Introduction
Rheumatoid arthritis (RA) is an autoimmune disease
characterized by chronic and progressive joint inflammation, which
can lead to irreversible destruction of articular cartilage,
eventually leading to joint deformity and physical disability
(1,2). It has been revealed that the
pathophysiological behaviors of fibroblast-like synoviocytes (FLSs)
serve crucial roles in the pathogenesis of RA (3). Abnormal secretion of inflammatory
cytokines is also an important cause of RA (4,5).
However, the pathogenesis of RA remains ill-defined.
Long non-coding RNAs (lncRNAs) are a class of ncRNA
>200 nucleotides (nts) long without protein-coding ability,
which have key roles in numerous biological processes, such as cell
differentiation, proliferation, migration and apoptosis (6,7).
Evidence has revealed that lncRNAs are associated with the
development of RA; for instance, Zou et al (8) demonstrated that one lncRNA, LERFS, was
decreased in RA, and that its elevation notably hindered RA
progression via inhibiting the proliferation, migration and
invasion of FLSs. Yue et al (9) reported that ITSN1-2 was highly
expressed in RA-FLSs, and that its depletion hampered RA-FLS growth
and inflammation, as well as inducing apoptosis in RA. These data
indicated that lncRNAs may serve dual roles in RA progression. A
previous report confirmed that zinc finger antisense 1 (ZFAS1)
contributed to the migration and invasion of RA-FLSs in RA
(10). However, the functions and
mechanisms of ZFAS1 have not been fully elucidated.
MicroRNAs (miRNAs/miRs) are a set of endogenous
ncRNAs ~22 nts in length, which modulate gene expression at the
post-transcriptional level (11).
miRNAs are also involved in the regulation of numerous biological
processes, and multiple studies have focused on the roles of miRNAs
in human diseases, including RA (12). For example, Philippe et al
(13) demonstrated that miR-20a was
dysregulated and affected the release of pro-inflammatory cytokines
in RA. Wang et al (14)
revealed that miR-573 decelerated the progression of RA and may be
a target for RA treatment. miR-410-3p has also been shown to
suppress cell viability and facilitate apoptosis in RA-FLSs
(15). A previous report revealed
that miR-3926 was decreased in the plasma of patients with RA
(16). Nevertheless, knowledge
about the underlying mechanisms of miR-3926 in RA is limited.
Follistatin-like protein 1 (FSTL1), also referred to
as follistatin-related protein gene, is a transforming growth
factor β1-inducible protein, containing follistatin-like and
extracellular calcium-binding domains, which is associated with RA
development (17). FSTL1 has been
reported to modulate inflammatory cytokine secretion and FLS
migration and invasion in RA (18,19).
However, to the best of our knowledge, there are no reports about
whether miR-3926 interacts with FSTL1 in RA.
The present study investigated the expression level
patterns of ZFAS1, miR-3926 and FSTL1 in RA. Furthermore, the roles
and mechanisms of ZFAS1, miR-3926 and FSTL1 in RA were further
investigated by gain- and loss-of-function experiments.
Materials and methods
Tissue collection
A total of 20 RA synovial tissues were harvested
from patients with RA (13 women and 7 men; age range, 41-66 years)
who underwent knee arthroplasty, and 20 non-arthritic control
tissues were collected from the knee joints of traumatic amputees
(11 women and 9 men; age range, 38-62 years) at the Second Hospital
of Shandong University (Jinan, China) between May 2013 and July
2018. RA was defined and classified according to the 2010 American
College of Rheumatology/European League against Rheumatism
classification criteria for RA (20). The experiments were approved by the
Ethics Committee of the Second Hospital of Shandong University and
written informed consent was provided by all participants. The
specimens were preserved at -80˚C for the isolation of total RNA or
protein.
Cell culture
FLSs were isolated from synovial tissues. After
being washed with PBS (Beijing Solarbio Science & Technology,
Co., Ltd.) and serum-free DMEM (HyClone; Cytiva), synovial tissues
were cut into small sections (1 mm3) and digested with 2
mg/ml collagenase I (Beijing Solarbio Science & Technology,
Co., Ltd.) for 2 h at 37˚C. The precipitate was resuspended in DMEM
containing 10% FBS (both HyClone; Cytiva) following centrifugation
at 200 x g for 10 min at 37˚C. FLSs were harvested and grown in
DMEM containing 10% FBS and 1% penicillin-streptomycin (all
HyClone; Cytiva) at 37˚C in a humidified atmosphere with 5%
CO2. FLSs at passages 3-6 were used for further
experiments.
Cell transfection
Small interfering RNA (siRNA) against ZFAS1
(si-ZFAS1; 5'-ACAAAACGGGAACTTACGGGC-3'), siRNA against FSTL1
(si-FSTL1; 5'-AGTAAATATCCTAGTTTTCTC-3') and scrambled siRNA
negative control (si-NC; 5'-TTCTCCGAACGTGTCACGTTT-3'); the
overexpression vector of ZFAS1 (pcDNA-ZFAS1) and empty pcDNA
(pcDNA-NC); miR-3926 mimics (miR-3926; 5'-UGGCCAAAAAGCAGGCAGAGA-3')
and scrambled miRNA control (miR-NC; 5'-UUCUCCGAACGUGUCACGUUU-3');
miR-3926 inhibitors (anti-miR-3926; 5'-UCUCUGCCUGCUUUUUGGCCA-3')
and inhibitor control (anti-miR-NC; 5'-CAGUACUUUUGUGUAGUACAA-3')
were all obtained from Guangzhou RiboBio Co., Ltd. RA-FLSs
(1.0x104 cells/well) were seeded into 24-well plates and
transfected with the oligonucleotides (50 nM) or vectors (2 µg)
using Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.). For miR-3926 + pcDNA-NC and miR-3926 +
pcDNA-ZFAS1 groups, cells were co-transfected with miR-3926 and
pcDNA-NC/pcDNA-ZFAS1. For si-FSTL1 + anti-miR-NC or si-FSTL1 +
anti-miR-3926 groups, cells were co-transfected with si-FSTL1 and
anti-miR-NC/anti-miR-3926. After transfection for 48 h, the cells
were collected for subsequent experiments.
