Introduction
Inflammatory bowel diseases (IBDs) primarily
comprise Crohn's disease (CD) and ulcerative colitis (UC) (1-3).
Although the pathology of IBDs has been widely investigated, the
exact causes of IBDs are remain largely unknown. Currently, there
are ~8 million patients with IBDs worldwide (4), and an improved understanding of such
diseases should improve the efficiency of clinical therapies and
benefit patients.
Intestinal epithelial cells (IECs) comprise a
variety of cell types, including Paneth cells and goblet cells, and
they facilitate the intestinal epithelial defense against pathogen
invasion (5-8).
Upon stimulation, IECs produce inflammatory cytokines, intestinal
barrier-associated molecules and antimicrobial peptides to maintain
intestinal homeostasis under pathophysiological conditions
(9,10). IEC dysfunction has been demonstrated
to disrupt intestinal homeostasis and induce or aggravate the
development of IBDs (9). For
instance, the specific knockout of STAT3 in IECs has been shown to
impair mucosal wound healing in IBD, causing mice to be highly
sensitive to experimental colitis (11). Other studies have shown that the
local delivery of IL-22 enhances the function of IECs in colitis
and alleviates the disease, while the local delivery of
IL-22-binding protein, which neutralizes IL-22 activity,
significantly inhibits the restitution of IECs and limits tissue
recovery following the induction of dextran sulfate sodium
(DSS)-induced colitis in mice (12,13).
Therefore, gaining a full understanding of the regulation of IEC
function is urgently necessary for the prevention and treatment of
IBD.
MicroRNAs (miRNAs/miRs) are small non-coding RNA
molecules that are not translated into proteins, but play critical
roles in the regulation of gene expression (14,15).
Previous studies have revealed that miRNAs serve important roles in
the development of IBD by regulating the function of cells in the
intestinal tract (15). For
example, in one study, miR-21 and miR-31 were shown to influence T
cell responses in patients with IBD and animal colitis models
(16). In another study, the
investigators found that compared with those of healthy controls,
the plasma levels of miR-199a-5p, miR-362-3p and miR-532-3p were
increased in patients with CD while those of miR-149 and
miRplus-F1065 were decreased (17).
Furthermore, the inhibition of certain miRNAs has been demonstrated
to directly affect the induction of colitis in mice. For example,
in 2,4,6-trinitrobenzenesulfonic acid (TNBS)-induced colitis in
mice, anti-miR-124 treatment alleviated the disease activity index
and inhibited proinflammatory cytokine expression via modulation of
the aryl hydrocarbon receptor (18). In another study, the inhibition of
miR-31 significantly promoted the expression of the
anti-inflammatory cytokine IL-25 in the colons of mice with
TNBS-induced colitis, exhibiting therapeutic effects on
TNBS-induced colitis and spontaneous colitis in IL-10-deficient
mice (19). However, whether miRNAs
regulate the function of IECs in IBD and the underlying mechanisms
remain largely unknown. Advances in this field are likely to
provide new information to improve understanding of the pathology
of IBD and the efficacy of IEC-based IBD therapies.
In the current study, the role of miR-452-5p in IBD
was explored. It has previously been demonstrated that miR-452-5p
post-transcriptionally abrogates SMAD4 expression, inhibiting the
downstream gene SMAD7(20). Also,
it has been suggested that genetic variants of several SMAD family
members, namely SMAD2/3/4/7, may alter the balance of
differentiation between T helper 17 and regulatory T cells,
resulting in the development of IBD, particularly UC (21). Additionally, the depletion of
epithelial SMAD4 upregulates inflammation and promotes
inflammation-associated cancer in mice with DSS-induced colitis
(22). Furthermore, SMAD4
epithelial protein levels are downregulated in patients with CD and
negatively correlated with disease activity (23). These findings suggest a key role for
SMAD4 or SMAD4/SMAD7 signaling in the pathology of IBD. As
miR-452-5p is associated with the upstream mechanism of SMAD4/SMAD7
signaling, we hypothesized that miR-452-5p may be involved in the
progression of IBD.
In the present study, the role of the
miR-452-5p/Mcl-1 axis in regulating the properties of IECs during
the pathology of colitis was investigated. The findings provide new
knowledge about the role of miRNAs in IBD pathogenesis and hold the
potential to improve future treatments for IBDs.
Materials and methods
Mice
A total of 40 C57BL/6 male mice (6-8 weeks old; 20±2
g) were purchased from Shanghai SLAC Laboratory Animal Co., Ltd.
and raised at the SPF animal facility of Huazhong University of
Science and Technology. The mice were housed at a constant room
temperature (23±2˚C) and relative humidity (50±10%) with free
access to food and water in a fixed 12-h light/dark cycle. All
animal experiments were performed according to the National
Institutes of Health Guide for the Care and Use of Laboratory
Animals (7th edition, revised 1996) and were approved by the Animal
Experimentation Ethics Committee of Huazhong University of Science
and Technology.
DSS-induced UC mouse model
A mouse model of UC was established using 3% DSS (3
g/100 ml; molecular weight 36,000-50,000; MP Biomedicals, LLC)
dissolved in sterile distilled water and provided ad libitum
from experimental days 1 to 5. The DSS solution was prepared fresh
every 2 days to ensure its effects were maintained. The same volume
of double-distilled water (DDW) was used as a control (28 mice in
the UC group and 12 mice in the control group). On day 5, the mice
were sacrificed by CO2 inhalation; the mice were placed
in a transparent polycarbonate euthanasia chamber covered with an
acrylic lid, with ports for gas inlet and outlet. The air in the
chamber was replaced with CO2 at a rate of 30% chamber
volume/min. The distal colon (2 cm) was removed and processed for
histological examination and protein isolation.
