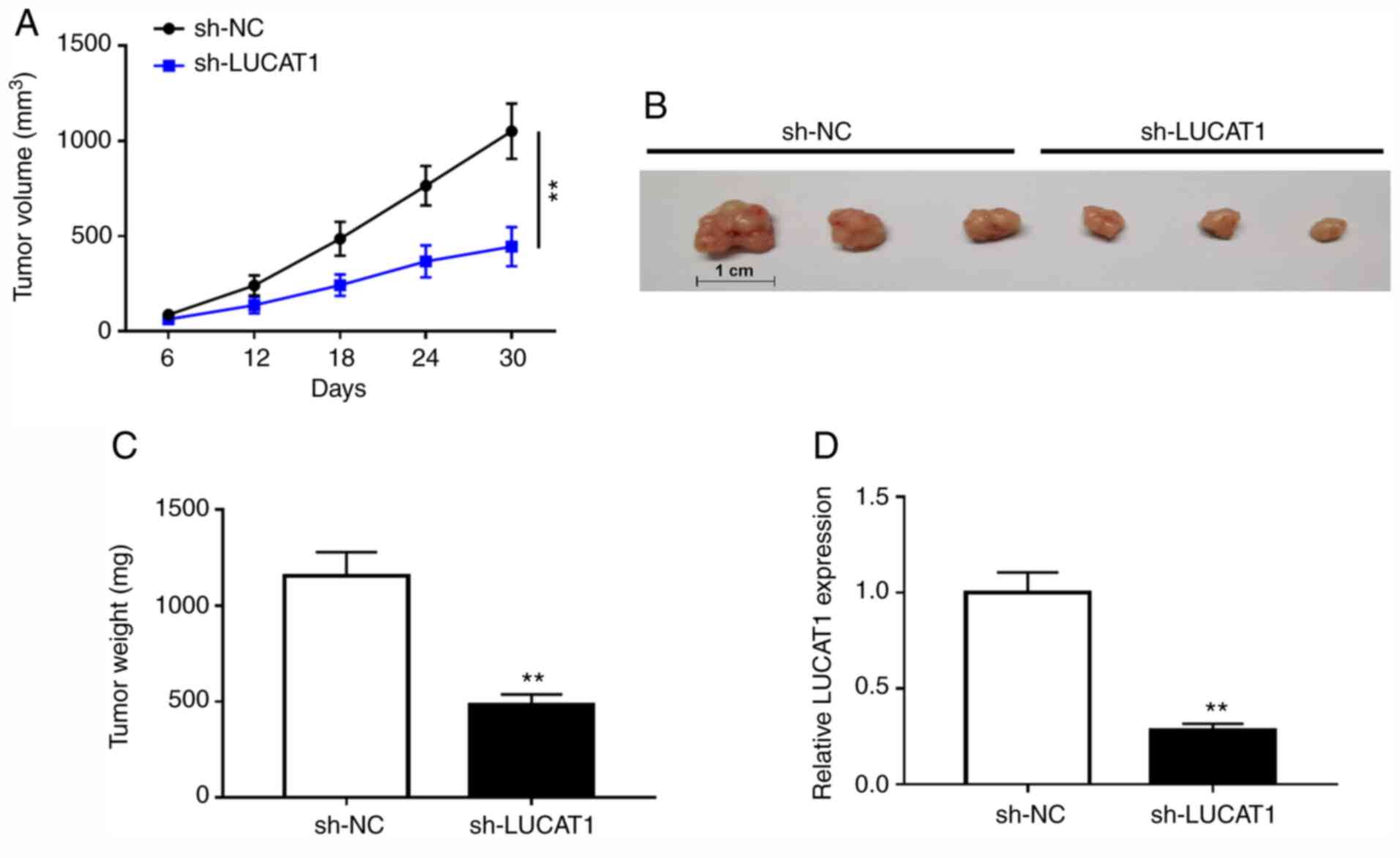
LUCAT1 deficiency suppresses ccRCC
tumor growth in vivo
A xenograft tumor mouse model of ccRCC was used to
verify the effect of LUCAT1 on tumor growth in vivo.
Compared with the mice injected with sh-NC, tumor volume and weight
were reduced in the sh-LUCAT1 group, indicating that LUCAT1
knockdown decreased the tumor growth of ccRCC in vivo
(Fig. 7A-C). Additionally, RT-qPCR
results verified that LUCAT1 was downregulated in the tumors of
mice injected with sh-LUCAT1-transfected ccRCC Caki-1 cells
(Fig. 7D). Taken together, these
data suggested that LUCAT1 silencing suppressed ccRCC tumor growth
in vivo.