Reverse transcription quantitative
(RT-q)PCR
Total RNA in synovial tissues and FLSs was extracted
using RNAiso Plus (Takara Biotechnology Co., Ltd.). Subsequently,
RNA was quantified using a NanoDrop 2000c spectrophotometer (Thermo
Fisher Scientific, Inc.). RT was conducted using M-MLV First Strand
(Takara Biotechnology Co., Ltd.) and miRNA 1st Strand cDNA
Synthesis kits (Vazyme Biotech Co., Ltd.) according to the
manufacturers' instructions. RT-qPCR was then performed using
BeyoFast™ SYBR Green qPCR Mix (Beyotime Institute of Biotechnology)
on an ABI 7900 Real-Time PCR system (Applied Biosystems; Thermo
Fisher Scientific, Inc.) under the following conditions: Initial
denaturation at 95˚C for 5 min; 40 cycles at 95˚C for 10 sec and
60˚C for 30 sec; 95˚C for 15 sec, 60˚C for 60 sec and 95˚C for 15
sec. The expression levels of ZFAS1, FSTL1 and miR-3926 were
analyzed via the 2-ΔΔCq method (21). GAPDH was used as the internal
reference for ZFAS1 and FSTL1, while small nuclear RNA U6 was
utilized as the internal reference for miR-3926. The primer
sequences were as follows: ZFAS1, forward
5'-CTATTGTCCTGCCCGTTAGAG-3' and reverse
5'-GTCAGGAGATCGAAGGTTGTAG-3'; miR-3926, forward
5'-GCCGAGTGGCCAAAAAGCA-3' and reverse 5'-CTCAACTGGTGTCGTGGA-3';
FSTL1, forward 5'-GAGCAATGCAAACCTCACAAG-3' and reverse
5'-CAGTGTCCATCGTAATCAACCTG-3'; GAPDH, forward
5'-GATTCCACCCATGGCAAATTC-3' and reverse
5'-CTGGAAGATGGTGATGGGATT-3'; and U6, forward
5'-CTTCGGCAGCACATATACT-3' and reverse
5'-AAAATATGGAACGCTTCACG-3'.
MTT assay
After transfection for 48 h, RA-FLSs were collected
and seeded into 96-well plates (5.0x103 cells/well).
Subsequently, 20 µl MTT (Beijing Solarbio Science & Technology,
Co., Ltd.) was added to each well at the indicated time points (0,
24, 48 or 72 h) and incubated for an additional 4 h at 37˚C. DMSO
(150 µl; Beijing Solarbio Science & Technology, Co., Ltd.) was
then added to dissolve the formazan crystals. Finally, the
absorbance at 490 nm was read via a microplate reader (Molecular
Devices, LLC).
Flow cytometric analysis
After transfection, Annexin V-FITC/propidium iodide
(PI) Apoptosis Detection kit (Beyotime Institute of Biotechnology)
was utilized to evaluate apoptosis of RA-FLSs. Briefly, transfected
RA-FLSs were harvested and resuspended in binding buffer at a
density of 1.0x106 cells/ml. Subsequently, RA-FLSs were
maintained in 5 µl Annexin V-FITC and 10 µl PI for 15 min at room
temperature in the dark. Finally, a FACScan® flow
cytometer (BD Biosciences) was used to assess cell apoptosis. The
results were analyzed by FlowJo 7.6.1. (FlowJo LLC).
Western blotting
Total protein was isolated by lysing synovial
tissues and FLSs in RIPA buffer (Beyotime Institute of
Biotechnology). The protein samples were quantified using a
NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific, Inc.) by
examining the OD value at 280 nm. Equal amounts of protein (25
µg/lane) were separated by 10% SDS-PAGE (Beijing Solarbio Science
& Technology, Co., Ltd.) and transferred onto PVDF membranes
(Pall Corporation). Subsequently, the membranes were blocked in 5%
skimmed milk for 2 h at room temperature and incubated overnight at
4˚C with primary antibodies (all BIOSS) against cleaved caspase-3
(C-caspase-3; cat. no. bs-0081R; 1:2,000), interleukin (IL)-6 (cat.
no. bs-4539R; 1:2,000), IL-1β (cat. no. bs-20448R; 1:2,000), tumor
necrosis factor (TNF)-α (cat. no. bs-10802R; 1:2,000), FSTL1 (cat.
no. bs-6050R; 1:2,000) and GAPDH (cat. no. bs-0755R; 1:5,000),
followed by incubation with a horseradish peroxidase-conjugated
secondary antibody (cat. no. bs-40296G-HRP; 1:10,000; BIOSS) for 1
h at room temperature. The bands were visualized via enhanced
chemiluminescence reagent (Vazyme Biotech Co., Ltd.) and analyzed
using ImageJ v1.8.0 (National Institutes of Health).
Transwell assay
For the detection of cell migration, Transwell
insert chambers (Corning Inc.) were used. Briefly, transfected
RA-FLSs (5.0x105 cells/ml) were suspended in serum-free
DMEM in the upper chamber, and DMEM (600 µl) containing 10% FBS
(HyClone; Cytiva) was added into the bottom chamber. After 24 h,
migrated cells were fixed in 75% methanol for 1 h at 37˚C and
stained with 0.1% crystal violet (Beijing Solarbio Science &
Technology, Co., Ltd.) for 15 min at 37˚C. The stained cells were
analyzed under a light microscope (Olympus Corporation). The same
method was used to detect cell invasion, with the exception that
the upper chamber was pre-coated with Matrigel (Beijing Solarbio
Science & Technology, Co., Ltd.).