Adenovirus (Ad) vector treatment
To explore the effects of miR-452-5p in IECs in the
UC mouse model, Ad-packaged vectors (Ad-anti-miR-452-5p and
Ad-Mock) were constructed using the pAdEasy/Track-CMV adenovirus
vector. A scrambled sequence served as the control. Viral packaging
was performed by Shanghai Kelei Biotechnology Co, Ltd. The Ad
vectors (100 µl normal saline containing 5.0x108 active
viral particles) were intracolonically administered to the mice 2
days prior to the administration of DSS. This involved
administering the adenovirus intracolonically via an 8-cm
polyethylene tube through the anus under mild anesthesia
(intraperitoneal injection of thiopental, 40 mg/kg). The mice were
maintained in a head-down position for ~1 min to prevent expulsion
of the solution. The mice were divided into 3 groups: Ad-Mock group
(mice injected with Ad-Mock and DDW; n=12), Ad-Mock+DSS group (mice
injected with Ad-Mock and DSS; n=14), and Ad-anti-miR-452-5p+DSS
group (mice injected with Ad-anti-miR-452-5p and DSS; n=14).
Evaluation of experimental UC
The symptoms of IBD, including loss of body weight,
occult blood, and diarrhea, were recorded every day. To calculate
the disease index, evaluations of body weight loss in comparison
with initial body weight, stool consistency and rectal bleeding
were performed. The scores were assigned as follows: For body
weight loss: No weight loss, 0; body weight loss 1-5%, 1; body
weight loss 6-10%, 2; body weight loss 11-20%, 3; and body weight
>20%, 4. For stool consistency: Well-formed pellets, 0; pasty
and semiformed stools, 2; and liquid stools, 4. For rectal
bleeding: No blood, 0; positive bleeding, 2; and gross bleeding, 4.
The total scores were the sum of the scores in the three
categories.
Histological analysis
Hematoxylin and eosin (H&E) staining was
performed to evaluate the pathology of the mouse model of UC. In
brief, colon tissues from normal mice and UC model mice were
collected and fixed in PBS containing 4% paraformaldehyde at room
temperature for 48 h. After dehydration with sequential 95 and 100%
ethanol followed by xylene, the tissues were embedded in paraffin
and sectioned at a thickness of 6 µm with a microtome (SM2500;
Leica Microsystems GmbH). These sections were stained with
hematoxylin (0.5%) for 5 min, followed by eosin (0.5%) for 5 min at
room temperature, and examined by light microscopy (Olympus
Corporation).
Reverse transcription-quantitative PCR
(RT-qPCR) analysis
Total RNA was extracted from the isolated IECs using
TRIzol reagent (Thermo Fisher Scientific, Inc.) and reverse
transcribed into cDNA at 55˚C for 10 min using a PrimeScript RT-PCR
kit (Takara Biotechnology Co., Ltd.). The mRNA levels were
quantified by qPCR using a 7500 ABI Prism system (Applied
Biosystems; Thermo Fisher Scientific, Inc.) using a SYBR-Green
Master Mix kit (Thermo Fisher Scientific, Inc.). The reaction
conditions were as follows: 94˚C for 3 min, followed by 30 cycles
at 94˚C for 45 sec, 57˚C for 45 sec and 72˚C for 45 sec, and final
extension at 72˚C for 10 min. The primers used for qPCR analysis
were as follows: Mouse TNFα forward, 5'-ACTGAACTTCGGGGTGATCG-3' and
reverse, 5'-GTTTGCTACGACGTGGGCTA-3'; mouse IL-8 forward,
5'-AGAGCTTGAGTGTGACGCC-3' and reverse, 5'-CCAGGTCAGTTAGCCTTGCC-3';
mouse IL-6 forward, 5'-GTCCTTCCTACCCCAATTTCCA-3' and reverse,
5'-TAACGCACTAGGTTTGCCGA-3'; mouse occludin (OCLN) forward,
5'-TTTCAGGTGAATGGGTCACCG-3' and reverse,
5'-GCTCCCAAGATAAGCGAACCT-3'; mouse mucin 2 (MUC-2) forward,
5'TTGTCACCTTCGATGGGCTC-3' and reverse, 5'-TCTCGTGGCGCACAATAAGT-3';
mouse zona occludens 1 (ZO-1) forward,
5'-AGAAAAAGAATGCACAGAGTTGT-3' and reverse,
5'-GAAATCGTGCTGATGTGCCA-3'; mouse myeloid cell leukemia 1 (Mcl-1)
forward, 5'-CACGTACAGGACCTAGAAGGC-3' and reverse,
5'-TAGTTTGGTGGCTGGAGCTTT-3'; mouse Bax forward,
5'-CTGCAGAGGATGATTGCTG-3' and reverse, 5'-ATCAGCAAACATGTCAGCT-3';
mouse Bcl-2 forward, 5'-CTGAGTACCTGAACCGGCAT-3' and reverse,
5'-TTGTGGCCCAGGTATGCAC-3'; mouse GAPDH forward,
5'-TCTTTTGCGTCGCCAGCC-3' and reverse,
5'-CCATGGGTGGAATCATATTGGAAC-3'; mouse miR-452-5p, forward
5'-UGUUUGCAGAGGAAAC-3' and reverse, 5'-AACGCTTCACGAATTTGCGT-3'; U6
forward, 5'-AACGCTTCACGAATTTGCGT-3' and reverse,
5'-CTCGCTTCGGCAGCACA-3'. The expression of miR-452-5p was
normalized to U6 and the other mRNAs were normalized to GAPDH using
the 2-∆∆Cq method (24).
