Introduction
Animal chronic pain models are usually classified as
peripheral nerve injury models or inflammatory pain models
(1). Both nerve injury and
inflammation can produce spontaneous pain, hyperalgesia and
abnormal pain, and both can affect spontaneous discharges from the
dorsal root ganglia (DRG) (2,3).
However, the potential mechanisms of DRG discharge differ between
nerve injury and chronic inflammation. Nerve injury typically leads
to overall downregulation of sodium channels and altered homotype
expression (4). By contrast,
chronic peripheral inflammation generally leads to the upregulation
of tetrodotoxin-resistant sodium channels (5). Inflammation is a factor in most pain
models, including those based on nerve injury (6). Macrophage infiltration, local
pro-inflammatory cytokine release (7), DRG glial cell activation and
retrograde transport to DRG are important triggers of hyperalgesia
(8). Inflammation also occurs under
clinical pain conditions, including postherpetic neuralgia and back
pain after lumbar disc herniation (9,10); the
substance released from the nucleus pulposus is immunogenic,
causing inflammation in adjacent DRGs. To learn more concerning the
contribution of inflammation to pathological pain, Wang et
al (11) developed the
localized inflammation of the DRG (LID) model. In this model the
cell bodies of sensory neurons are directly stimulated by the
immune activator zymosan, without nerve injury. Considering the
association between inflammatory processes and states of
inflammatory and neuropathic chronic pain, the present study aimed
to explore gene activity and expression changes induced by local
inflammation of DRG.
Strong et al (12) submitted the GSE38859 dataset to the
Gene Expression Omnibus (GEO) database. They screened
behavior-related gene expression changes after DRG inflammation and
demonstrated that immune-related genes were the largest category
altered, including members of the complement system and several
upregulated chemokine ligands, such as C-X-C motif ligand (CXCL)9,
CXCL10 and CXCL16(12). However,
their study only focused on gene function and the role of numerous
differentially expressed genes (DEGs) was not explored further. In
the present study, in order to identify the key candidate genes and
pathway changes in DRG inflammation various bioinformatics
technologies were used to reanalyze the microarray data in the GEO
database. CXCL9, complement component 3 (C3), and matrix
metallopeptidase 9 (MMP9) were found to have strong interactive
relationships with other genes, suggesting that they may be
potential targets for the treatment of DRG inflammation-induced
pain. These findings may provide greater insight into the genetic
mechanisms underlying DRG inflammation pain and present potential
therapeutic targets for the treatment of lower back pain.
Materials and methods
Microarray data set collection and
identification of DEGs
The microarray expression dataset GSE38859 was
obtained from the Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo/). Exon expression
profiling was based on the Agilent GPL6543 platform (Affymetrix Rat
Exon 1.0 ST Array) and provided 6 sham and 6 inflamed DRG tissues.
The probes were converted to the corresponding gene symbols
according to the annotation information in the raw data. To reduce
multiple testing, each corresponded to a unique gene symbol and
only the probe set with the highest average expression was
considered when multiple probe sets were associated with the same
gene. Pre-treatment standardization on gene expression data for
each experiment was performed using the R/Bioconductor Limma
package. R is the language and operating environment for
statistical analysis and drawing. DEGs were uploaded to omicstudio
(https://www.omicstudio.cn/tool?order=complex), a
visual analytics platform for principal component analysis. After
linear model fitting, the Bayesian linear model of the limma
package was estimated to identify DEGs. Statistically significant
DEGs were defined with P<0.05 and |logFC|>1 as a cut-off
criterion. Heatmap and volcano plots visualizations were performed
using the R packages ‘pheatmap’ and ‘ggplot2’, respectively.
Enrichment analyses of DEGs
In the present study, Gene Ontology (GO) enrichment
and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis
of DEGs were carried out using the R package. P<0.01 was chosen
as the cutoff criteria. Gene Set Enrichment Analysis (GSEA) was
also performed using the ‘clusterProfiler’ package in R (13), and all visualization was handled in
R using the ggplot2 graphics package. The whole gene expression
values of the samples were analyzed based on the h.all.v
7.0.entrez.gmt [Hallmarks] gene set database (https://www.gsea-msigdb.org/gsea/msigdb/collections.jsp#H).
Significant enrichment pathways were defined by FDR <0.25 and
P<0.05.
Module screening from the
protein-protein interaction (PPI) network
Comprehensive information on the proteins was
identified and the Search Tool for Retrieval of Interacting Genes
(STRING; v11.0; https://string-db.org/), a search tool for retrieving
interacting genes/proteins, was used to evaluate protein-PPI
information. Interaction between proteins within a cell facilitates
our understanding of how proteins operate in a coordinated manner
in the cell (14). Subsequently,
the PPI network was constructed and visualized by Cytoscape
software (version 3.7.1; https://cytoscape.org/). Molecular Complex Detection
(MCODE) analysis, an app in Cytoscape, was then used to select the
most significant PPI network modules. The criteria for selection
were as follows: MCODE score >3; degree cutoff, 2; node score
cut-off, 0.2; and max depth, 100.
Animals and local inflammation of the
DRG (LID) models
Adult male Sprague Dawley rats purchased from the
Experimental Animal Center of Zhejiang Province were used in this
study. Rats used for experiments were aged 6-8 weeks, weighed
250-350 g and were sex-matched. A total of 16 rats were used in
this present study. A sample size of 8 rats was used per experiment
to ensure repeatability. The animals were housed in a
temperature-controlled animal facility (room temperature, 25˚C;
humidity, 40-60%) on a 12 h light–dark cycle and food and water was
freely available. All procedures were approved by the Animal Care
and Use Committee of Ningbo University following the Guidelines for
the Care and Use of Laboratory Animals from the National Institutes
of Health (NIH) (15).
An intraperitoneal injection was used to anesthetize
the selected animals with sodium pentobarbital (50 mg/kg), before a
longitudinal incision was made in the middle of the S1 to L4 spine.
The back skin and muscular fasciae were bluntly isolated, exposing
the L5 intervertebral foramen. With the needle still inside, 10 µl
of the immune activator zymosan (2 mg/ml in incomplete Freund's
adjuvant; Sigma-Aldrich; Merck KGaA) was slowly injected into the
L5 intervertebral foramen, above the DRG. The needle remained in
place for an additional 3 min after injection to avoid leakage. A
control group, sham animals, experienced the same surgery process
without the final step of injecting zymosan.
Reverse transcription-quantitative PCR
(RT-qPCR)
RT-qPCR analysis was performed as previously
described (16). After 3 days
following LID- or sham- surgery, rats were decapitated following an
overdose of pentobarbital sodium (150 mg/kg). Freshly isolated DRG
tissues were collected on ice and immersed in TRIzol®
reagent (Invitrogen; Thermo Fisher Scientific, Inc.) and
immediately stored at -80˚C until the time of RNA extraction.
Complementary DNA (cDNA) was synthesized with the reverse
transcription enzyme SuperScript II (Invitrogen; Thermo Fisher
Scientific, Inc.) together with reverse transcription primers at
50˚C for 15 min and 85˚C for 5 sec. cDNA was then amplified using a
HiFiScript cDNA Synthesis Kit (CoWin Biosciences) using an ABI Q5
RT-PCR System (Applied Biosystems; Thermo Fisher Scientific, Inc.).