Dual-luciferase reporter assay
The binding sites between ZFAS1 and miR-3926 were
predicted using the online software LncBase Predicted V.2
(http://carolina.imis.athena-innovation.gr/diana_tools/web/index.php?r=lncbasev2/index-predicted),
and the binding sites between miR-3926 and FSTL1 were predicted
using TargetScan (http://www.targetscan.org/vert_71/). Subsequently,
dual-luciferase reporter assays were used to verify these
predictions. The sequences of ZFAS1 or FSTL1 3'untranslated region
(UTR) including wild-type (WT) or mutant (MUT) binding sequences of
miR-3926 were cloned and inserted into pmirGLO vectors (Promega
Corporation), in order to generate ZFAS1 WT, ZFAS1 MUT, FSTL1 3'UTR
WT and FSTL1 3'UTR MUT luciferase reporter vectors, respectively.
Subsequently, RA-FLSs (2.0x104 cells/well) were seeded
into 24-well plates and co-transfected with miR-3926 or miR-NC and
corresponding luciferase reporter vectors using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturers' instructions.
After co-transfection for 48 h, luciferase activity was evaluated
using a dual-luciferase reporter assay kit (Promega Corporation)
according to the manufacturers' instructions. Renilla
luciferase activity was used to normalize firefly luciferase
activity.
Statistical analysis
The data were analyzed using GraphPad Prism 7
software (GraphPad, Inc.) and are presented as the mean ± standard
deviation of three independent experiments. Significant differences
between two groups were analyzed by unpaired Student's t-test,
whereas those between three groups were analyzed by one-way ANOVA
followed by Tukey's post hoc test. The linear associations between
ZFAS1, miR-3926 and FSTL1 in RA tissues were analyzed by Pearson's
correlation analysis. P<0.05 was considered to indicate a
statistically significant difference.
Results
ZFAS1 is highly expressed in RA
synovial tissue and RA-FLSs
Firstly, the expression levels of ZFAS1 in RA and
healthy synovial tissues were evaluated via RT-qPCR. ZFAS1
expression levels were markedly elevated in RA synovial tissue
compared with those in non-arthritic control tissue (Fig. 1A). Additionally, the expression
levels of ZFAS1 in RA-FLSs and non-arthritic control
tissue-extracted FLSs (N-FLSs) were measured. The data demonstrated
that ZFAS1 was more highly expressed in RA-FLSs than in N-FLSs
(Fig. 1B). These findings indicated
that ZFAS1 was abnormally expressed in RA.
Silencing of ZFAS1 inhibits cell
proliferation, migration, invasion and inflammatory cytokine
expression, and promotes cell apoptosis in RA-FLSs
In order to identify the potential roles of ZFAS1 in
RA, loss-of-function experiments were performed via transfecting
si-ZFAS1 into RA-FLSs. The knockdown efficiency was determined via
RT-qPCR, which demonstrated that ZFAS1 expression levels were
significantly decreased in RA-FLSs following si-ZFAS1 transfection
(Fig. 2A). The MTT assay indicated
that si-ZFAS1 transfection significantly suppressed cell
proliferation in RA-FLSs compared with that in cells transfected
with si-NC (Fig. 2B). Flow
cytometric analysis demonstrated that compared with in the si-NC
group, cell apoptosis was significantly induced in RA-FLSs
transfected with si-ZFAS1 (Fig.
2C). Furthermore, the expression levels of the
apoptosis-associated protein C-caspase-3 were increased in RA-FLSs
transfected with si-ZFAS1, as indicated by western blotting
(Fig. 2D). Transwell assay
demonstrated that ZFAS1 knockdown significantly hampered cell
migration and invasion in RA-FLSs (Fig.
2E and F). In addition, the
expression levels of inflammatory cytokines, IL-6, IL-1β and TNF-α,
were examined by western blotting. The data showed that the
expression levels of IL-6, IL-1β and TNF-α were decreased in
RA-FLSs post-transfection with si-ZFAS1 compared with in the si-NC
group (Fig. 2G). Taken together,
these data indicated that ZFAS1 knockdown suppressed the
progression of RA.
 | Figure 2ZFAS1 downregulation suppresses cell
proliferation, migration, invasion and inflammatory cytokine
expression, and facilitates cell apoptosis in RA-FLSs. RA-FLSs were
transfected with si-NC or si-ZFAS1. (A) Expression levels of ZFAS1
were detected via reverse transcription-quantitative PCR. (B)
Proliferation of RA-FLSs was evaluated via MTT assay. (C) Apoptosis
of RA-FLSs was assessed via flow cytometric analysis. (D) Protein
expression levels of C-caspase-3 were measured via western
blotting. (E) Migration and (F) invasion of RA-FLSs were examined
via Transwell assay (magnification, x100). (G) Expression levels of
IL-6, IL-1β and TNF-α were measured via western blotting.
*P<0.05 vs. si-NC. ZFAS1, zinc finger antisense 1;
RA-FLSs, rheumatoid arthritis-fibroblast-like synoviocytes; si,
small interfering RNA; NC, negative control; C-caspase-3,
cleaved-caspase-3; IL, interleukin; TNF-α, tumor necrosis factor-α;
PI, propidium iodide. |
ZFAS1 negatively modulates miR-3926
expression levels by directly targeting miR-3926 in RA-FLSs
In order to investigate the mechanism underlying the
effects of ZFAS1 on RA, online software LncBase Predicted V.2 was
utilized to predict the potential targets of ZFAS1. A total of four
miRNAs (miR-548a-3p, miR-3926, miR-27a-3p and miR-4701-5p) were
selected for their low expression in RA. Subsequently, RT-qPCR was
used to determine the expression levels of these miRNAs in RA-FLSs
following ZFAS1 knockdown or overexpression. miR-3926 was selected
as the study object for its marked change in RA-FLSs following
ZFAS1 knockdown or ZFAS1 overexpression compared with other miRNAs
(Table SI). The complementary
sequences between ZFAS1 and miR-3926 are presented in Fig. 3A. To verify these predictions, a
dual-luciferase reporter assay was performed. The results
demonstrated that the luciferase activity in RA-FLSs co-transfected
with ZFAS1 WT and miR-3926 was significantly inhibited compared
with that in RA-FLSs co-transfected with ZFAS1 WT and miR-NC,
whereas the luciferase activity was not affected in the ZFAS1 MUT
group (Fig. 3B). Subsequently, the
expression levels of miR-3926 in RA synovial tissue and RA-FLSs
were determined via RT-qPCR. The results indicated that miR-3926
expression levels were significantly decreased in RA synovial
tissue and RA-FLSs compared with those in non-arthritic control
tissue and N-FLSs (Fig. 3C and
D). As evidenced by Pearson's
correlation analysis, miR-3926 expression levels were inversely
correlated with ZFAS1 expression levels in RA synovial tissue
(Fig. 3E).