IEC isolation
IECs were isolated as previously described (25). Briefly, colon samples were
collected, and the contents were removed by washing with PBS 3
times. After the removal of Peyer's patches and mesenteric fat, the
samples were cut into 1-cm pieces and placed into Hank's Balanced
Salt Solution (Thermo Fisher Scientific, Inc.) supplemented with 1
mM dithiothreitol, 0.5 mM EDTA and 10% FBS (Gibco; Thermo Fisher
Scientific, Inc.), and shaken at 37˚C for 30 min. The cell
suspension was collected, filtered through a 40-µm strainer to
remove debris, centrifuged at 4˚C for 5 min at 700 x g, and then
resuspended in 25% Percoll. The cell suspension was gently added to
the top of 40% Percoll in a 15-ml tube. The tubes were centrifuged
at 400 x g at room temperature for 20 min, and IECs were collected
from the interphase for analysis by RT-qPCR and western
blotting.
Cell treatment and transfection
IEC-6 cells (BeNa Culture Collection) were cultured
in McCoy's 5A medium (Sigma-Aldrich; Merck KGaA) supplemented with
10% FBS (Thermo Fisher Scientific, Inc.), 100 U/ml penicillin G
potassium and 100 µg/ml streptomycin at 37˚C in a humidified
atmosphere with 5% CO2. LPS (1 µg/ml; Sigma-Aldrich;
Merck KGaA) was used to stimulate IEC-6 cells to induce
inflammation for 4 h at 37˚C. Silencing of Mcl-1 was performed by
cloning short hairpin RNA (shRNA) oligonucleotides targeting Mcl-1
into the pCMV vector (Shanghai GenePharma Co., Ltd.), and its
scrambled negative control (sh-NC) was also purchased from Shanghai
GenePharma Co., Ltd. Mock refers to untreated cells. The miR-452-5p
mimics (miR-452-5p; 5'-UGUUUGCAGAGGAAACUGAGAC-3') and non-targeting
control mimics (NC mimics; 5'-UUUGUACUACACAAAAGUACUG-3'),
miR-452-5p inhibitor (anti-miR-452-5p;
5'-GUCUCAGUUUCCUCUGCAAACA-3') and its non-targeting control
(anti-miR-NC; 5'-CAGUACUUUUGUGUAGUACAAA-3') were also purchased
from Shanghai GenePharma Co., Ltd. IEC-6 cells were seeded into
24-well plates at a density of 2.0x104 cells/well,
following which 50 nM synthetic oligonucleotides or 2 µg vectors
were transfected into the cells using Lipofectamine®
2000 (Thermo Fisher Scientific, Inc.) for 48 h at 37˚C according to
the manufacturer's instructions. Cells were collected after 48 h
for further experiments.
Western blotting
Colon tissues and in vitro cultured cells
were collected, and the samples were lysed with RIPA lysis buffer
(Beyotime Institute of Biotechnology). containing protease
inhibitors at a mass/volume ratio of 100 mg/ml. Protein was then
extracted and protein concentration was detected using a BCA kit
(Beyotime Institute of Biotechnology). At least 10 µg protein
extracts were used for 10% SDS-PAGE. After running the gels, the
protein samples were transferred onto nitrocellulose membranes,
followed by blocking with 5% skimmed milk at 4˚C overnight. The
membranes were sequentially incubated with the following primary
antibodies: Mcl-1 (1:500; cat. no. ab32087; Abcam), TNFα (1:1,000;
cat. no. ab183218; Abcam), IL-8 (1:1,000; cat. no. AMM02601G; Santa
Cruz Biotechnology, Inc.), IL-6 (1:1,000; cat. no. ab229381;
Abcam), OCLN (1:1,000; cat. no. ab216327; Abcam), OCLN (1:1,000;
cat. no. ab221547; Abcam), MUC-2 (1:1,000; cat. no. ab272692;
Abcam) and β-actin (1:2,000; cat. no. AMM04710G; Santa Cruz
Biotechnology, Inc.) overnight at 4˚C, followed by incubation with
HRP-conjugated goat anti-rabbit IgG H&L antibodies (1:2,000;
cat. no. ab6721; Abcam) for 1 h at room temperature. GAPDH served
as an internal control. After washing with TBS with 0.1% Tween 20
three times, the immunoreactive bands were detected using enhanced
chemiluminescence (Pierce; Thermo Fisher Scientific, Inc.) and
analyzed using ImageJ v1.8.0 software (National Institutes of
Health).