All primers were synthesized by BGI Genomics Co., Ltd. (Table I). The synthesized cDNA was used
qPCR to determine the expression changes in corresponding genes.
The reaction mixture was: 10 µl 2X SYBR Premix Ex Taq TMII, 1 µl 10
µM forward primer, 1 µl 10 µM reverse primer, 7 µl ddH2O and 1 µl
cDNA, in a total volume of 20 µl. The thermocycling conditions
were: 95˚C for 2 min, followed by 40 cycles of 95˚C for 10 sec,
60˚C for 50 sec. GAPDH or Actin were used as internal reference
genes, and the relative expression changes in target genes were
calculated by the 2-∆∆Cq method (17).
 | Table IPrimer sequences used in the present
study. |
Table I
Primer sequences used in the present
study.
| Gene | Sequence |
|---|
| CXCL9 | F:
5'-GACCCAGATTCAGCAAGGGT-3' |
| | R:
5'-CTTTGACTCCGGATGGTGGG-3' |
| MMP-9 | F:
5'-GGTGATTGACGACGCCTTTG-3' |
| | R:
5'-CTGGATGACGATGTCTGCGT-3' |
| C3 | F:
5'-GCGGTACTACCAGACCATCG-3' |
| | R:
5'-CTTCTGGCACGACCTTCAGT-3' |
| GAPDH | F:
5'-AAGGTCGGTGAACGGATT-3' |
| | R:
5'-TGAACTTGCCGTGGGTAGAG-3' |
Western blotting
The fresh tissues of mice were lysed in precooled
RIPA buffer (EMD Millipore), and then homogenized using an
automatic rapid sample grinder. The tissue homogenate was
transferred to a sterile centrifuge tube, centrifuged at 4˚C and
10,000 x g for 10 min, and the supernatant was transferred to a new
centrifuge tube. The protein concentration was determined using the
BCA method (Thermo Fisher Scientific, Inc.). Total protein (20-30
µg) was separated via SDS-PAGE electrophoresis (concentrated gel,
5%; separation gel, 10-12%). After SDS-PAGE electrophoresis, the
electrophoresis was carried out at a constant current of 200 mA for
2 h. At the end of membrane transfer, the nitrocellulose membrane
was rinsed with TBS-0.02% Tween-20 (TBST) and sealed at room
temperature for 1-2 h with blocking solution (TBST containing 3-5%
skimmed milk powder). Rabbit anti-C3 (cat. no. 97425; Cell
Signaling Technology, Inc.; 1:1,000), mouse anti-actin (cat. no.
3700; Cell Signaling Technology, Inc.; 1:10,000), rabbit anti- MMP9
(cat. no. 13667; Cell Signaling Technology, Inc.; 1:1000) and mouse
anti-CXCL9 (cat. no. 93556; Cell Signaling Technology, Inc.;
1:1,000) were diluted with a primary antibody diluent [TBST
solution containing 3% BSA (Sigma-Aldrich; Merck KGaA) and 0.01%
sodium azide] and incubated at 4˚C for 15-18 h. Subsequently, the
membranes were washed five times with TBST for 5 min each time.
Horseradish peroxidase-conjugated secondary antibody (goat
anti-mouse IgG: cat. no. E030110-01; EarthOx, LLC; 1:10,000; goat
anti-rabbit IgG: cat. no. E030120-01; EarthOx, LLC; 1:10,000) was
diluted in TBST solution and incubated at room temperature for 2 h.
Subsequently, TBST was used to rinse the membrane 5 times and TBS
was used to rinse the membrane once, each time for 5 min. After
rinsing, SuperSignal™ West Atto Ultimate Sensitivity ECL substrate
(cat. no. A38555; Thermo Fisher Scientific, Inc.) was added and
then incubated in the dark for 1-5 min, before exposure to the
chemiluminescence imaging system. The protein expression detected
via western blotting was quantified using ImageJ v1.8.0 (NIH).
Statistical analysis
Data are presented as the mean ± SEM. Analyses were
performed using Microsoft Excel (version 16.44; Microsoft
Corporation). Student's t-test was used to analyze statistical
differences. P<0.05 was considered to indicate a statistically
significant difference.
Results
Data normalization
The primary purpose of normalization was to
eliminate technical and systematic variability from the data
compared between different samples. After microarray data
normalization, biological variability between different samples was
assessed by plotting a principal component analysis graph (Fig. 1A). DRG-inflamed (n=6) and DRG-sham
(n=6) overall had distinct, non-overlapping expression profiles.
The density plot results demonstrated that the distribution of the
sample intensities were generally consistent and could be used for
downstream analysis (Fig. 1B). A
box plot indicates each sample's gene expression and the black
lines in the boxes were almost on the same straight line,
indicating that the raw data were normalized successfully, which
ensures the accuracy of the data (Fig.
1C).
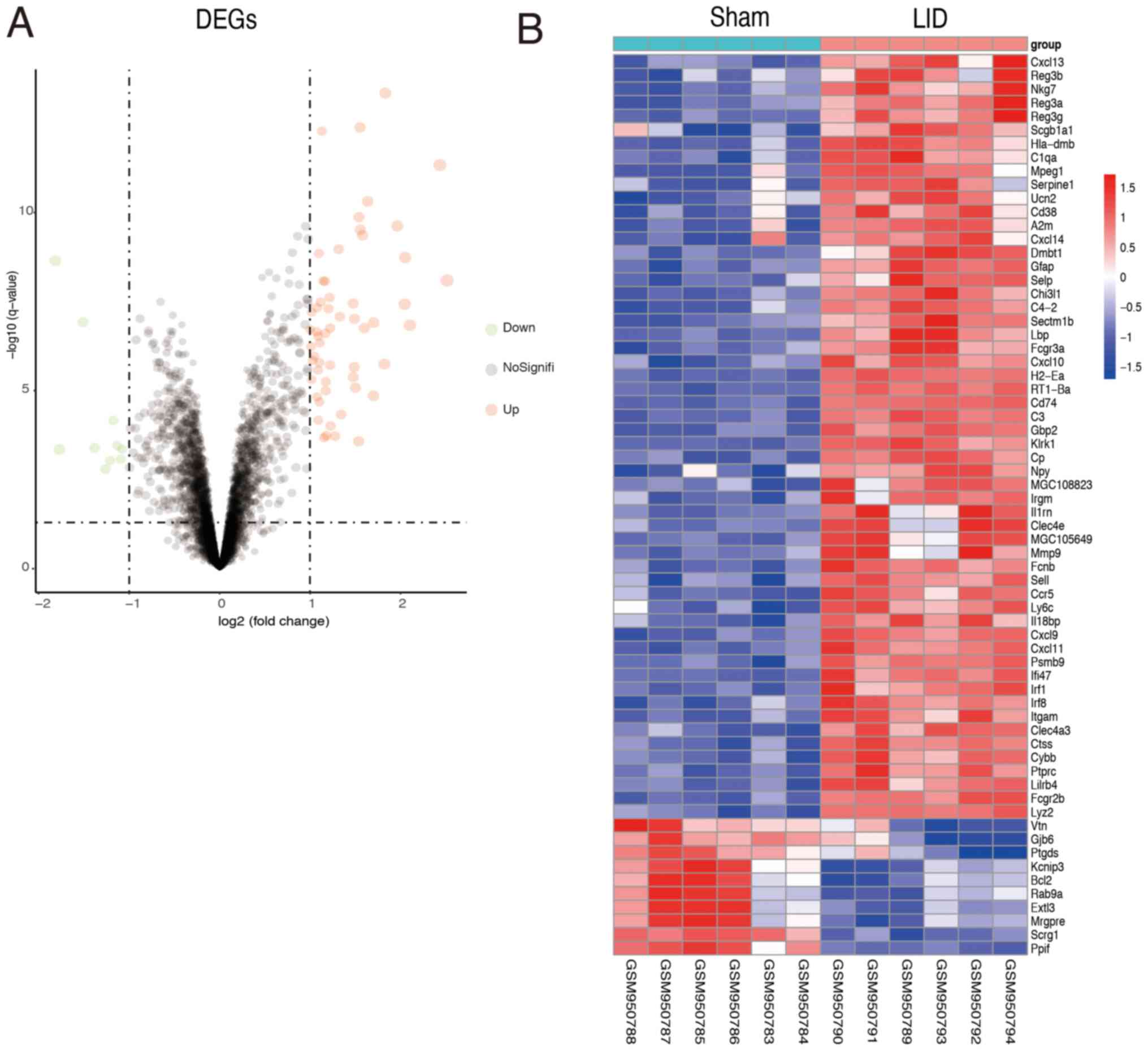
Identification of DEGs
A total of 66 DEGs were screened, including 56
upregulated and 10 downregulated DEGs. In addition, the volcano
plot of the DEGs is presented in Fig.