 | Figure 3ZFAS1 directly binds miR-3926 and
negatively regulates miR-3926 expression levels in RA-FLSs. (A)
Predicted binding sites between ZFAS1 and miR-3926. (B) Luciferase
activity in RA-FLSs transfected with ZFAS1 WT or ZFAS1 MUT together
with miR-3926 or miR-NC was analyzed via dual-luciferase reporter
assay. Expression levels of miR-3926 in (C) RA synovial and
non-arthritic control tissue, and (D) RA-FLSs and N-FLSs were
assessed via RT-qPCR. (E) Correlation between expression levels of
ZFAS1 and miR-3926 in RA-FLSs was analyzed via Pearson's
correlation analysis. (F) Expression levels of ZFAS1 in RA-FLSs
transfected with pcDNA-ZFAS1 or pcDNA-NC was examined via RT-qPCR
analysis. (G) RA-FLSs were transfected with si-NC, si-ZFAS1,
pcDNA-NC or pcDNA-ZFAS1, then the expression levels of miR-3926
were determined via RT-qPCR. (H) Expression levels of miR-3926 in
RA-FLSs transfected with anti-miR-NC or anti-miR-3926 were
determined via RT-qPCR. *P<0.05 vs. miR-NC. ZFAS1,
zinc finger antisense 1; miR, microRNA; RA-FLSs, rheumatoid
arthritis-fibroblast-like synoviocytes; WT, wild type; MUT, mutant;
NC, negative control; N, normal; RT-qPCR, reverse
transcription-quantitative PCR; si, small interfering RNA. |
pcDNA-ZFAS1 was successfully transfected into
RA-FLSs to elevate the expression levels of ZFAS1 (Fig. 3F). ZFAS1 depletion markedly enhanced
the expression levels of miR-3926, whereas ZFAS1 overexpression
resulted in decreased expression levels of miR-3926 in RA-FLSs
(Fig. 3G). miR-3926 inhibitor
transfection also significantly downregulated the expression levels
of miR-3926 in RA-FLSs, which indicated that the miR-3926 inhibitor
was successfully transfected into RA-FLSs (Fig. 3H). These data indicated that ZFAS1
may directly interact with and negatively regulate miR-3926
expression in RA-FLSs.
Overexpression of ZFAS1 abolishes
effects of miR-3926 overexpression on cell proliferation,
apoptosis, migration, invasion and inflammatory cytokine production
in RA-FLSs
Given that the present study suggested that ZFAS1
may target miR-3926 to modulate miR-3926 expression in RA-FLSs, the
present study investigated whether ZFAS1 could regulate the
promotion of RA via targeting miR-3926. RA-FLSs were transfected
with miR-NC, miR-3926, miR-3926 + pcDNA-NC or miR-3926 +
pcDNA-ZFAS1, and miR-3926 expression levels were measured via
RT-qPCR. Overexpression of miR-3926 caused by miR-3926 transfection
was partly abolished by pcDNA-ZFAS1 transfection in RA-FLSs
(Fig. 4A). miR-3926 overexpression
led to a marked inhibition in cell proliferation in RA-FLSs,
whereas this inhibition was partially reversed following the
overexpression of ZFAS1, as evidenced by the MTT assay (Fig. 4B). Flow cytometric analysis
indicated that the apoptotic rate of RA-FLSs was significantly
increased by miR-3926, whereas pcDNA-ZFAS1 counteracted this effect
(Fig. 4C). Furthermore, the
expression levels of the pro-apoptotic protein C-caspase3 were
markedly elevated by miR-3926 overexpression; however, ZFAS1
overexpression significantly weakened this elevation in RA-FLSs
(Fig. 4D). As demonstrated by the
Transwell assay, there was a significant decrease in the number of
migratory and invasive cells transfected with miR-3926; this
decrease was rescued following transfection with pcDNA-ZFAS1 in
RA-FLSs (Fig. 4E and F). The results of western blotting
demonstrated that miR-3926 overexpression suppressed the production
of IL-6, IL-1β and TNF-α in RA-FLSs, whereas ZFAS1 overexpression
partly attenuated the effects in RA-FLSs (Fig. 4G). These data indicated that the
inhibitory effect of miR-3926 on RA development was markedly
overturned by ZFAS1.