Discussion
Accumulating evidence has suggested that lncRNAs
emerged as essential regulators in tumorigenesis and potential
biomarkers for the diagnosis or prognosis of ccRCC (22). LUCAT1 is located on chromosome
5q14.3, and was first identified in airway epithelial cells
(23). It has been demonstrated
that LUCAT1 was upregulated in ccRCC tissues and promoted the
proliferation and invasion of ccRCC cells (10). However, the potential role and the
regulatory mechanism of LUCAT1 in ccRCC progression requires
further elucidation. YAP1, a transcriptional co-activator, has been
indicated to promote tumorigenesis in various types of cancer,
including ccRCC (18,19).
In the present study, LUCAT1 and YAP1 were markedly
upregulated in ccRCC tissues and cell lines. LUCAT1 expression was
positively correlated with that of YAP1 in ccRCC tissues. In
addition, LUCAT1 expression was closely associated with tumor
stage, tumor size and lymph node metastasis in patients with ccRCC.
Moreover, Kaplan-Meier survival curves suggested a negative
association between high LUCAT1 expression and the overall survival
of patients with ccRCC.
An interaction between lncRNAs and miRNAs has been
identified in various cancers, such as bladder cancer, gastric
cancer and ccRCC (11,24,25).
In the present study, miR-375 was revealed to be a direct target of
LUCAT1. miR-375 has been reported to be a tumor suppressor in
various types of cancer, including gastric cancer, glioma and ccRCC
(14-17).
For example, Wang and Sun (17)
reported that miR-375 inhibited cell proliferation, migration and
invasion via regulating 3-phosphoinositide-dependent protein kinase
1 expression in renal cancer. Consistent with a previously
published study (17), the results
of the present study indicated that miR-375 was significantly
downregulated in ccRCC tissues and cell lines compared with healthy
tissues and normal cells, respectively, and miR-375 expression
level was negatively correlated with that of LUCAT1 in ccRCC
tissues. Furthermore, it was observed that LUCAT1 suppressed the
expression of miR-375 and promoted cell proliferation, migration
and invasion by targeting miR-375. Moreover, the tumor-suppressive
role of LUCAT1 knockdown in ccRCC was verified in vivo.
It has been hypothesized that lncRNAs served as
ceRNAs or miRNA sponges, regulating the expression of target mRNAs
(26). Previous studies have
demonstrated that miR-375 was associated with the progression of
human cancers by regulating the expression of its target genes,
such as SP1, PI3K-subunit α and HER2 (27-29).
In the present study, YAP1 was revealed to be a target of miR-375
in RCC cells and was negatively regulated by miR-375. YAP1 has been
indicated to be closely associated with cell proliferation,
invasion and migration in prostate and colorectal cancer (30,31).
YAP1 overexpression reversed the inhibitory effects of miR-375 on
the proliferation, migration and invasion of RCC cells, suggesting
that miR-375 inhibited tumor progression by targeting YAP1. These
data were consistent with previous studies in ovarian cancer
(32), gastric carcinogenesis
(14) and osteosarcoma (33). In addition, the results of the
current study demonstrated that LUCAT1 may regulate YAP1 expression
by sponging miR-375. Overexpression of YAP1 abolished the
inhibitory effects of LUCAT1 knockdown on cell proliferation,
invasion and migration in RCC cells, supporting the regulatory role
of the LUCAT1/miR-375/YAP1 axis in RCC progression. However, due to
the limited experimental conditions, the lack of immunofluorescence
experiments was a limitation to the present study.
In conclusion, the present study indicated that
LUCAT1 functioned as a sponge for miR-375 to upregulate YAP1
expression, thereby regulating the development of ccRCC. Our
results revealed that LUCAT1 silencing suppressed the growth of
ccRCC in vivo and in vitro. The present study also
revealed that the LUCAT1/miR-375/YAP1 axis may be a promising
therapeutic target for patients with ccRCC.
Supplementary Material
H&E images of healthy kidney and
clear cell renal cell carcinoma tissues (magnification, x200).
Transfection efficiency of miR-375
mimics and inhibitor and YAP1 overexpressing vector. The expression
of miR-375 was determined in Caki-1 and A498 cells transfected with
(A) miR-NC mimics or miR-375 mimics and (B) miR-NC or miR-375
inhibitor. Caki-1 and A498 cells were transfected with empty vector
or YAP1 overexpressing vector, and (C) the mRNA and (D) protein
levels of YAP1 were detected by reverse transcription-quantitative
PCR or western blotting, respectively. **P<0.01. NC,
negative control; miR, microRNA; YAP1, yes-associated protein
1.
