Introduction
Breast cancer is one of the most frequently
diagnosed malignancies among women (1). During the past few decades, the
incidence of breast cancer has continued to increase, and a total
of 2,088,849 newly diagnosed cases of breast cancer were reported
in 2018 worldwide (2,3). The majority of patients with breast
cancer are curable at the non-metastatic stage; however, current
therapeutics exhibit minimal efficacy against advanced-stage
metastatic disease. Based on transcriptional profiling studies,
breast cancer has been established as a heterogeneous malignancy
comprising different subtypes, including ductal carcinoma, lobular
carcinoma, fibroadenoma and ductal carcinoma in situ
(4,5). Based on the molecular characteristics,
breast cancer may be divided into hormone receptor-positive breast
cancer, in which breast cancer cells express estrogen receptor (ER)
or progesterone receptor (PR), or triple-negative breast cancer
(TNBC), in which breast cancer cells are negative for ER, PR and
human epidermal growth factor receptor 2 (HER2) expression.
Although TNBC only accounts for 15-20% of all breast cancer cases,
its aggressive nature and the current lack of effective therapies
make it one of the most lethal malignancies. Therefore, further
understanding the underlying mechanisms involved in TNBC may help
with the development of effective therapeutic strategies.
MicroRNAs (miRNAs/miRs) are small non-coding RNAs,
19-26 nucleotides in length, that have no protein-coding ability.
miRNAs bind to target mRNAs to suppress their translation (6). Accumulating evidence indicates that
miRNAs serve an important role in tumorigenesis, as they are
commonly found to be aberrantly expressed in cancer tissues. The
dysregulated expression of miRNAs results in uncontrolled cell
proliferation, growth and migration in numerous different types of
cancer through regulating the expression of downstream tumor
suppressor genes or oncogenes. miR-301a-3p has been recognized as a
regulator of the T-helper 17 cell immune response in autoimmune
demyelinating diseases (7). In
addition, miR-301a-3p was previously demonstrated to play an
oncogenic role in various types of cancer, including breast cancer
(8-13).
However, to the best of our knowledge, the downstream target genes
of miR-301a-3p in TNBC have not been identified to date.
The present study was undertaken to determine the
significance of miR-301a-3p in TNBC. The expression levels of
miR-301a-3p were analyzed in normal breast MCF-10A cells and in
MDA-MB-231 TNBC cells. The effects of miR-301a-3p on the viability
and migratory ability of MDA-MB-231 cells were then examined, and
the role of mesenchyme homeobox 2 (MEOX2) in this process was also
investigated. The aim was to determine whether miR-301a-3p acts as
an oncogenic miRNA in TNBC and whether its effects are mediated by
regulating MEOX2 expression.
Materials and methods
Patient studies
Human breast cancer tissues were collected from
patients who were diagnosed with TNBC and non-TNBC by three
independent pathologists. The patients were enrolled between March
2016 and May 2020 at the Beijing Obstetrics and Gynecology Hospital
(Beijing, China). The protocol of the present study was approved by
the Clinical Research Ethics Committee of Beijing Obstetrics and
Gynecology Hospital, and written, informed consent was obtained
from each patient for the use of their tissue prior to
participation.
The Cancer Genome Atlas (TCGA) data
analysis
Normalized transcriptome expression datasets for
breast cancer from the TCGA were analyzed using the ENCORI
Pan-Cancer Analysis Platform (http://starbase.sysu.edu.cn/index.php). Briefly, a
total of 1,085 breast cancer tissues and 104 normal tissues were
available for miR-310a-3p expression level analysis.
Cell lines and culture
MCF-10A normal human breast epithelial cells and
MDA-MB-231 TNBC cells were purchased from the American Type Culture
Collection. MDA-MB-231 and MCF-10A cells were cultured in RPMI-1640
and DMEM/F12 medium (HyClone; Cytiva), respectively, supplemented
with FBS (Gibco; Thermo Fisher Scientific, Inc.) and
penicillin-streptomycin solution (Corning, Inc.). The cells were
maintained at 37˚C in an atmosphere containing 5%
CO2.