Flow cytometry
After transfection for 48 h, an Annexin
V-fluorescein isothiocyanate (FITC)/propidium iodide (PI) Apoptosis
Detection kit (BD Biosciences) was used to analyze the apoptosis of
the IEC-6 cells. The cells were collected with trypsin, washed with
PBS, and resuspended in 500 µl HEPES buffer solution
(Sigma-Aldrich; Merck KGaA). The cells were then incubated with 5
µl Annexin V-FITC and 5 µl PI at room temperature for 15 min in the
dark. Finally, a FACSVerse™ flow cytometer with FACSCanto II FACP
Array™ software (v.3.0; both BD Biosciences) was used to analyze
apoptosis.
Luciferase reporter assay
The binding sites for miR-452-5p and Mcl-1 were
predicted using the starBase 3.0 website (http://starbase.sysu.edu.cn/). The luciferase reporter
assay was performed as previously described (26). The 3'-untranslated version (3'UTR)
of Mcl-1 containing the predicted wild-type (Wt) miR-452-5p-binding
sequence and a mutant (Mut) version of this sequence were amplified
by Shanghai GenePharma Co., Ltd. and inserted into the luciferase
reporter gene of the pmirGLO vector (Promega Corporation), which
produced the reporter plasmids Mcl-1-Wt and Mcl-1-Mut,
respectively. IEC-6 cells were co-transfected with miR-452-5p
mimics, anti-miR-452-5p or Mock (NC mimics) and the Mcl-1-Wt/Mut
luciferase constructs (100 ng) and the Renilla luciferase
control plasmid pRL-SV40 plasmid (2 ng) using Lipofectamine 2000.
pRL-SV40 was used to standardize the transfection efficiency and
exclude experimental errors caused by differences in transfection
efficiencies. The ratio of the firefly and Renilla
reniformis luciferase activities was used as an indicator of
the luciferase activity. Luciferase activity in the IECs was
analyzed using a Dual-Luciferase® reporter assay system
(Promega Corporation) after 24 h of incubation.
Statistical analysis
All experiments were performed at least three times.
Data are presented as the mean ± SEM. Differences between two
groups were evaluated by two-tailed Student's t-test. Statistical
significance among three or more groups was assessed by one-way
ANOVA followed by post hoc Dunnett's test (for comparisons with one
control) or Tukey's test (for comparisons among various groups).
Statistical significance for the disease activity index was
analyzed using the Mann-Whitney test. The correlation between
miR-452-5p and Mcl-1 expression was analyzed using Spearman's
correlation test. P<0.05 was considered to indicate a
statistically significant difference.
Results
miR-452-5p regulates the functions of
IECs during the development of UC
The expression of miR-452-5p was assessed in
IECs isolated from healthy mice and mice with DSS-induced UC.
H&E staining was also conducted. The results indicated that
there was severe mucosal injury in DSS-induced mice, characterized
by epithelial cell disruption, massive bowel edema and distorted
architecture of crypts (Fig. 1A).
Based on the RT-qPCR results, it was observed that compared with
healthy colon-derived IECs, the IECs in DSS-induced colitis tissue
expressed significantly higher levels of miR-452-5p (Fig. 1B). This indicates that miR-452-5p
may act as a regulator of IBD development and exert its effects by
regulating the function of IECs. To confirm this hypothesis, mice
were treated with Ad-Mock or Ad-anti-miR-452-5p. After confirming
the downregulation of miR-452-5p in the IECs of mice to which
Ad-anti-miR-452-5p was administered (Fig. 2A), the susceptibility of the mice to
DSS-induced UC was assessed. It was found that anti-miR-452-5p
exhibited beneficial effects on UC model mice, as revealed by a
decreased disease activity index (Fig.
2B) and attenuated loss of colon length (Fig. 2C). Notably, the inhibition of
miR-452-5p in IECs significantly restrained the expression of the
inflammatory cytokines TNF-α, IL-6 and IL-8, and maintained normal
expression levels of the integrity-associated molecules OCLN, ZO-1
and MUC-2 in the IECs of the mice exposed to DSS (Fig. 2D and E). Collectively, these results indicate
that miR-452-5p participates in the development of IBD by
regulating the function of IECs.