LUCAT1 overexpression promotes the
proliferation, migration and invasion of renal cell carcinoma
cells. Caki.1 and A498 cells were transfected with vector or LUCAT1
overexpression vector. (A) The expression level of LUCAT1 was
measured by reverse transcription.quantitative PCR. Cell
proliferation was examined using the Cell Counting Kit.8 assay in
transfected (B) Caki.1 and (C) A498 cells. (D) Cell migratory and
(E) invasive capabilities were assessed by Transwell assay in
transfected Caki.1 and A498 cells. **P<0.01. LUCAT1,
lung cancer.associated transcript 1; OD, optical density.
Association of clinicopathological
features with YAP1 expression in patients with clear cell renal
cell carcinoma.
Association of clinicopathological
features with miR-375 expression in patients with clear cell renal
cell carcinoma.
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
XW conceived and designed the study and prepared the
first draft of the manuscript. XW, HO, LZ, HL, HZ and XL performed
the experiments, reviewed and agreed to the final submission of the
manuscript. HO, LZ, HL, HZ and XL analyzed and interpreted the
data. XW and HO confirm the authenticity of the raw data. All
authors read and approved the final manuscript.
Ethics approval and consent to
participate
The human study was approved by the Ethics Committee
of Chenzhou No. 1 People's Hospital (Chenzhou, China) and all
participants provided written informed consent. The animal study
was approved by the Animal Ethics Committee of Chenzhou No. 1
People's Hospital.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Moch H, Cubilla AL, Humphrey PA, Reuter VE
and Ulbright TM: The 2016 WHO classification of tumours of the
urinary system and male genital organs-part A: Renal, penile, and
testicular tumours. Eur Urol. 70:93–105. 2016.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Ricketts CJ, De Cubas AA, Fan H, Smith CC,
Lang M, Reznik E, Bowlby R, Gibb EA, Akbani R, Beroukhim R, et al:
The cancer genome atlas comprehensive molecular characterization of
renal cell carcinoma. Cell Rep. 23:313–326.e5. 2018.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Keegan KA, Schupp CW, Chamie K, Hellenthal
NJ, Evans CP and Koppie TM: Histopathology of surgically treated
renal cell carcinoma: Survival differences by subtype and stage. J
Urol. 188:391–397. 2012.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Wang L and Lee BR: Novel agents and
approaches for advanced renal cell carcinoma. J Urol.
188(716)2012.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Capitanio U and Montorsi F: Renal cancer.
Lancet. 387:894–906. 2016.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Wu R, Su Y, Wu H, Dai Y, Zhao M and Lu Q:
Characters, functions and clinical perspectives of long non-coding
RNAs. Mol Genet Genomics. 291:1013–1033. 2016.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Liu G, Zhao X, Zhou J, Cheng X, Ye Z and
Ji Z: lncRNA TP73-AS1 promotes cell proliferation and inhibits cell
apoptosis in clear cell renal cell carcinoma through repressing
KISS1 expression and inactivation of PI3K/Akt/mTOR signaling
pathway. Cell Physiol Biochem. 48:371–384. 2018.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Hirata H, Hinoda Y, Shahryari V, Deng G,
Nakajima K, Tabatabai ZL, Ishii N and Dahiya R: Long noncoding RNA
MALAT1 promotes aggressive renal cell carcinoma through Ezh2 and
interacts with miR-205. Cancer Res. 75:1322–1331. 2015.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Xiao H, Bao L, Xiao W, Ruan H, Song Z, Qu
Y, Chen K, Zhang X and Yang H: Long non-coding RNA Lucat1 is a poor
prognostic factor and demonstrates malignant biological behavior in
clear cell renal cell carcinoma. Oncotarget. 8:113622–113634.
2017.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Zheng Z, Zhao F, Zhu D, Han J, Chen H, Cai
Y, Chen Z and Xie W: Long non-coding RNA LUCAT1 promotes
proliferation and invasion in clear cell renal cell carcinoma
through AKT/GSK-3β signaling pathway. Cell Physiol Biochem.
48:891–904. 2018.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Wang LN, Zhu XQ, Song XS and Xu Y: Long
noncoding RNA lung cancer associated transcript 1 promotes
proliferation and invasion of clear cell renal cell carcinoma cells
by negatively regulating miR-495-3p. J Cell Biochem. 119:7599–7609.
2018.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Bartel DP: Metazoan microRNAs. Cell.
173:20–51. 2018.PubMed/NCBI View Article : Google Scholar
|
|
13
|
He YH, Chen C and Shi Z: The biological
roles and clinical implications of microRNAs in clear cell renal
cell carcinoma. J Cell Physiol. 233:4458–4465. 2018.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Kang W, Huang T, Zhou Y, Zhang J, Lung
RWM, Tong JHM, Chan AWH, Zhang B, Wong CC, Wu F, et al: miR-375 is
involved in Hippo pathway by targeting YAP1/TEAD4-CTGF axis in
gastric carcinogenesis. Cell Death Dis. 9(92)2018.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Ji CX, Fan YH, Xu F, Lv SG, Ye MH, Wu MJ,
Zhu XG and Wu L: MicroRNA-375 inhibits glioma cell proliferation
and migration by downregulating RWDD3 in vitro. Oncol Rep.