2A and heatmap plots in Fig.
2B.
Functional enrichment analysis of
DRGs
To further investigate the function of DEGs, GO term
and KEGG pathway analyses were displayed in R. DEGs were divided
into three major functional categories, namely biological processes
(BPs), molecular functions (MFs) and cellular components (CCs).
Collectively, the data showed that DEGs were associated with GO
terms related to immunity, including ‘innate immune response’,
‘activation of immune response’ and ‘activation of inflammatory
response’ (Fig. 3A and Table II). In the MF category, DEGs were
enriched in the terms ‘carbohydrate binding’, ‘G protein-coupled
receptor binding’ and ‘heparin binding’ (Fig. 3A and Table III). In the CC analysis, the DEGs
were enriched in the terms ‘external side of plasma membrane’,
‘side of membrane’ and ‘rough endoplasmic reticulum’ (Fig. 3A and Table IV). In these candidate DEGs, 14
signaling pathways were enriched in pathways in the KEGG database,
including ‘cytokine-cytokine receptor interaction’, ‘viral protein
interaction with cytokine and cytokine receptor’ and ‘chemokine
signaling pathway' pathways (Fig.
3B and Table V). In addition,
GSEA was performed for all genes on the microarray. The results of
GSEA suggested that the LID model expression profiles were enriched
in genes associated with the terms ‘interferon-gamma response’ and
‘inflammatory response’ (Fig.
3C).
 | Figure 3GO, KEGG and GSEA enrichment analysis
of the DEGs were performed using ClusterProfiler. (A) GO analysis
for LID models DEGs in the BP (top), CC (middle) and MF (bottom)
categories. (B) KEGG pathways enrichment analysis of DEGs. (C) GSEA
of a gene signature associated with the ‘interferon gamma response’
and the ‘inflammatory response’. GO, Gene Ontology; KEGG, Kyoto
Encyclopedia of Genes and Genomes; GSEA, Gene Set Enrichment
Analysis; DEGs, differentially expressed genes; LID, localized
inflammation of the dorsal root ganglion; BP, biological processes;
CC, cellular component; MF, molecular function. |
 | Table IITop 20 GO biological process terms
associated with differentially expressed genes. |
Table II
Top 20 GO biological process terms
associated with differentially expressed genes.
| Term | Description | Genes | Counts | P-value |
|---|
| GO:0032496 | Response to
lipopolysaccharide |
Gbp2/Klrk1/Fcgr2b/Cxcl9/Cxcl11/Ccr5/Lbp/Cxcl10/Irf8/Fcgr3a/Il18bp/Selp/Il1rn/Serpine1/Mmp9/Scgb1a1/Gjb6 | 16 |
5.41x10-15 |
| GO:0002237 | Response to
molecule of bacterial origin |
Gbp2/Klrk1/Fcgr2b/Cxcl9/Cxcl11/Ccr5/Lbp/Cxcl10/Irf8/Fcgr3a/Il18bp/Selp/Il1rn/Serpine1/Mmp9/Scgb1a1/Gjb6 | 16 |
1.03x10-14 |
| GO:0002685 | Regulation of
leukocyte migration |
Cd74/Klrk1/Cxcl9/Cxcl11/Lbp/Cxcl10/Sell/Selp/Cxcl13/Serpine1/Mmp9/Cxcl14 | 12 |
4.72x10-14 |
| GO:0050900 | Leukocyte
migration |
Cd74/Cxcl9/Cxcl11/Ptprc/Ccr5/Lbp/Cxcl10/Sell/Selp/Cxcl13/Serpine1/Mmp9/Cxcl14/Bcl2/Vtn | 16 |
4.81x10-14 |
| GO:0030335 | Positive regulation
of cell migration |
Cd74/Cxcl9/Cxcl11/Ptprc/Ccr5/Lbp/Cxcl10/Sell/Selp/Cxcl13/Serpine1/Mmp9/Cxcl14/Bcl2/Vtn | 15 |
7.88x10-13 |
| GO:0002688 | Regulation of
leukocyte chemotaxis |
Cd74/Klrk1/Cxcl9/Cxcl11/Lbp/Cxcl10/Sell/Cxcl13/Serpine1/Cxcl14 | 10 |
3.46x10-13 |
| GO:0098542 | Defense response to
other organism |
Gbp2/Klrk1/Cxcl9/Lyz2/Ptprc/Irf1/Ccr5/Lbp/Cxcl10/Irf8/Reg3g/Cxcl13/Serpine1/Clec4e/Reg3b/Bcl2 | 16 |
6.33x10-13 |
| GO:0045087 | Innate immune
response |
RT1-Ba/C3/Gbp2/Klrk1/Fcnb/Irf1/Cybb/Lbp/Reg3g/C1qa/Irgm/A2m/Clec4e | 13 |
2.01x10-9 |
| GO:2000147 | Positive regulation
of cell motility |
Cd74/Cxcl9/Cxcl11/Ptprc/Ccr5/Lbp/Cxcl10/Sell/Selp/Cxcl13/Serpine1/Mmp9/Cxcl14/Bcl2/Vtn | 15 |
1.19x10-12 |
| GO:0051272 | Positive regulation