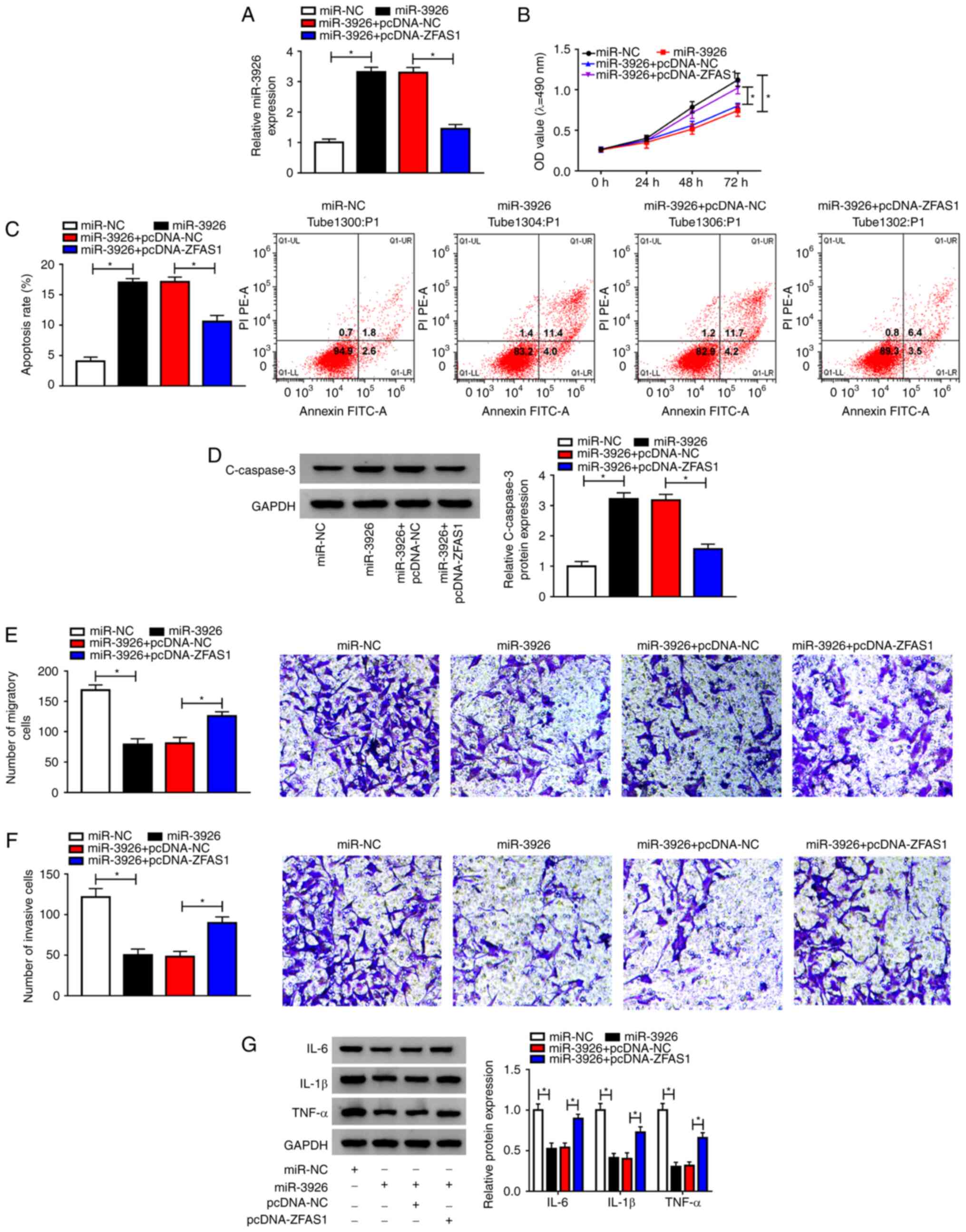 | Figure 4ZFAS1 alters cell proliferation,
apoptosis, migration, invasion and inflammatory cytokine expression
via binding to miR-3926 in RA-FLSs. RA-FLSs were transfected with
miR-NC, miR-3926, miR-3926 + pcDNA-NC or miR-3926 + pcDNA-ZFAS1.
(A) Expression levels of miR-3926 in RA-FLSs were measured via
reverse transcription-quantitative PCR. (B) RA-FLS proliferation
was determined via MTT assay. (C) RA-FLS apoptosis was analyzed via
flow cytometric analysis. (D) Expression levels of C-caspase-3 in
RA-FLSs were determined via western blotting. (E) Migration and (F)
invasion of RA-FLS were assessed via Transwell assay
(magnification, x100). (G) Expression levels of IL-6, IL-1β and
TNF-α were determined via western blotting assay.
*P<0.05. ZFAS1, zinc finger antisense 1; miR,
microRNA; RA-FLSs, rheumatoid arthritis-fibroblast-like
synoviocytes; NC, negative control; C-caspase-3, cleaved-caspase-3;
IL, interleukin; TNF-α, tumor necrosis factor-α. |
FSTL1 is a direct target of miR-3926
in RA-FLSs
Using TargetScan online software, FSTL1 was
identified as a target gene of miR-3926 (Fig. 5A). Dual-luciferase reporter assay
revealed a significant suppression in the luciferase activity of
RA-FLSs co-transfected with FSTL1 3'UTR WT and miR-3926 compared
with the FSTL1 3'UTR WT and miR-NC co-transfected group, whereas
the luciferase activity was not affected in the FSTL1 3'UTR MUT
group (Fig. 5B). The mRNA and
protein expression levels of FSTL1 in RA synovial tissue and
healthy control tissue were determined via RT-qPCR and western
blotting. The data demonstrated that the mRNA and protein
expression levels of FSTL1 were significantly increased in RA
synovial tissues compared with non-arthritic control tissues
(Fig. 5C and D). Similarly, the mRNA and protein
expression levels of FSTL1 were higher in RA-FLSs than those in
N-FLSs (Fig. 5E and F). Pearson's correlation analysis
indicated that there was an inverse correlation between the
expression levels of FSTL1 and miR-3926 in RA synovial tissue
(Fig. 5G). Furthermore, miR-3926
overexpression significantly suppressed the mRNA and protein
expression levels of FSTL1 in RA-FLSs, whereas miR-3926 inhibition
markedly promoted the mRNA and protein expression levels of FSTL1
in RA-FLSs (Fig. 5H and I). Taken together, these results indicated
that miR-3926 targeted FSTL1 and negatively modulated FSTL1
expression levels in RA-FLSs.
 | Figure 5miR-3926 directly targets FSTL1 and
negatively regulates its expression in RA-FLSs. (A) Potential
binding sites between miR-3926 and FSTL1 were predicted via
TargetScan. (B) Dual-luciferase reporter assay was performed to
determine luciferase activity in RA-FLSs co-transfected with FSTL1
3'UTR WT or FSTL1 3'UTR MUT and miR-3926 or miR-NC. (C) mRNA and
(D) protein expression levels of FSTL1 in RA synovial and
non-arthritic control tissue were analyzed via RT-qPCR and western
blotting, respectively. (E) mRNA and (F) protein expression levels
of FSTL1 in RA-FLSs and N-FLSs were assessed via RT-qPCR and
western blotting, respectively. (G) Pearson's correlation analysis
was utilized to analyze the correlation between miR-3926 and FSTL1
in RA synovial tissue. RA-FLSs were transfected with miR-NC,
miR-3926, anti-miR-NC or anti-miR-3926 and the (H) mRNA and (I)
protein expression levels of FSTL1 were determined via RT-qPCR and
western blotting assay, respectively. *P<0.05 vs.