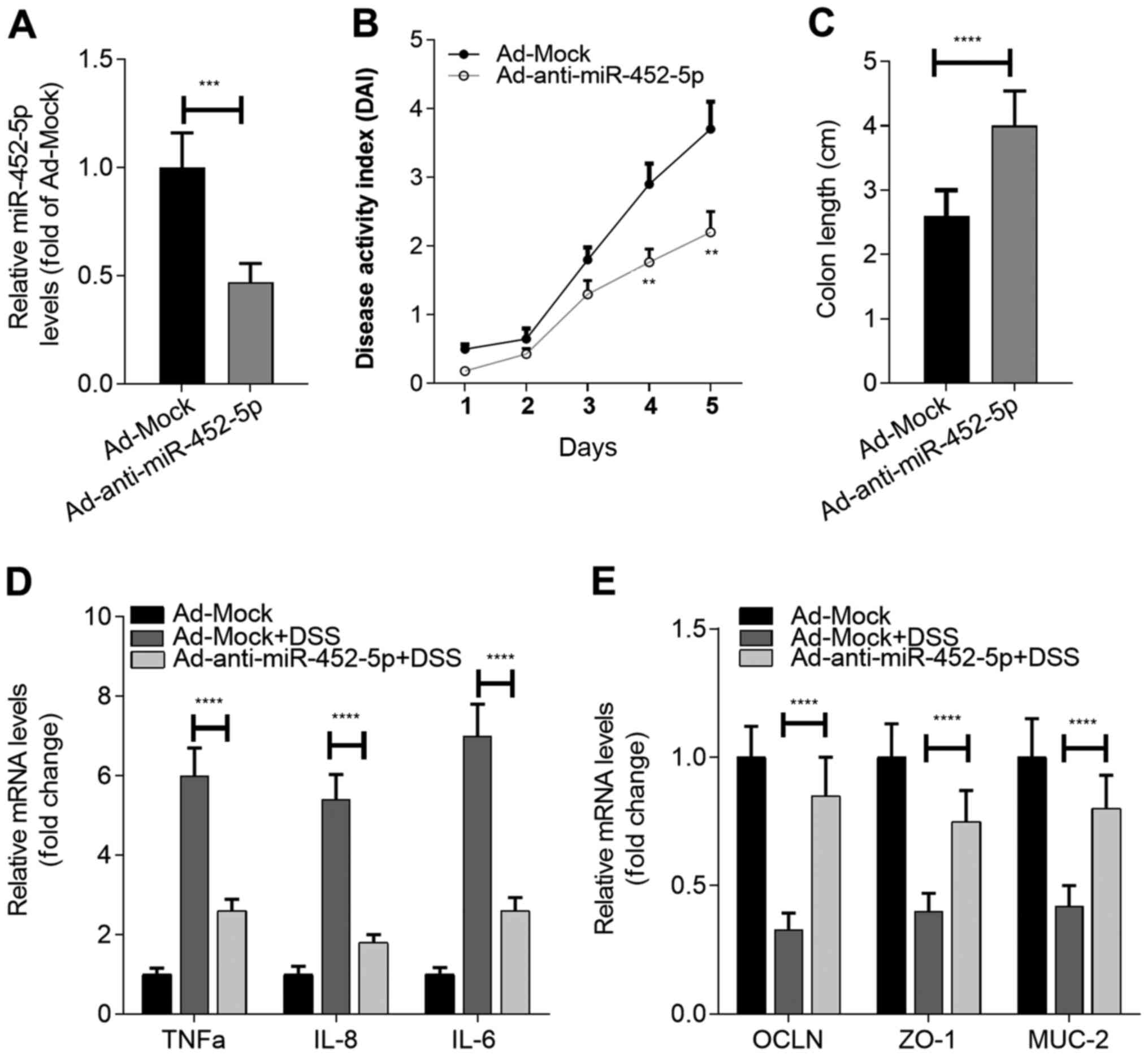 | Figure 2Inhibition of miR-452-5p alleviates
the symptoms of IBD. (A) The efficiency of miR-452-5p knockdown in
the IECs of mice. (B) Effect of miR-452-5p knockdown on the
development of IBD. (C) Effect of miR-452-5p knockdown on colon
shortening during IBD. Effects of miR-452-5p knockdown on the RNA
expression of (D) inflammatory cytokines and (E)
integrity-associated molecules in IECs during IBD.
**P<0.01, ***P<0.001 and
****P<0.0001 vs. Ad-Mock or as indicated. miR,
microRNA; IBD, inflammatory bowel disease; IECs, intestinal
epithelial cells; OCLN, occludin; ZO-1, zona occludens 1; MUC-2,
mucin-2; Ad, adenovirus; DDW, double-distilled water; DSS, dextran
sulfate sodium. |
miR-452-5p negatively regulates the
expression of Mcl-1 in IECs
The starBase analysis revealed that numerous mRNAs,
including Mcl-2, contain putative binding sites for miR-452-5p.
Based on the potential role of Mcl-1 in IBD (27), the relationship between Mcl-1 and
miR-452-5p in colitis was explored. The results revealed that the
expression of Mcl-1 was significantly downregulated in the IECs of
mice with DSS-induced colitis compared with those from the healthy
controls (Fig. 3A-C). Spearman's
correlation analysis (Fig. 3D)
revealed that miR-452-5p and Mcl-1 were negatively correlated,
suggesting that miR-452-5p negatively regulates the expression of
Mcl-1 in IECs. Consistent with this, anti-miR-452-5p significantly
promoted the expression of Mcl-1 in the IECs of the Ad-miR-452-5p
group compared with the Ad-Mock group (Fig. 3E and F).
To further confirm the effects of miR-452-5p on
Mcl-1 expression in IECs, miR-452-5p mimics and anti-miR-452-5p
were transfected into IEC-6 cells. The transfection efficiencies of
anti-miR-452-5p and miR-452-5p mimics in these IECs were tested
using RT-qPCR. The results demonstrated that the expression of
miR-452-5p was decreased by anti-miR-452-5p and elevated by
miR-452-5p mimics (Fig. S1A).
Further analysis revealed that miR-452-5p mimics significantly
inhibited the expression of Mcl-1 in the IEC-6 cell line, while the
knockdown of miR-452-5p had the opposite effect (Fig. 4A and B). Furthermore, a potential binding site
of miR-452-5p and the 3'UTR of Mcl-1 was predicted using the
starBase database. To determine whether miR-452-5p directly
regulates Mcl-1, luciferase reporter assays were performed in cells
containing the full-length 3'UTR of Wt Mcl-1. The miR-452-5p mimic
reduced the activity of the Mcl-1-Wt luciferase reporter whereas
anti-miR-452-5p increased luciferase reporter activity.