of cellular component movement |
Cd74/Cxcl9/Cxcl11/Ptprc/Ccr5/Lbp/Cxcl10/Sell/Selp/Cxcl13/Serpine1/Mmp9/Cxcl14/Bcl2/Vtn | 15 |
1.65x10-12 |
| GO:0040017 | Positive regulation
of locomotion |
Cd74/Cxcl9/Cxcl11/Ptprc/Ccr5/Lbp/Cxcl10/Sell/Selp/Cxcl13/Serpine1/Mmp9/Cxcl14/Bcl2/Vtn | 15 |
2.44x10-12 |
| GO:0006935 | Chemotaxis |
Cd74/C3/Klrk1/Cxcl9/Cxcl11/Ccr5/Lbp/Cxcl10/Itgam/Sell/Cxcl13/Serpine1/Cxcl14 | 13 |
1.31x10-9 |
| GO:0060326 | Cell
chemotaxis |
Cd74/Klrk1/Cxcl9/Cxcl11/Ccr5/Lbp/Cxcl10/Itgam/Sell/Cxcl13/Serpine1/Cxcl14 | 12 |
3.59x10-12 |
| GO:0030595 | Leukocyte
chemotaxis |
Cd74/Klrk1/Cxcl9/Cxcl11/Lbp/Cxcl10/Itgam/Sell/Cxcl13/Serpine1/Cxcl14 | 11 |
2.76x10-12 |
| GO:0002253 | Activation of
immune response |
C3/Klrk1/Fcnb/Ptprc/Irf1/Lbp/Reg3g/C1qa/A2m/Cd38/Bcl2 | 11 |
8.13x10-10 |
| GO:0002687 | Positive regulation
of leukocyte migration |
Cd74/Cxcl9/Cxcl11/Lbp/Cxcl10/Sell/Selp/Cxcl13/Serpine1/Mmp9/Cxcl14 | 11 |
7.89x10-14 |
| GO:0042742 | Defense response to
bacterium |
Gbp2/Klrk1/Lyz2/Ccr5/Lbp/Irf8/Reg3g/Cxcl13/Serpine1/Clec4e/Reg3b | 11 |
5.31x10-11 |
| GO:0050921 | Positive regulation
of chemotaxis |
Cd74/Cxcl9/Cxcl11/Ccr5/Lbp/Cxcl10/Sell/Cxcl13/Serpine1/Cxcl14 | 10 |
5.20x10-12 |
| GO:0002526 | Acute inflammatory
response |
C3/Fcgr2b/Ccr5/Lbp/Reg3a/Reg3g/A2m/Il1rn/Reg3b | 9 |
1.70x10-10 |
| GO:0006953 | Acute-phase
response |
Ccr5/Lbp/Reg3a/Reg3g/A2m/Il1rn/Reg3b | 7 |
1.30x10-10 |
 | Table IIITop 20 GO molecular function terms
associated with differentially expressed genes. |
Table III
Top 20 GO molecular function terms
associated with differentially expressed genes.
| Term | Description | Genes | Counts | P-value |
|---|
| GO:0048248 | CXCR3 chemokine
receptor binding |
Cxcl9/Cxcl11/Cxcl10/Cxcl13 | 4 |
6.27x10-10 |
| GO:0030246 | Carbohydrate
binding |
Klrk1/Fcnb/Chi3l1/Reg3a/Reg3g/Sell/Selp/Clec4e/Reg3b/Vtn | 10 |
1.01x10-8 |
| GO:0001664 | G protein-coupled
receptor binding |
C3/Cxcl9/Cxcl11/Fcnb/Ccr5/Cxcl10/Cxcl13/Npy/Ucn2/Cxcl14 | 10 |
1.37x10-8 |
| GO:0042379 | Chemokine receptor
binding |
Cxcl9/Cxcl11/Ccr5/Cxcl10/Cxcl13/Cxcl14 | 6 |
1.39x10-8 |
| GO:0045236 | CXCR chemokine
receptor binding |
Cxcl9/Cxcl11/Cxcl10/Cxcl13 | 4 |
8.78x10-8 |
| GO:0008009 | Chemokine
activity |
Cxcl9/Cxcl11/Cxcl10/Cxcl13/Cxcl14 | 5 |
1.40x10-7 |
| GO:0008201 | Heparin
binding |
Cxcl11/Ptprc/Cxcl10/Itgam/Selp/Cxcl13/Vtn | 7 |
3.29x10-7 |
| GO:0005539 | Glycosaminoglycan
binding |
Cxcl11/Ptprc/Cxcl10/Itgam/Selp/Cxcl13/Vtn | 7 |
1.96x10-6 |
| GO:1901681 | Sulfur compound
binding |
Cxcl11/Ptprc/Cxcl10/Itgam/Selp/Cxcl13/Vtn | 7 | 1.08
x10-8 |
| GO:0019955 | Cytokine
binding |
Cd74/Ccr5/Il18bp/A2m/Il1rn | 5 |
1.13x10-8 |
| GO:0005126 | Cytokine receptor
binding |
Cxcl9/Cxcl11/Ccr5/Cxcl10/Cxcl13/Il1rn/Cxcl14 | 7 |
2.21x10-5 |
| GO:0022804 | Transmembrane
transporter activity |
Ctss/Ptprc/Ccr5/Itgam/Selp | 5 |
3.42x10-5 |
| GO:0005125 | Cytokine
activity |
Cxcl9/Cxcl11/Cxcl10/Cxcl13/Il1rn/Cxcl14 | 6 |
3.80x10-5 |
| GO:0043394 | Proteoglycan
binding |
Ctss/Ptprc/Itgam | 3 |
1.47x10-4 |
| GO:0019966 | Interleukin-1
binding | A2m/Il1rn | 2 |
1.73x10-4 |
| GO:0033691 | Sialic acid
binding | Fcnb/Selp | 2 |
2.41x10-4 |
| GO:0019864 | IgG binding | Fcgr2b/Fcgr3a | 2 |
6.27x10-4 |
| GO:0002020 | Protease
binding |
Sell/A2m/Serpine1/Bcl2 | 4 |
6.98x10-4 |
| GO:0042165 | Neurotransmitter
binding |
Chrna2/Chat/Chrna1/Slc6a12 | 4 |
7.87x10-4 |
| GO:0015294 | Solute: cation
symporter activity |
Slc45a3/Slc28a1/Slc13a3/Slc13a4/Slc6a12 | 5 |
8.38x10-4 |
 | Table IVTop 20 GO cellular components terms
associated with differentially expressed genes. |
Table IV
Top 20 GO cellular components terms
associated with differentially expressed genes.