miR-NC. miR, microRNA; FSTL1, follistatin-like protein 1; UTR,
untranslated region; WT, wild type; MUT, mutant; NC, negative
control; RT-qPCR, reverse transcription-quantitative PCR; RA,
rheumatoid arthritis; FLSs, fibroblast-like synoviocytes; N,
normal. |
Depletion of miR-3926 rescues the
effects of FSTL1 knockdown on cell proliferation, apoptosis,
migration, invasion and inflammatory cytokine production in
RA-FLSs
Based on the aforementioned results, it was
hypothesized that miR-3926 may regulate the development of RA via
targeting FSTL1. Thus, si-NC, si-FSTL1, si-FSTL1 + anti-miR-NC or
si-FSTL1 + anti-miR-3926 were transfected into RA-FLSs to
investigate the association between miR-3926 and FSTL1 in the
regulation of RA progression. si-FSTL1 transfection significantly
suppressed the mRNA and protein expression levels of FSTL1 in
RA-FLSs, whereas anti-miR-3926 transfection partly overturned these
effects (Fig. 6A and B). The MTT assay demonstrated that FSTL1
knockdown significantly suppressed the proliferation of RA-FLSs,
whereas miR-3926 inhibition reversed this suppressive effect
(Fig. 6C). Flow cytometric analysis
indicated that apoptosis of RA-FLSs was significantly promoted by
FSTL1 knockdown, whereas this promotion was abolished by the
inhibition of miR-3926 in RA-FLSs (Fig.
6D). Furthermore, the protein expression levels of C-caspase-3
were significantly elevated in RA-FLSs transfected with si-FSTL1,
whereas anti-miR-3926 transfection significantly abrogated this
elevation (Fig. 6E). As indicated
by the results of the Transwell assay, cell migration and invasion
of RA-FLSs transfected with si-FSTL1 were hampered, whereas these
effects were partially weakened following transfection with
anti-miR-3926 (Fig. 6F and G). Western blotting revealed that FSTL1
knockdown led to a decrease in the expression levels of IL-6, IL-1β
and TNF-α in RA-FLSs, whereas miR-3926 depletion attenuated this
effect (Fig. 6H). These findings
indicated that miR-3926 modulated cell proliferation, apoptosis,
migration, invasion and inflammatory cytokine expression by
interacting with FSTL1 in RA-FLSs.
 | Figure 6Inhibition of miR-3926 restores the
inhibitory effect of FSTL1 knockdown on RA-FLSs development.
RA-FLSs were transfected with si-NC, si-FSTL1, si-FSTL1 +
anti-miR-NC or si-FSTL1 + anti-miR-3926. (A) mRNA and (B) protein
expression levels of FSTL1 in RA-FLSs were determined via reverse
transcription-quantitative PCR and western blotting, respectively.
(C) Cell proliferation in RA-FLSs was assessed via MTT assay. (D)
Cell apoptosis in RA-FLSs was evaluated via flow cytometric
analysis. (E) Expression levels of C-caspase-3 in RA-FLSs were
determined via western blotting. Cell (F) migration and (G)
invasion of RA-FLSs were determined by Transwell assay
(magnification, x100). (H) Expression levels of IL-6, IL-1β and
TNF-α in RA-FLSs were assessed via western blotting.
*P<0.05. miR, microRNA; FSTL1, follistatin-like
protein 1; RA-FLSs, rheumatoid arthritis-fibroblast-like
synoviocytes; si, small interfering RNA; NC, negative control;
C-caspase-3, cleaved-caspase-3; IL, interleukin; TNF-α, tumor
necrosis factor-α. |
ZFAS1 regulates FSTL1 expression
levels via sponging miR-3926 in RA-FLSs
As indicated by Pearson's correlation analysis,
FSTL1 expression levels were positively correlated with ZFAS1
expression levels in RA synovial tissue (Fig. 7A). In order to further determine the
association between ZFAS1, miR-3926 and FSTL1 in RA-FLSs, si-NC,
si-ZFAS1, si-ZFAS1 + anti-miR-NC or si-ZFAS1 + anti-miR-3926 was
transfected into RA-FLSs, and RT-qPCR and western blotting were
performed to measure the mRNA and protein expression levels of
FSTL1, respectively. The data revealed that the mRNA and protein
expression levels of FSTL1 were decreased in RA-FLSs transfected
with si-ZFAS1, whereas anti-miR-3926 transfection partly rescued
these effects (Fig. 7B and C). Taken together, these results indicated
that ZFAS1 positively altered the expression levels of FSTL1 via
sponging miR-3926 in RA-FLSs.
Discussion
Research is increasingly focusing on the association
between lncRNAs and human disease development. The functional roles
of ZFAS1 have been explored in numerous human diseases, such as
non-small cell lung cancer (NSCLC) (22), T-cell acute lymphoblastic leukemia
(23), hepatocellular carcinoma
(24) and osteoarthritis (25). In the present study, ZFAS1 was
significantly elevated in RA synovial tissues and RA-FLSs.
Furthermore, ZFAS1 modulated the growth, apoptosis, migration,
invasion and inflammation of FLSs via the miR-3926/FSTL1 axis in
RA.
It has been confirmed that ZFAS1 mediates the
regulation of cell growth, metastasis and apoptosis in a number of
diseases. In NSCLC, ZFAS1 has been reported to be highly expressed,
whereas silencing of ZFAS1 inhibited NSCLC cell growth via
interacting with miR-193-3p (22).
Furthermore, Ye et al (10)
demonstrated that ZFAS1 was elevated in patients with RA, and
contributed to RA-FLS migration and invasion by targeting miR-27a.
In the present study, ZFAS1 was significantly elevated in RA
synovial tissue and RA-FLSs. Depletion of ZFAS1 hampered RA-FLS
growth, migration and invasion, and facilitated RA-FLS apoptosis.