Furthermore, mutagenesis of the miR-452-5p binding site in the
Mcl-1 3'UTR abolished these effects (Fig. 4C and D). These results demonstrate that
miR-452-5p directly targets Mcl-1.
miR-452-5p/Mcl-1 axis influences the
responsiveness of IECs upon activation
The roles of the miR-452-5p/Mcl-1 axis in the
regulation of IEC function under activated conditions were further
analyzed. Consistent with the in vivo data, the RT-qPCR
results showed that increased levels of miR-452-5p were associated
with decreased levels of Mcl-1 upon LPS stimulation (Fig. 5A). Further experiments were
conducted by transfecting the IECs with sh-Mcl-1, the transfection
efficiency of which was tested by RT-qPCR. The results indicated
that Mcl-1 expression was successfully decreased by sh-Mcl-1
(Fig. S1B). As shown in Fig. 5B-H, treatment with LPS significantly
promoted the levels of TNFα, IL-6 and IL-8, inhibited the levels of
OCLN, ZO-1 and MUC-2, and increased apoptosis compared with those
in the control group. The inhibition of miR-452-5p significantly
inhibited the expression of TNFα, IL-6 and IL-8 induced by LPS
activation, while the knockdown of Mcl-1 abrogated the effects of
miR-452-5p inhibition on these inflammatory cytokines in IECs
(Fig. 5B and D). Furthermore, the inhibition of
miR-452-5p reversed the downregulation of OCLN, ZO-1 and MUC-2
induced by LPS stimulation, and Mcl-1 knockdown abolished these
effects (Fig. 5C and E). As Mcl-1 also has effects on apoptosis,
whether the miR-452-5p/Mcl-1 axis modulates IEC apoptosis was
examined. The results showed that miR-452-5p knockdown inhibited
IEC apoptosis and Mcl-1 knockdown attenuated this effect,
indicating that miR-452-5p promotes the apoptosis of IECs by
inhibiting Mcl-1 expression (Fig.
5F and G). These data indicate
that miR-452-5p regulates the responsiveness of IECs to LPS
activation in an Mcl-1-dependent manner.
Collectively, these data demonstrate that miR-452-5p
negatively regulates the expression of Mcl-1 in IECs, exacerbating
the progression of colitis. This information will help with
understanding the pathology of IBD and may facilitate improvements
in the efficacy of clinical strategies.
Discussion
IBDs are chronic inflammatory disorders that affect
intestinal tissues, and fully understanding IBD pathology is
critical for the development of efficient strategies to combat
these conditions (1). Previous
studies have demonstrated that IECs serve critical roles in the
pathogenesis of IBDs (28,29). In the present study, it was
demonstrated that upon the induction of UC in mice using DSS, IECs
expressed increased levels of inflammatory cytokines, namely TNFα,
IL-8 and IL-6, and reduced levels of intestinal
integrity-associated molecules, namely OCLN, ZO-1 and MUC-2. These
observations indicate that the function of the IEC barrier is
largely impaired during IBD progression, and these dysfunctional
IECs also participate in the initiation and maintenance of chronic
inflammation. Thus, developing therapies to target IECs holds great
potential for the treatment of IBD.
In the present study, RT-qPCR analysis revealed that
the expression of miR-452-5p was upregulated in the mouse model of
UC and in LPS-treated IECs. The data also demonstrated that
miR-452-5p is an important regulator of the expression of
inflammatory cytokines and intestinal integrity-associated
molecules. Furthermore, the knockdown of miR-452-5p significantly
alleviated the symptoms of IBD in the mouse model. These results
indicate that miR-452-5p plays a key role in the pathogenesis of
IBD. Thus, further investigations to elucidate the function of
other miRNAs in IECs or other types of intestinal cells are likely
to provide more potential therapeutic targets for IBD
treatment.
Mcl-1 is a member of the Bcl-2 family and is
associated with apoptosis (30). A
previous study indicated that the intestinal pathology associated
with IEC-specific Mcl-1 deficiency exhibited hallmark features of
IBD, including barrier dysfunction, chronic inflammation, increased
IEC apoptosis, hyperproliferation and impaired IEC differentiation,
demonstrating the crucial role of Mcl-1 in the maintenance of
intestinal homeostasis (27). Mcl-1
has also been reported to be downregulated in tissue samples from
patients with fibrotic CD (31),
suggesting a potential role of Mcl-1 in IBD. Additionally, Mcl-1
has been shown to be involved in the regulation of the LPS-induced
inflammatory response. For example, in one study, Mcl-1
overexpression alleviated LPS-induced IL-1β, IL-6, IL-8, and TNFα
expression in ATDC5 murine chondrogenic cells (32). In another study, Mcl-1 knockdown
promoted LPS-induced apoptosis and the release of inflammatory
cytokines in C28/I2 chondrocytes (33). In the present study, bioinformatics
analysis predicted that Mcl-1 contains a binding site for
miR-452-5p, and a luciferase reporter assay confirmed this binding
capacity. Furthermore, the overexpression of miR-452-5p inhibited
the expression of Mcl-1 in IECs, and the knockdown of Mcl-1
abrogated the effects of anti-miR-452-5p on the expression of
inflammatory cytokines and integrity-associated molecules. Although
the present study confirmed that miR-452-5p regulates the
expression of Mcl-1, further investigation into whether other
molecules and signaling pathways are regulated by miR-452-5p and
participate in the miR-452-5p-mediated regulation of IECs is
merited.