| Term | Description | Genes | Counts | P-value |
|---|
| GO:0009897 | External side of
plasma membrane |
RT1-Ba/Cd74/Klrk1/Fcgr2b/Cxcl9/Fcnb/Ptprc/Ccr5/Cxcl10/Itgam/Fcgr3a/Sell/Selp | 13 | 4.42
x10-4 |
| GO:0098552 | Side of
membrane |
RT1-Ba/Cd74/Klrk1/Fcgr2b/Cxcl9/Fcnb/Ptprc/Ccr5/Cxcl10/Itgam/Fcgr3a/Sell/Selp | 6 |
5.88x10-4 |
| GO:0005791 | Rough endoplasmic
reticulum |
Lyz2/Cybb/Scgb1a1/Vtn/Ptgds | 5 |
7.82x10-4 |
| GO:0030670 | Phagocytic vesicle
membrane |
Dmbt1/Irgm/Rab9a | 3 |
1.13x10-3 |
| GO:0048237 | Rough endoplasmic
reticulum lumen | Lyz2/Vtn | 2 |
1.17x10-3 |
| GO:0030666 | Endocytic vesicle
membrane |
Dmbt1/Irgm/Rab9a | 3 |
1.17x10-3 |
| GO:0072562 | Blood
microparticle | C3/Cp/A2m/Vtn | 3 |
1.17x10-3 |
| GO:0030141 | Secretory
granule |
Lyz2/Selp/Dmbt1/Serpine1/Reg3b/Scgb1a1 | 6 |
1.78x10-3 |
| GO:0062023 | Collagen-containing
extracellular matrix |
Igf1/Elane/Srpx2/Loxl1/Colec12/Igfbp6/Col9a2/Omd/Lamc2/Adamts2/Col8a2 | 11 |
2.11x10-3 |
| GO:0030016 | Myofibril |
Myo18b/Rpl4/Nrap/Tnnt2/Myh2/Ryr1/Capn3/Tnnc2 | 8 |
2.33x10-3 |
| GO:0005861 | Troponin
complex | Tnnt2/Tnnc2 | 2 |
3.19x10-3 |
| GO:0043292 | Contractile
fiber |
Myo18b/Rpl4/Nrap/Tnnt2/Myh2/Ryr1/Capn3/Tnnc2 | 8 |
3.45x10-3 |
| GO:0070382 | Exocytic
vesicle |
Hspa8/Igf1/Syt10/Sept1/Cplx3/Sphk1/Sytl1/Wfs1 | 8 |
5.87x10-3 |
| GO:0002177 | Manchette | Spef2/Iqcg | 2 |
6.14x10-3 |
| GO:0043198 | Dendritic
shaft |
Hspa8/Hcn1/Rgs7bp/Ntsr1 | 4 |
6.25x10-3 |
| GO:0098684 | Photoreceptor
ribbon synapse | Hspa8/Cplx3 | 2 |
7.32x10-3 |
| GO:0099026 | Anchored component
of presynaptic membrane | Rgs7bp/Cplx3 | 2 |
7.32x10--3 |
| GO:0001772 | Immunological
synapse |
Cd3e/Rhoh/Zap70 | 3 |
8.22x10-3 |
| GO:0030133 | Transport
vesicle |
Hspa8/Igf1/Syt10/Sept1/Lyz1/Cplx3/Sphk1/Sytl1/Wfs1 | 9 |
9.49x10-3 |
| GO:0042101 | T cell receptor
complex | Cd3e/Zap70 | 2 |
1.14x10-3 |
 | Table VKEGG pathway analysis. |
Table V
KEGG pathway analysis.
| ID | Description | Genes | Counts | P-value |
|---|
| rno05152 | Tuberculosis |
RT1-Ba/Cd74/C3/Fcgr2b/Ctss/Lbp/Itgam/Fcgr3a/Clec4e/Bcl2 | 10 |
1.56x10-8 |
| rno05150 | Staphylococcus
aureus infection |
RT1-Ba/C3/Fcgr2b/Itgam/Fcgr3a/Selp/C1qa | 7 |
6.21x10-7 |
| ron04145 | Phagosome |
RT1-Ba/C3/Fcgr2b/Itgam/Fcgr3a/Ctss/Cybb | 7 |
3.44x10-6 |
| rno04060 | Cytokine-cytokine
receptor interaction |
Cxcl9/Cxcl11/Ccr5/Cxcl10/Cxcl13/Il1rn/Cxcl14 | 7 |
2.82x10-5 |
| rno04061 | Viral protein
interaction with cytokine and cytokine receptor |
Cxcl9/Cxcl11/Ccr5/Cxcl10/Cxcl13/Cxcl14 | 6 |
3.65x10-5 |
| rno04062 | Chemokine signaling
pathway |
Cxcl9/Cxcl11/Ccr5/Cxcl10/Cxcl13/Cxcl14 | 6 |
4.14x10-5 |
| rno05133 | Pertussis |
C3/Irf1/Irf8/Itgam/C1qa | 5 |
6.53x10-5 |
| rno05140 | Leishmaniasis |
RT1-Ba/C3/Cybb/Itgam/Fcgr3a | 5 |
2.26x10-4 |
| rno04610 | Complement and
coagulation cascades |
C3/Itgam/C1qa/Serpine1/Vtn | 5 |
2.62x10-4 |
| rno05145 | Toxoplasmosis |
RT1-Ba/Ccr5/Ppif/Irgm/Bcl2 | 5 |
3.00x10-4 |
PPI network and module analysis
To investigate interaction between the DEGs, a PPI
network was constructed in STRING, which consisted of 182 edges and
62 nodes. STRING analysis showed that a total of 54 genes were
filtered into the DEG PPI network complex. The network was
visualized using the software tool Cytoscape (Fig. 4A). Moreover, three significant
models were screened out from the PPI network by module analysis
using MCODE in Cytoscape (Fig. 4A
and Table VI). A total of 9 genes
(MMP9, CXCL3, C3, Ptprc, CXCL11, CXCL9, CCr5, CXCL10 and C1qa) were
screened out based on the high degree of connectivity (≥10;
Table VI). Furthermore, MMP9,
CXCL3 and C3 overlapped within the top 10 DEGs. The sham DRG on the
third day was compared with the LID DRG to verify the relevance of
the screened genes via RT-qPCR. This time point was selected due to
the frequent spontaneous activity of the DRG. Consistent with the
bioinformatics results, the expression of MMP9, C3 and CXCL9
increased significantly after LID surgery (Fig. 4D).