The present study also determined the impact of ZFAS1 knockdown on
the expression of inflammatory cytokines. The data showed that the
expression levels of IL-6, IL-1β and TNF-α were notably decreased
following ZFAS1 knockdown in RA-FLSs, indicating that ZFAS1
knockdown inhibited inflammation in RA. Taken together, these data
suggested that ZFAS1 may serve a key role in the progression of
RA.
The interaction between lncRNAs and miRNAs has
become a focus of research (25).
In human diseases, ZFAS1 has been demonstrated to function as a
sponge for multiple miRNAs, such as miR-135a (26), miR-150(27), miR-10a (28) and miR-329(29). However, the association between
ZFAS1 and miR-3926 has not been fully elucidated. To the best of
our knowledge, the present study was the first to confirm that
miR-3926 may be a direct target of ZFAS1. Wang et al
(16) demonstrated that miR-3926
was decreased in RA and may be a biomarker for RA diagnosis. Fu
et al (30) showed that
miR-3926 was markedly decreased in RA, and that elevated expression
levels of miR-3926 markedly suppressed RA synovial fibroblast
growth and the secretion of inflammatory cytokines (IL-6, IL-1β and
TNF-α) by targeting Toll-like receptor 5. Consistent with these
data, the present study demonstrated that miR-3926 expression
levels were significantly lower and negatively regulated by ZFAS1
in patients with RA compared with healthy controls. Furthermore,
transfection with miR-3926 led to notable decreases in RA-FLS
growth, migration, invasion and inflammatory cytokine expression,
as well as a marked enhancement in RA-FLS apoptosis, whereas these
effects were all abrogated by pcDNA-ZFAS1 transfection.
Further analysis identified FSTL1 as a target gene
of miR-3926. FSTL1 expression levels were increased in RA and
inversely altered by miR-3926. Ehara et al (31) demonstrated that FSTL1 was
overexpressed in RA. Mattiotti et al (32) revealed that high expression levels
of FLST1 were associated with clinical features of RA. Clutter
et al (33) showed that
FSTL1 accelerated the progression of arthritis via increasing
expression levels of IFN-γ. A study by Shi et al (19) discovered that FSTL1 was increased in
RA; moreover, FSTL1 was targeted by miR-27a and was involved in the
regulation of RA-FLS migration and invasion. The present study
elucidated that deficiency of FSTL1 suppressed cell growth,
migration, invasion and inflammation, and induced cell apoptosis in
RA-FLSs, whereas miR-3926 inhibition attenuated these effects,
indicating that miR-3926 may target FSTL1 to regulate the
progression of RA.
In human diseases, ZFAS1 has been reported to act as
a sponge for numerous miRNAs, such as miR-135a (26), miR-150(27), miR-10a (28) and miR-329(29). Further investigation is required to
elucidate the associations between ZFAS1, FLST1 and these miRNAs in
RA regulation.
In conclusion, the present study demonstrated that
ZFAS1 was increased in patients with RA, and that ZFAS1 knockdown
relieved the development of RA by modulating the miR-3926/FSTL1
axis. These results suggested that ZFAS1 may be a potential
candidate for RA treatment.
Supplementary Material
Predicted miRNA targets of ZFAS1
following ZFAS1 knockdown or overexpression.
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
PC and XY conceptualized the study and designed the
experiments. JL and WZ collected and analyzed data. QW and MG
performed the experiments. QW, PC and XY wrote, reviewed and edited
the manuscript. QW and PC confirm the authenticity of all the raw
data. All authors read and approved the final version of the
manuscript.
Ethics approval and consent to
participate
The present study was approved by the ethical review
committee of the Second Hospital of Shandong University. Written
informed consent was obtained from all enrolled patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Araki Y and Mimura T: The mechanisms
underlying chronic inflammation in rheumatoid arthritis from the
perspective of the epigenetic landscape. J Immunol Res.
2016(6290682)2016.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Bottini N and Firestein GS: Duality of
fibroblast-like synoviocytes in RA: Passive responders and
imprinted aggressors. Nat Rev Rheumatol. 9:24–33. 2013.PubMed/NCBI View Article : Google Scholar
|
|
4
|
McInnes IB and Schett G: The pathogenesis
of rheumatoid arthritis. N Engl J Med. 365:2205–2219.
2011.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Brennan FM and McInnes IB: Evidence that
cytokines play a role in rheumatoid arthritis. J Clin Invest.
118:3537–3545. 2008.PubMed/NCBI View
Article : Google Scholar
|
|
6
|
Gibb EA, Brown CJ and Lam WL: The
functional role of long non-coding RNA in human carcinomas. Mol
Cancer. 10(38)2011.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Mercer TR, Dinger ME and Mattick JS: Long
non-coding RNAs: Insights into functions. Nat Rev Genet.
10:155–159. 2009.PubMed/NCBI View
Article : Google Scholar
|
|
8
|
Zou Y, Xu S, Xiao Y, Qiu Q, Shi M, Wang J,
Liang L, Zhan Z, Yang X, Olsen N, et al: Long noncoding RNA LERFS
negatively regulates rheumatoid synovial aggression and
proliferation. J Clin Invest. 128:4510–4524. 2018.PubMed/NCBI View
Article : Google Scholar
|
|
9
|
Yue T, Fan X, Zhang Z, Liu Z, Guo M, Bai
F, Gong X, Gao C and Xiao L: Downregulation of lncRNA ITSN1-2
correlates with decreased disease risk and activity of rheumatoid
arthritis (RA), and reduces RA fibroblast-like synoviocytes
proliferation and inflammation via inhibiting NOD2/RIP2 signaling
pathway. Am J Transl Res. 11:4650–4666. 2019.PubMed/NCBI
|
|
10
|
Ye Y, Gao X and Yang N: LncRNA ZFAS1
promotes cell migration and invasion of fibroblast-like
synoviocytes by suppression of miR-27a in rheumatoid arthritis.
Human Cell. 31:14–21. 2018.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Mendell JT and Olson EN: MicroRNAs in
stress signaling and human disease. Cell. 148:1172–1187.