By comparing the levels of IEC-expressed miR-452-5p
between normal mice and mice with experimental UC, the present
study indicated that miR-452-5p may represent a promising target
for IBD treatment. The knockdown of miR-452-5p in the IECs of the
mice demonstrated that miR-452-5p inhibition significantly relieved
the symptoms of IBD. Furthermore, the data suggested that
miR-452-5p promoted inflammation and impaired intestinal integrity
by negatively regulating the expression of Mcl-1 in IECs, as the
knockdown of Mcl-1 abrogated the effects of miR-452-5p knockdown in
IECs in vitro. Overall, the present study demonstrated that
miR-452-5p regulates the responsiveness of IECs in IBD by
inhibiting Mcl-1 expression. These findings provide new information
on the pathogenesis of IBD and may be of benefit to future clinical
treatments.
Supplementary Material
Examination of transfection efficiency
in IECs. (A) Knockdown efficiency of anti-miR-452-5p and
overexpression efficiency of miR-452-5p vector in IEC-6 cells was
tested by RT-qPCR. (B) Knockdown efficiency of sh-Mcl-1 in IEC-6
cells was examined by RT-qPCR. ****P<0.0001. IECs,
intestinal epithelial cells; miR, microRNA; NC, negative control;
sh, short hairpin; Mcl-1, myeloid cell leukemia 1; RT-qPCR, reverse
transcription-quantitative PCR.
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
MD performed the experiments and analyzed the data.
JH, RT, HG and XL helped to perform the experiments and analyzed
the data. YL designed the study, wrote the manuscript and provided
material support. MD and YL confirmed the authenticity of all the
raw data. All the authors read and approved the final
manuscript.
Ethics approval and consent to
participate
The study was approved by the Animal Experimentation
Ethics Committee of Huazhong University of Science and
Technology.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Uhlig HH and Powrie F: Translating
immunology into therapeutic concepts for inflammatory bowel
disease. Annu Rev Immunol. 36:755–781. 2018.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Ungaro R, Mehandru S, Allen PB,
Peyrin-Biroulet L and Colombel JF: Ulcerative colitis. Lancet.
389:1756–1770. 2017.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Torres J, Mehandru S, Colombel JF and
Peyrin-Biroulet L: Crohn's disease. Lancet. 389:1741–1755.
2017.PubMed/NCBI View Article : Google Scholar
|
|
4
|
GBD 2017 Inflammatory Bowel Disease
Collaborators. The global, regional, and national burden of
inflammatory bowel disease in 195 countries and territories,
1990-2017: A systematic analysis for the Global Burden of Disease
Study 2017. Lancet Gastroenterol Hepatol. 5:17–30. 2020.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Hegyi P, Maleth J, Walters JR, Hofmann AF
and Keely SJ: Guts and gall: Bile acids in regulation of intestinal
epithelial function in health and disease. Physiol Rev.
98:1983–2023. 2018.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Saavedra PHV, Huang L, Ghazavi F, Kourula
S, Vanden Berghe T, Takahashi N, Vandenabeele P and Lamkanfi M:
Apoptosis of intestinal epithelial cells restricts Clostridium
difficile infection in a model of pseudomembranous colitis. Nat
Commun. 9(4846)2018.PubMed/NCBI View Article : Google Scholar
|
|
7
|
VanDussen KL, Stojmirovic A, Li K, Liu TC,
Kimes PK, Muegge BD, Simpson KF, Ciorba MA, Perrigoue JG, Friedman
JR, et al: Abnormal small intestinal epithelial microvilli in
patients with Crohn's disease. Gastroenterology. 155:815–828.
2018.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Geng H, Bu HF, Liu F, Wu L, Pfeifer K,
Chou PM, Wang X, Sun J, Lu L, Pandey A, et al: In inflamed
intestinal tissues and epithelial cells, interleukin 22 signaling
increases expression of H19 long noncoding RNA, which promotes
mucosal regeneration. Gastroenterology. 155:144–155.
2018.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Peterson LW and Artis D: Intestinal
epithelial cells: Regulators of barrier function and immune
homeostasis. Nat Rev Immunol. 14:141–153. 2014.PubMed/NCBI View
Article : Google Scholar
|
|
10
|
Turner JR: Intestinal mucosal barrier
function in health and disease. Nat Rev Immunol. 9:799–809.
2009.PubMed/NCBI View
Article : Google Scholar
|
|
11
|
Pickert G, Neufert C, Leppkes M, Zheng Y,
Wittkopf N, Warntjen M, Lehr HA, Hirth S, Weigmann B, Wirtz S, et
al: STAT3 links IL-22 signaling in intestinal epithelial cells to
mucosal wound healing. J Exp Med. 206:1465–1472. 2009.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Sugimoto K, Ogawa A, Mizoguchi E,
Shimomura Y, Andoh A, Bhan AK, Blumberg RS, Xavier RJ and Mizoguchi
A: IL-22 ameliorates intestinal inflammation in a mouse model of
ulcerative colitis. J Clin Invest. 118:534–544. 2008.PubMed/NCBI View
Article : Google Scholar
|
|
13
|
Wang Y, Mumm JB, Herbst R, Kolbeck R and
Wang Y: IL-22 increases permeability of intestinal epithelial tight
junctions by enhancing claudin-2 expression. J Immunol.