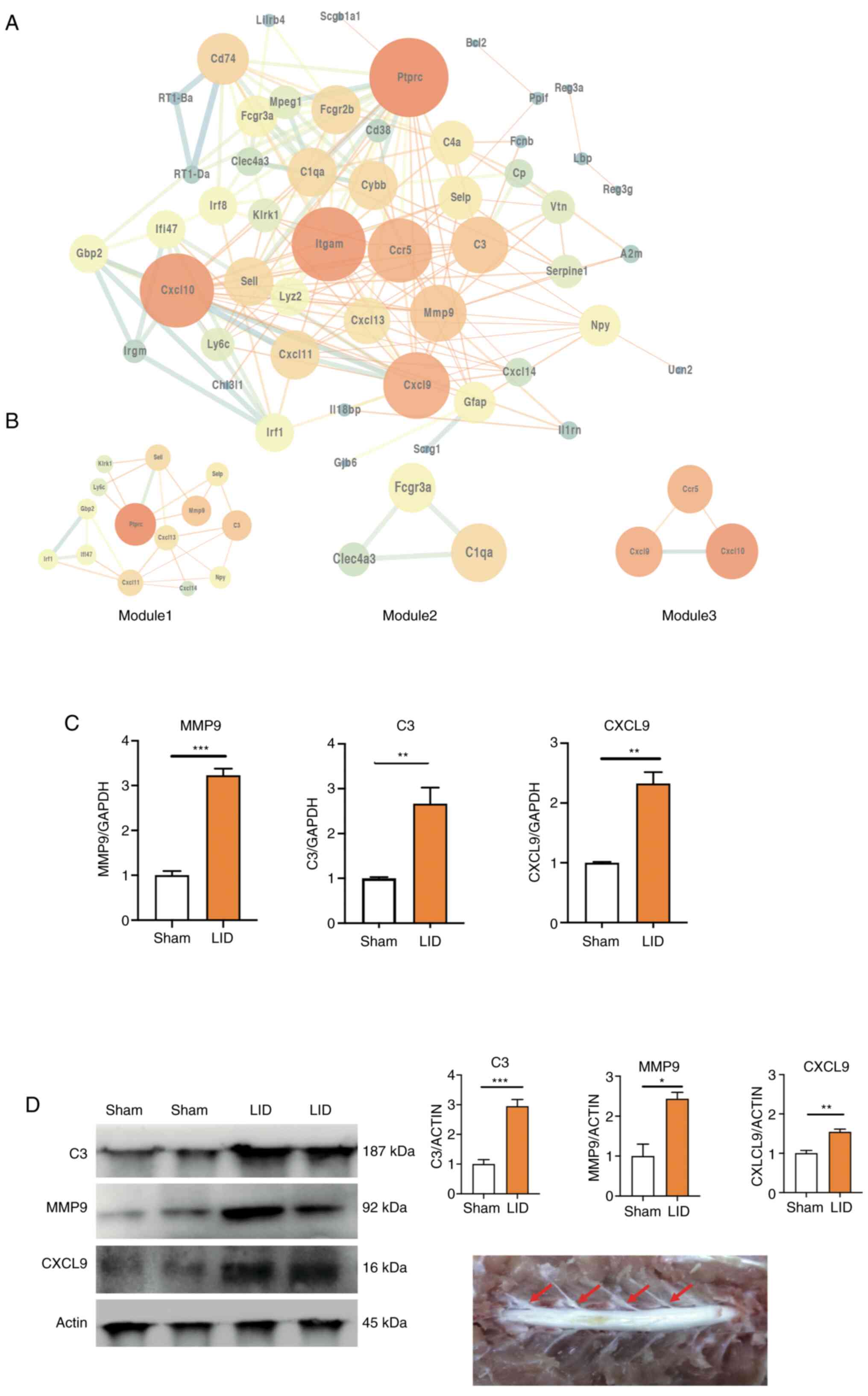 | Figure 4PPI network of DEGs. (A) PPI network
of DEGs. (B) Identification of a sub-network. (C) Gene expression
of CXCL9, MMP9, C3 in DRG tissue after surgery were validated by
reverse transcription-quantitative PCR. (D) The level of DRG, C3,
MMP9 and CX3CL9 protein after sham and LID surgery and isolated DRG
tissues. *P<0.05, **P<0.01,
***P<0.001. PPI, protein-protein interaction; DEGs,
differentially expressed genes; DRG, dorsal root ganglia; LID,
localized inflammation of the dorsal root ganglion; CXCL9, C-X-C
motif ligand 9; MMP9, matrix metallopeptidase 9; C3, complement
component 3. |
 | Table VIModule analysis of differentially
expressed genes using Cytoscape. |
Table VI
Module analysis of differentially
expressed genes using Cytoscape.
| Module | Gene | MCODE_Score | Degree | Topological
coefficient |
|---|
| Module1 | Ptprc | 5.2 | 23 | 0.386188069 |
| | Mmp9 | 6 | 14 | 0.671360536 |
| | C3 | 5.78 | 14 | 0.46776785 |
| | Sell | 5.2 | 11 | 0.594848482 |
| | Cxcl11 | 5.78 | 11 | 0.576136362 |
| | Cxcl13 | 5.06 | 10 | 0.629525 |
| | Selp | 5 | 9 | 0.630740735 |
| | Npy | 6 | 8 | 0.597535717 |
| | Ifi47 | 5 | 7 | 0.670634926 |
| | Irf1 | 5 | 7 | 0.670634926 |
| | Gbp2 | 5 | 7 | 0.62438424 |
| | Klrk1 | 6 | 6 | 0.868472224 |
| | Ly6c | 6 | 6 | 0.868472224 |
| | Cxcl14 | 5 | 5 | 1.000.999.996 |
| Module2 | CXCL9 | 4.3 | 18 | 1 |
| | CCr5 | 4.08 | 17 | 1 |
| | CXCL10 | 4.08 | 21 | 0.88148148 |
| Module2 | Clec4a3 | 3.73 | 5 | 1 |
| | C1qa | 3.88 | 11 | 0.74955445 |
| | Fcgr3a | 3.42 | 8 | 0.69727273 |
Discussion
In the present study, 66 DEGs were screened from
GSE38859 and used for further analyses in which potential targets
were identified that may be useful for the treatment and diagnosis
of LID. The DRG inflammation process must be more extensively
understood in order to identify the most promising genes among a
large list of candidate genes. The top 10 DEGs (H2-Ea, RT1-Ba,
MMP9, C3, Gbp2, Klrk1, Fcgr2b, Ifi4, CXCL9 and Lyz2) were
considered the most promising candidates likely to affect the DRG
inflammation process. The PPI network showed interactions among the
identified DEGs. The key nodes in the network may play critical
roles in the pathological process of LID. In the PPI network, 9
DEGs (MMP9, CXCL3, C3, Ptprc, CXCL11, CXCL9, CCR5, CXCL10 and C1qa)
were classified as hub genes, and 3 of these were among the top 10
DEGs (CXCL9, MMP9 and C3).
As localized inflammation of the L5 DRG induced a
marked increase in spontaneous bursting activity, the material was
sampled for RT-qPCR on the third day (18). The qPCR and western blotting results
were consistent with the bioinformatics results, showing that
CXCL9, MMP9 and C3 were increased significantly in a LID model in
comparison with a sham.
Chemokine-mediated neuroinflammation plays a
critical role in neuropathic pain pathogenesis (19,20).
CXCL9, also known as monokine induced by interferon-γ, is a CXC
family chemokine (21). CXCL9 is
produced by interferon-γ-stimulated macrophages and glial cells
(22). A recent article reported
that CXCL9 is primarily expressed in calcitonin gene-related
peptide-positive and isolectin B4-positive DRG neurons and
participates in the development of cancer-induced pain (23). However, a different study noted that
spinal CXCL9 does not contribute to neuropathic pain despite its
upregulation in the spinal cord after spinal nerve injury (24). This suggests that the mechanism of
neuropathic pain differs from pain models and that CXCL9 has
different functions in different tissues and distinct tissue
specificity. CXCL9 was identified as a seed gene in the present PPI
analysis. Seed gene are most closely related to disease genes, and
these genes may become new disease-related targets. In light of
these observations, it was concluded that CXCL9 may be a good
candidate biomarker for diagnosing DRG inflammation.