Cell transfection
MDA-MB-231 cells were plated into 6-well plates and,
upon reaching 70-80% confluence, the cells were transfected with 20
µM miR-301a-3p mimic, mimic negative control (NC), miR-301a-3p
inhibitor and inhibitor NC (all purchased from Guangzhou RiboBio
Co. Ltd.) using Lipofectamine® 3000 reagent (Invitrogen;
Thermo Fisher Scientific, Inc.). Following 48 h of transfection,
cells were collected for use in further experiments. The sequences
of the oligonucleotides were as follows: miR-301a-3p mimic,
5'-GCUCUGACUUUAUUGCACUACU-3'; mimic NC,
5'-UCACAACCUCCUAGAAAGAGUAGA-3'; miR-301a-3p inhibitor,
5'-AGUAGUGCAAUAAAGUCAGAGC-3'; and inhibitor NC,
5'-UCUACUCUUUCUAGGAGGUUGUGA-3'. For MEOX2 knockdown, cells were
transfected with 40 nM small interfering (si)RNA targeted to MEXO2
(5'-AAGGUAGGACAUGUGGUCAGAUCUU-3') and NC
(5'-AAUUAGCGGAGGCUAUAAGAUGUUC-3'), all purchased from Guangzhou
RiboBio Co., Ltd. using RNAiMAX reagent (Invitrogen; Thermo Fisher
Scientific, Inc.). The full-length open reading frame of the human
MEOX2 gene (NM_005924.5) was synthesized and cloned into pcDNA3.1
plasmid. pcDNA3.1 MEOX2 or empty vector (3 µg for 6-well plates)
was transfected into MDA-MB-231 cells using
Lipofectamine® 3000 reagent (Invitrogen; Thermo Fisher
Scientific, Inc.). Following 48 h of transfection, cells were
collected for use in further experiments.
Reverse transcription-quantitative PCR
(RT-qPCR) analysis
MDA-MB-231 cells were washed with PBS and lysed in
0.5 ml RNAiso Plus (Takara Biotechnology Co., Ltd.). Following
incubation for 10 min on ice, 0.1 ml trichloromethane was added to
the lysates and total RNA was isolated. Total RNA was
reverse-transcribed into cDNA, using the PrimeScript RT-PCR kit
according to the manufacturer's instructions (Takara Biotechnology
Co., Ltd.). qPCR was subsequently performed using a SYBR Green PCR
kit (Takara Biotechnology Co., Ltd.). The thermocycling conditions
were as follows: 94°C for 3 min, followed by 40 cycles
of denaturation at 94°C for 30 sec, annealing at
60°C for 30 sec, and extension at 72°C for 30
sec, with a final extension step at 72°C for 10 min. U6
and β-actin served as internal controls for miR-301a-3p and MEOX2
expression levels, respectively. The following primer sequences
were used for the qPCR: MEOX2 forward,
5'-TCAGAAGTCAACAGCAAACCCAG-3' and reverse,
5'-TTCACCAGTTCCTTTTCCCGAG-3'; β-actin forward,
5'-GACCTGACTGACTACCTCATGAAGAT-3' and reverse,
5'-GTCACACTTCATGATGGAGTTGAAGG-3'; miR-301a-3p, forward
5'-CGTGCGAAGCTCAGGAGGG-3' and reverse, 5'-TGGCTGTCGTGGACTGCG-3';
and U6 forward 5'-CTCGCTTCGGCAGCACA-3' and reverse,
5'-AACGCTTCACGAATTTGCGT-3'. RNA expression levels were normalized
to U6 and β-actin. The 2-ΔΔCq method was performed to
determine the relative expression (14).
Western blotting
Total protein was extracted from MDA-MB-231 cells
using RIPA lysis buffer (Beyotime Institute of Biotechnology).