199:3316–3325. 2017.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Garofalo M and Croce CM: microRNAs: Master
regulators as potential therapeutics in cancer. Annu Rev Pharmacol
Toxicol. 51:25–43. 2011.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Visone R, Petrocca F and Croce CM:
Micro-RNAs in gastrointestinal and liver disease. Gastroenterology.
135:1866–1869. 2008.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Mohammadnia-Afrouzi M, Hosseini AZ,
Khalili A, Abediankenari S, Amari A, Aghili B and Nataj HH: Altered
microRNA expression and immunosuppressive cytokine production by
regulatory T cells of ulcerative colitis patients. Immunol Invest.
45:63–74. 2016.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Wu F, Guo NJ, Tian H, Marohn M, Gearhart
S, Bayless TM, Brant SR and Kwon JH: Peripheral blood microRNAs
distinguish active ulcerative colitis and Crohn's disease. Inflamm
Bowel Dis. 17:241–250. 2011.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Zhao Y, Ma T, Chen W, Chen Y, Li M, Ren L,
Chen J, Cao R, Feng Y, Zhang H and Shi R: MicroRNA-124 promotes
intestinal inflammation by targeting aryl hydrocarbon receptor in
Crohn's disease. J Crohns Colitis. 10:703–712. 2016.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Shi T, Xie Y, Fu Y, Zhou Q, Ma Z, Ma J,
Huang Z, Zhang J and Chen J: The signaling axis of
microRNA-31/interleukin-25 regulates Th1/Th17-mediated inflammation
response in colitis. Mucosal Immunol. 10:983–995. 2017.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Zhai W, Li S, Zhang J, Chen Y, Ma J, Kong
W, Gong D, Zheng J, Xue W and Xu Y: Sunitinib-suppressed miR-452-5p
facilitates renal cancer cell invasion and metastasis through
modulating SMAD4/SMAD7 signals. Mol Cancer. 17(157)2018.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Yamashita A, Inamine T, Suzuki S, Fukuda
S, Unoike M, Kawafuchi Y, Machida H, Isomoto H, Nakao K and
Tsukamoto K: Genetic variants of SMAD2/3/4/7 are associated with
susceptibility to ulcerative colitis in a Japanese genetic
background. Immunol Lett. 207:64–72. 2019.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Means AL, Freeman TJ, Zhu J, Woodbury LG,
Marincola-Smith P, Wu C, Meyer AR, Weaver CJ, Padmanabhan C, An H,
et al: Epithelial Smad4 deletion up-regulates inflammation and
promotes inflammation-associated cancer. Cell Mol Gastroenterol
Hepatol. 6:257–276. 2018.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Klausen P, Karstensen JG, Coskun M,
Săftoiu A, Vilmann P, Cowland JB and Riis LB: SMAD4 protein
expression is downregulated in ileal epithelial cells from patients
with Crohn's disease with significant inverse correlation to
disease activity. Gastroenterol Res Pract.
2018(9307848)2018.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Roulis M, Armaka M, Manoloukos M,
Apostolaki M and Kollias G: Intestinal epithelial cells as
producers but not targets of chronic TNF suffice to cause murine
Crohn-like pathology. Proc Natl Acad Sci USA. 108:5396–5401.
2011.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Ito Y, Inoue A, Seers T, Hato Y, Igarashi
A, Toyama T, Taganov KD, Boldin MP and Asahara H: Identification of
targets of tumor suppressor microRNA-34a using a reporter library
system. Proc Natl Acad Sci USA. 114:3927–3932. 2017.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Healy ME, Boege Y, Hodder MC, Böhm F,
Malehmir M, Scherr AL, Jetzer J, Chan LK, Parrotta R, Jacobs K, et
al: MCL1 is required for maintenance of intestinal homeostasis and
prevention of carcinogenesis in mice. Gastroenterology.
159:183–199. 2020.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Ramanan D and Cadwell K: Intrinsic defense
mechanisms of the intestinal epithelium. Cell Host Microbe.
19:434–441. 2016.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Odenwald MA and Turner JR: The intestinal
epithelial barrier: A therapeutic target? Nat Rev Gastroenterol
Hepatol. 14:9–21. 2017.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Kotschy A, Szlavik Z, Murray J, Davidson
J, Maragno AL, Le Toumelin-Braizat G, Chanrion M, Kelly GL, Gong
JN, Moujalled DM, et al: The MCL1 inhibitor S63845 is tolerable and
effective in diverse cancer models. Nature. 538:477–482.
2016.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Nijhuis A, Curciarello R, Mehta S, Feakins
R, Bishop CL, Lindsay JO and Silver A: MCL-1 is modulated in
Crohn's disease fibrosis by miR-29b via IL-6 and IL-8. Cell Tissue
Res. 368:325–335. 2017.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Wang Y and Kong D: MicroRNA-136 promotes
lipopolysaccharide-induced ATDC5 cell injury and inflammatory
cytokine expression by targeting myeloid cell leukemia 1. J Cell
Biochem. 119:9316–9326. 2018.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Zhao C, Wang Y, Jin H and Yu T: Knockdown
of microRNA-203 alleviates LPS-induced injury by targeting MCL-1 in
C28/I2 chondrocytes. Exp Cell Res. 359:171–178. 2017.PubMed/NCBI View Article : Google Scholar
|



