39:1825–1834. 2018.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Liu S, Song L, Yao H, Zhang L, Xu D, Gao F
and Li Q: miR-375 Is epigenetically downregulated by HPV-16 E6
mediated DNMT1 upregulation and modulates EMT of cervical cancer
cells by suppressing lncRNA MALAT1. PLoS One.
11(e0163460)2016.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Wang J and Sun X: MicroRNA-375 inhibits
the proliferation, migration and invasion of kidney cancer cells by
triggering apoptosis and modulation of PDK1 expression. Environ
Toxicol Pharmacol. 62:227–233. 2018.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Rybarczyk A, Klacz J, Wronska A,
Matuszewski M, Kmiec Z and Wierzbicki PM: Overexpression of the
YAP1 oncogene in clear cell renal cell carcinoma is associated with
poor outcome. Oncol Rep. 38:427–439. 2017.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Xiao J, Shi Q, Li W, Mu X, Peng J, Li M,
Chen M, Huang H, Wang C, Gao K and Fan J: ARRDC1 and ARRDC3 act as
tumor suppressors in renal cell carcinoma by facilitating YAP1
degradation. Am J Cancer Res. 8:132–143. 2018.PubMed/NCBI
|
|
20
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Karreth FA and Pandolfi PP: ceRNA
cross-talk in cancer: When ce-bling rivalries go awry. Cancer
Discov. 3:1113–1121. 2013.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Gayed BA, Youssef RF, Bagrodia A, Kapur P,
Darwish OM, Krabbe LM, Sagalowsky A, Lotan Y and Margulis V:
Prognostic role of cell cycle and proliferative biomarkers in
patients with clear cell renal cell carcinoma. J Urol.
190:1662–1667. 2013.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Thai P, Statt S, Chen CH, Liang E,
Campbell C and Wu R: Characterization of a novel long noncoding
RNA, SCAL1, induced by cigarette smoke and elevated in lung cancer
cell lines. Am J Respir Cell Mol Biol. 49:204–211. 2013.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Wieczorek E and Reszka E: MRNA, microRNA
and lncRNA as novel bladder tumor markers. Clin Chim Acta.
477:141–153. 2018.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Zhang G, Li S, Lu J, Ge Y, Wang Q, Ma G,
Zhao Q, Wu D, Gong W, Du M, et al: lncRNA MT1JP functions as a
ceRNA in regulating FBXW7 through competitively binding to
miR-92a-3p in gastric cancer. Mol Cancer. 17(87)2018.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Salmena L, Poliseno L, Tay Y, Kats L and
Pandolfi PP: A ceRNA hypothesis: The Rosetta Stone of a hidden RNA
language? Cell. 146:353–358. 2011.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Liu XH, Wang J and Dong YH: The inhibitory
effect of miR-375 targeting sp1 in colorectal cancer cell
proliferation. Eur Rev Med Pharmacol Sci. 22:405–411.
2018.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Wang Y, Tang Q, Li M, Jiang S and Wang X:
MicroRNA-375 inhibits colorectal cancer growth by targeting PIK3CA.
Biochem Biophys Res Commun. 444:199–204. 2014.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Cheng L, Zhan B, Luo P and Wang B:
miRNA375 regulates the cell survival and apoptosis of human
non-small cell carcinoma by targeting HER2. Mol Med Rep.
15:1387–1392. 2017.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Shi Y, Cao T, Sun Y, Xia J, Wang P and Ma
J: Nitidine Chloride inhibits cell proliferation and invasion via
downregulation of YAP expression in prostate cancer cells. Am J
Transl Res. 11:709–720. 2019.PubMed/NCBI
|
|
31
|
Zhou Z, Zhang HS, Zhang ZG, Sun HL, Liu
HY, Gou XM, Yu XY and Huang YH: Loss of HACE1 promotes colorectal
cancer cell migration via upregulation of YAP1. J Cell Physiol.
234:9663–9672. 2019.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Yan H, Li H, Li P, Li X, Lin J, Zhu L,
Silva MA, Wang X, Wang P and Zhang Z: Long noncoding RNA MLK7-AS1
promotes ovarian cancer cells progression by modulating
miR-375/YAP1 axis. J Exp Clin Cancer Res. 37(237)2018.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Liu G, Huang K, Jie Z, Wu Y, Chen J, Chen
Z, Fang X and Shen S: CircFAT1 sponges miR-375 to promote the
expression of Yes-associated protein 1 in osteosarcoma cells. Mol
Cancer. 17(170)2018.PubMed/NCBI View Article : Google Scholar
|