Kawasaki et al (25) found increased MMP9 levels shortly
after nerve injury in injured DRG primary sensory neurons.
Moreover, treatment with an MMP9 inhibitor delayed allodynia and
hyperalgesia for 11 days (26,27),
implying that MMP9 participates in the onset rather than the
maintenance of neuropathic pain. Liou et al (26) used a spinal nerve ligation (SNL)
model to demonstrate that MMP9 concentrations were upregulated
after nerve injury and then returned to the normal ranges within 14
days. MMP9 is significantly associated with the onset of
neuropathic pain rather than its maintenance (28). However, specific molecular
mechanisms related to MMP9 and DRG inflammation pain are lacking;
this mechanism should be further investigated, as MMP9 may be a
novel candidate biomarker for DRG inflammation pain.
Complement is a key component of the innate immune
system, and mounting evidence suggests that ongoing complement
activation may lead to pain after inflammation and injury (29). C5a and C3a can activate and
sensitize skin nociceptors (30).
C3 knockout rats have reduced intradermal nerve fiber density after
paclitaxel treatment and reduced mechanical allodynia (31). However, the role of complement in
LID remains unclear. C3 may be a potential candidate biomarker for
LID.
GO and KEGG analyses of the DEGs were performed to
further understand the molecular basis for DRG inflammatory pain
mechanisms. GO BPs were mainly enriched in inflammation and
immunity terms, including ‘response to lipopolysaccharide’,
‘response to molecule of bacterial origin’ and ‘acute inflammatory
response’. KEGG pathway enrichment analysis suggested that these
DEGs were related to the terms ‘chemokine signaling pathway’ and
‘cytokine-cytokine receptor interactions’.
Neuropathic pain can cause central sensitization
(32,33), and its mechanisms include cytokine
and chemokine release by spinal cord glial cells (19,34,35).
Increasing evidence indicates that chemokine signals are crucial
players in neuropathic pain (20,34,36-38).
CXCL10 promotes neuropathic pain by increasing the permeability of
the blood-spinal cord barrier (20). The activation of C-C chemokine
receptor type 5 reduces the analgesic function of opioid receptors
and enhances pain at the inflammation site (39,40).
The aforementioned findings are consistent with the
KEGG analysis in the present study; however, these studies were
based on spared nerve injury or SNL models. Therefore, the specific
molecular mechanism of chemokines in DRG inflammation pain needs
further study.
To verify the consistency of the GO and KEGG pathway
enrichment analyses, GSEA analysis was performed on all genes. The
analysis showed that the LID model was closely related to the terms
‘inflammatory response’, ‘interferon-γ response’ and interferon-α
response pathways, supporting the GO and KEGG analyses'
results.
The molecular mechanisms of lower back pain caused
by DRG inflammatory pain and nerve root pain may differ. The
present study's main purpose was to identify candidate genes
related to local DRG inflammation, providing potential targets for
the treatment or monitoring of certain forms of lower back pain
that are unrelated to mechanical oppression. Although the genes
identified in the present study were initially confirmed in a
previous study, further studies are needed to explore these genes
and pathways' specific regulatory mechanisms.
In summary, 66 DEGs were subjected to extensive
bioinformatics analyses and CXCL9, MMP9, and C3 were identified as
the most promising biomarkers or therapeutic targets for DRG
inflammation.
Acknowledgements
Not applicable.
Funding
Funding: This study was funded by a grant from the Medical and
Health Science and Technology project of Zhejiang Province (grant
no. 2019KY569).
Availability of data and materials
The datasets generated and/or analyzed during the
current study are available in the Gene Expression Omnibus
repository, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE38859.
Authors' contributions
LC, JZ and ZY contributed to animal experiments,
analysis and interpretation of the data and drafted the manuscript.
LC, PW, WC and YW contributed to experiments, analysis and
interpretation of the data and writing of the manuscript. PW
supervised the study and contributed to the conception and design
of the study, the analysis and interpretation of the data and
writing of the manuscript. All authors read and approved the final
version of the manuscript. LC and PW confirm the authenticity of
all the raw data. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
All animal procedures were approved by the Animal
Care and Use Committee of Ningbo University.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Burma NE, Leduc-Pessah H, Fan CY and Trang
T: Animal models of chronic pain: Advances and challenges for
clinical translation. J Neurosci Res. 95:1242–1256. 2017.PubMed/NCBI View Article : Google Scholar
|
|
2
|
North RY, Li Y, Ray P, Rhines LD, Tatsui
CE, Rao G, Johansson CA, Zhang H, Kim YH, Zhang B, et al:
Electrophysiological and transcriptomic correlates of neuropathic
pain in human dorsal root ganglion neurons. Brain. 142:1215–1226.
2019.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Chen G, Kim YH, Li H, Luo H, Liu DL, Zhang
ZJ, Lay M, Chang W, Zhang YQ and Ji RR: PD-L1 inhibits acute and
chronic pain by suppressing nociceptive neuron activity via PD-1.
Nat Neurosci. 20:917–926. 2017.PubMed/NCBI View
Article : Google Scholar
|
|
4
|
Alles SRA and Smith PA: Etiology and
pharmacology of neuropathic pain. Pharmacol Rev. 70:315–347.
2018.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Villarreal CF, Sachs D, Funez MI, Parada
CA, de Queiroz Cunha F and Ferreira SH: The peripheral
pro-nociceptive state induced by repetitive inflammatory stimuli
involves continuous activation of protein kinase A and protein
kinase C epsilon and its Na(V)1.8 sodium channel functional
regulation in the primary sensory neuron. Biochem Pharmacol.
77:867–877. 2009.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Sommer C, Leinders M and Üçeyler N:
Inflammation in the pathophysiology of neuropathic pain. Pain.
159:595–602. 2018.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Wang H, Xu H, Wu LJ, Kim SS, Chen T, Koga
K, Descalzi G, Gong B, Vadakkan KI, Zhang X, et al: Identification
of an adenylyl cyclase inhibitor for treating neuropathic and
inflammatory pain. Sci Transl Med. 3(65ra3)2011.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Todd AJ: Neuronal circuitry for pain
processing in the dorsal horn. Nat Rev Neurosci. 11:823–836.
2010.PubMed/NCBI View
Article : Google Scholar
|
|
9
|
Kennedy PGE and Gershon AA: Clinical
features of varicella-zoster virus infection. Viruses.
10(10)2018.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Cunha C, Silva AJ, Pereira P, Vaz R,
Gonçalves RM and Barbosa MA: The inflammatory response in the
regression of lumbar disc herniation. Arthritis Res Ther.
20(251)2018.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Wang JG, Strong JA, Xie W and Zhang JM:
Local inflammation in rat dorsal root ganglion alters excitability
and ion currents in small-diameter sensory neurons. Anesthesiology.
107:322–332. 2007.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Strong JA, Xie W, Coyle DE and Zhang JM:
Microarray analysis of rat sensory ganglia after local inflammation
implicates novel cytokines in pain. PLoS One.
7(e40779)2012.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Yu G, Wang LG, Han Y and He QY:
clusterProfiler: An R package for comparing biological themes among
gene clusters. OMICS. 16:284–287. 2012.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Lin JS and Lai EM: Protein-protein
interactions: Co-immunoprecipitation. Methods Mol Biol.