2012.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Furer V, Greenberg JD, Attur M, Abramson
SB and Pillinger MH: The role of microRNA in rheumatoid arthritis
and other autoimmune diseases. Clin Immunol. 136:1–15.
2010.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Philippe L, Alsaleh G, Pichot A, Ostermann
E, Zuber G, Frisch B, Sibilia J, Pfeffer S, Bahram S, Wachsmann D
and Georgel P: MiR-20a regulates ASK1 expression and TLR4-dependent
cytokine release in rheumatoid fibroblast-like synoviocytes. Ann
Rheum Dis. 72:1071–1079. 2013.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Wang L, Song G, Zheng Y, Wang D, Dong H,
Pan J and Chang X: miR-573 is a negative regulator in the
pathogenesis of rheumatoid arthritis. Cell Mol Immunol. 13:839–848.
2016.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Wang Y, Jiao T, Fu W, Zhao S, Yang L, Xu N
and Zhang N: miR-410-3p regulates proliferation and apoptosis of
fibroblast-like synoviocytes by targeting YY1 in rheumatoid
arthritis. Biomed Pharmacother. 119(109426)2019.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Wang W, Zhang Y, Zhu B, Duan T, Xu Q, Wang
R, Lu L and Jiao Z: Plasma microRNA expression profiles in Chinese
patients with rheumatoid arthritis. Oncotarget. 6:42557–42568.
2015.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Shibanuma M, Mashimo JI, Mita A, Kuroki T
and Nose K: Cloning from a mouse osteoblastic cell line of a set of
transforming-growth-factor-beta1-regulated genes, one of which
seems to encode a follistatin-related polypeptide. Eur J Biochem.
217:13–19. 1993.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Chaly Y, Marinov AD, Oxburgh L, Bushnell
DS and Hirsch R: FSTL1 promotes arthritis in mice by enhancing
inflammatory cytokine/chemokine expression. Arthritis Rheum.
64:1082–1088. 2012.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Shi DL, Shi GR, Xie J, Du XZ and Yang H:
MicroRNA-27a inhibits cell migration and invasion of
fibroblast-like synoviocytes by targeting follistatin-like protein
1 in rheumatoid arthritis. Mol Cells. 39:611–618. 2016.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Aletaha D, Neogi T, Silman AJ, Funovits J,
Felson DT, Bingham CO III, Birnbaum NS, Burmester GR, Bykerk VP,
Cohen MD, et al: 2010 rheumatoid arthritis classification criteria:
An American college of rheumatology/european league against
rheumatism collaborative initiative. Arthritis Rheum. 62:2569–2581.
2010.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2011.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Ge H, Chen S, Huang S and Zhu J: Long
noncoding RNA ZFAS1 acts as an oncogene by targeting miR-193a-3p in
human non-small cell lung cancer. Eur Rev Med Pharmacol Sci.
23:6516–6523. 2019.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Liu Q, Ma H, Sun X, Liu B, Xiao Y, Pan S,
Zhou H, Dong W and Jia L: The regulatory ZFAS1/miR-150/ST6GAL1
crosstalk modulates sialylation of EGFR via PI3K/Akt pathway in
T-cell acute lymphoblastic leukemia. J Exp Clin Cancer Res.
38(199)2019.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Li T, Xie J, Shen C, Cheng D, Shi Y, Wu Z,
Deng X, Chen H, Shen B, Peng C, et al: Amplification of long
noncoding RNA ZFAS1 promotes metastasis in hepatocellular
carcinoma. Cancer Res. 75:3181–3191. 2015.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Paraskevopoulou MD and Hatzigeorgiou AG:
Analyzing miRNA-lncRNA interactions. Methods Mol Biol.
1402:271–286. 2016.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Zhao Z, Lin X, Tong Y and Li W: Silencing
lncRNA ZFAS1 or elevated microRNA-135a represses proliferation,
migration, invasion and resistance to apoptosis of osteosarcoma
cells. Cancer Cell Int. 19(326)2019.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Wu T, Wu D, Wu Q, Zou B, Huang X, Cheng X,
Wu Y, Hong K, Li P, Yang R, et al: Knockdown of long non-coding
RNA-ZFAS1 protects cardiomyocytes against acute myocardial
infarction via anti-apoptosis by regulating miR-150/CRP. J Cell
Biochem. 118:3281–3289. 2017.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Dong D, Mu Z, Wei N, Sun M, Wang W, Xin N,
Shao Y and Zhao C: Long non-coding RNA ZFAS1 promotes proliferation
and metastasis of clear cell renal cell carcinoma via targeting
miR-10a/SKA1 pathway. Biomed Pharmacother. 111:917–925.
2019.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Wang JS, Liu QH, Cheng XH, Zhang WY and
Jin YC: The long noncoding RNA ZFAS1 facilitates bladder cancer
tumorigenesis by sponging miR-329. Biomed Pharmacother.
103:174–181. 2018.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Fu D, Xiao C, Xie Y, Gao J and Ye S:
MiR-3926 inhibits synovial fibroblasts proliferation and
inflammatory cytokines secretion through targeting toll like
receptor 5. Gene. 687:200–206. 2019.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Ehara Y, Sakurai D, Tsuchiya N, Nakano K,
Tanaka Y, Yamaguchi A and Tokunaga K: Follistatin-related protein
gene (FRP) is expressed in the synovial tissues of rheumatoid
arthritis, but its polymorphisms are not associated with genetic
susceptibility. Clin Exp Rheumatol. 22:707–712. 2004.PubMed/NCBI
|
|
32
|
Mattiotti A, Prakash S, Barnett P and van
den Hoff MJB: Follistatin-like 1 in development and human diseases.
Cell Mol Life Sci. 75:2339–2354. 2018.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Clutter SD, Wilson DC, Marinov AD and
Hirsch R: Follistatin-like protein 1 promotes arthritis by
up-regulating IFN-gamma. J Immunol. 182:234–239. 2009.PubMed/NCBI View Article : Google Scholar
|





