1615:211–219. 2017.PubMed/NCBI View Article : Google Scholar
|
|
15
|
National Research Council (US): Committee
for the Update of the Guide for the Care and Use of Laboratory
Animals: Guide for the Care and Use of Laboratory Animals. 8th
edition. National Academies Press, Washington, DC, 2011.
|
|
16
|
Xu F, Yang J, Lu F, Liu R, Zheng J, Zhang
J, Cui W, Wang C, Zhou W, Wang Q, et al: Fast green FCF alleviates
pain hypersensitivity and down-regulates the levels of spinal P2X4
expression and pro-inflammatory cytokines in a rodent inflammatory
pain model. Front Pharmacol. 9(534)2018.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Xie W, Strong JA, Kim D, Shahrestani S and
Zhang JM: Bursting activity in myelinated sensory neurons plays a
key role in pain behavior induced by localized inflammation of the
rat sensory ganglion. Neuroscience. 206:212–223. 2012.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Oh SB, Tran PB, Gillard SE, Hurley RW,
Hammond DL and Miller RJ: Chemokines and glycoprotein120 produce
pain hypersensitivity by directly exciting primary nociceptive
neurons. J Neurosci. 21:5027–5035. 2001.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Li HL, Huang Y, Zhou YL, Teng RH, Zhou SZ,
Lin JP, Yang Y, Zhu SM, Xu H and Yao YX: C-X-C motif chemokine 10
contributes to the development of neuropathic pain by increasing
the permeability of the blood-spinal cord barrier. Front Immunol.
11(477)2020.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Müller M, Carter S, Hofer MJ and Campbell
IL: Review: The chemokine receptor CXCR3 and its ligands CXCL9,
CXCL10 and CXCL11 in neuroimmunity - a tale of conflict and
conundrum. Neuropathol Appl Neurobiol. 36:368–387. 2010.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Tokunaga R, Zhang W, Naseem M, Puccini A,
Berger MD, Soni S, McSkane M, Baba H and Lenz HJ: CXCL9, CXCL10,
CXCL11/CXCR3 axis for immune activation - A target for novel cancer
therapy. Cancer Treat Rev. 63:40–47. 2018.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Sun RM, Wei J, Wang SS, Xu GY and Jiang
GQ: Upregulation of lncRNA-NONRATT021203.2 in the dorsal root
ganglion contributes to cancer-induced pain via CXCL9 in rats.
Biochem Biophys Res Commun. 524:983–989. 2020.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Wu XB, He LN, Jiang BC, Shi H, Bai XQ,
Zhang WW and Gao YJ: Spinal CXCL9 and CXCL11 are not involved in
neuropathic pain despite an upregulation in the spinal cord
following spinal nerve injury. Mol Pain: May 22, 2018.
|
|
25
|
Kawasaki Y, Xu ZZ, Wang X, Park JY, Zhuang
ZY, Tan PH, Gao YJ, Roy K, Corfas G, Lo EH, et al: Distinct roles
of matrix metalloproteases in the early- and late-phase development
of neuropathic pain. Nat Med. 14:331–336. 2008.PubMed/NCBI View
Article : Google Scholar
|
|
26
|
Liou JT, Sum DC, Liu FC, Mao CC, Lai YS
and Day YJ: Spatial and temporal analysis of nociception-related
spinal cord matrix metalloproteinase expression in a murine
neuropathic pain model. J Chin Med Assoc. 76:201–210.
2013.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Rojewska E, Popiolek-Barczyk K, Jurga AM,
Makuch W, Przewlocka B and Mika J: Involvement of pro- and
antinociceptive factors in minocycline analgesia in rat neuropathic
pain model. J Neuroimmunol. 277:57–66. 2014.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Zhao L, Song C, Huang Y, Lei W and Sun J:
MMP-9 regulates CX3CL1/CX3CR1 in the early phase of neuropathic
pain in chronic sciatic nerve constriction injury (CCI) rats. Ann
Palliat Med. 9:2020–2027. 2020.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Fritzinger DC and Benjamin DE: The
complement system in neuropathic and postoperative pain. Open Pain
J. 9:26–37. 2016.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Jang JH, Clark DJ, Li X, Yorek MS, Usachev
YM and Brennan TJ: Nociceptive sensitization by complement C5a and
C3a in mouse. Pain. 148:343–352. 2010.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Xu J, Zhang L, Xie M, Li Y, Huang P,
Saunders TL, Fox DA, Rosenquist R and Lin F: Role of complement in
a rat model of paclitaxel-induced peripheral neuropathy. J Immunol.
200:4094–4101. 2018.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Basbaum AI, Bautista DM, Scherrer G and
Julius D: Cellular and molecular mechanisms of pain. Cell.
139:267–284. 2009.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Woolf CJ: Central sensitization:
Implications for the diagnosis and treatment of pain. Pain. 152
(Suppl 3):S2–S15. 2011.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Dawes JM, Calvo M, Perkins JR, Paterson
KJ, Kiesewetter H, Hobbs C, Kaan TK, Orengo C, Bennett DL and
McMahon SB: CXCL5 mediates UVB irradiation-induced pain. Sci Transl
Med. 3(90ra60)2011.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Slade GD, Conrad MS, Diatchenko L, Rashid
NU, Zhong S, Smith S, Rhodes J, Medvedev A, Makarov S, Maixner W,
et al: Cytokine biomarkers and chronic pain: Association of genes,
transcription, and circulating proteins with temporomandibular
disorders and widespread palpation tenderness. Pain. 152:2802–2812.
2011.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Padi SSV, Shi XQ, Zhao YQ, Ruff MR,
Baichoo N, Pert CB and Zhang J: Attenuation of rodent neuropathic
pain by an orally active peptide, RAP-103, which potently blocks
CCR2- and CCR5-mediated monocyte chemotaxis and inflammation. Pain.
153:95–106. 2012.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Liou JT, Lee CM and Day YJ: The immune
aspect in neuropathic pain: Role of chemokines. Acta Anaesthesiol
Taiwan. 51:127–132. 2013.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Matsushita K, Tozaki-Saitoh H, Kojima C,
Masuda T, Tsuda M, Inoue K and Hoka S: Chemokine (C-C motif)
receptor 5 is an important pathological regulator in the
development and maintenance of neuropathic pain. Anesthesiology.
120:1491–1503. 2014.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Szabo I, Chen XH, Xin L, Adler MW, Howard
OM, Oppenheim JJ and Rogers TJ: Heterologous desensitization of
opioid receptors by chemokines inhibits chemotaxis and enhances the
perception of pain. Proc Natl Acad Sci USA. 99:10276–10281.
2002.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Akgün E, Javed MI, Lunzer MM, Powers MD,
Sham YY, Watanabe Y and Portoghese PS: Inhibition of inflammatory
and neuropathic pain by targeting a Mu opioid receptor/chemokine
receptor5 heteromer (MOR-CCR5). J Med Chem. 58:8647–8657.
2015.PubMed/NCBI View Article : Google Scholar
|