Total protein was quantified using a BCA assay kit, and protein
lysates were then suspended in loading buffer and boiled at 98˚C
for 10 min. Subsequently, 30 µg protein/lane was separated via
10-12% SDS-PAGE and separated proteins were transferred onto PVDF
membranes, which were then blocked in 5% skimmed milk at
4°C overnight. After washing with TBS, the membranes
were incubated with the following primary antibodies at 4-8˚C for
≥12 h: Anti-MEOX2 (1:1,000; cat. no. P50222; RayBiotech, Inc.) and
β-actin (1:1,000; cat. no. GTX11003, ProteinTech Group, Inc.).
Following primary antibody incubation, the membranes were washed
with PBS-Tween-20 (0.1%) thrice and incubated with the
corresponding secondary antibodies [HRP-conjugated goat anti-mouse
IgG (H + L), 1:2,000; cat. no. AS003; ABclonal Biotech Co., Ltd.]
at room temperature for 2 h. Protein bands were visualized using an
ECL-Plus kit (Thermo Fisher Scientific, Inc.).
Cell viability assay
MDA-MB-231 cell viability was determined using an
MTT assay. Briefly, 5x103 cells/well were seeded into
96-well plates and cultured for 72 h. Subsequently, MTT (5 mg/ml)
was added into each well and incubated for a further 3 h. The
culture medium was subsequently discarded and 100 µl DMSO was added
to each well to dissolve the purple formazan crystals. The optical
density value was measured at a wavelength of 595 nm to analyze
cell viability.
Flow cytometric analysis of
apoptosis
Cell apoptosis was analyzed using a propidium iodide
(PI)/Annexin V-FITC kit (Beyotime Institute of Biotechnology).
Briefly, MDA-MB-231 cells were digested with EDTA-free trypsin and
centrifuged at 157 x g for 5 min at room temperature, then the cell
pellet was washed twice with PBS. Subsequently,
0.5-1x105 cells were dissolved in binding buffer and
incubated with 5 µl Annexin V-FITC and 10 µl PI. Apoptotic cells
were analyzed using a BD Accuri™ C6 Plus flow cytometer (BD
Biosciences) using BD Accuri™ C6 software (BD
Biosciences), after which the flow cytometric data were analyzed
using a Guava easyCyte flow cytometer (EMD Millipore).
Cell invasion and migration
assays
Transwell plates with or without Matrigel precoating
were used to determine cell invasion and migration, respectively.
The upper surface of the chamber was precoated with 15 µl of
Matrigel (BD Biosciences) at room temperature for the cell invasion
assay. A total of 1.5x105 MDA-MB-231 cells were plated
into 8.0-µm pore Transwell chambers (BD Biosciences) and cultured
at 37˚C with 5% CO2. The upper chamber was incubated
with FBS free RPMI-1640 and DMEM/F12 medium, repsctively, whereas a
total of 500 µl 10% FBS medium was added in the lower chamber.
Following 24 h of incubation, cells in the upper chamber were
removed by cotton-tipped swabs, while cells in the lower chamber
were fixed with 100% methanol for 30 min at room temperature and
stained with 0.2% crystal violet solution at room temperature. The
chambers were then washed with PBS and dried at room temperature.
Migratory or invasive cells were visualized under an inverted
microscope (magnification, x200; Olympus Corporation).
Wound healing assay
Wound healing assay was performed to analyze the
cell migratory ability. Briefly, 2x105 MDA-MB-231 cells
were seeded into 6-cm culture plates and cultured in RPMI-1640
medium supplemented with 10% FBS for 48 h. Upon cells reaching 90%
confluence, a 200-µl pipette tip was used to create a single linear
scratch in the middle of the cell monolayer. The medium was
removed, and the cells were incubated with RPMI-1640 medium
containing 1% FBS. The wound was visualized using an inverted
microscope (magnification, x200; Olympus Corporation) and images
were captured at 0 and 24 h.
Dual luciferase reporter assay
Wild-type (WT) or mutant (MUT) 3'-untranslated
region (UTR) sequences of MEOX2 were cloned into psi-CHECK vectors
(Promega Corporation). Cells were subsequently transfected with the
aforementioned vectors and co-transfected with miR-301a-3p mimic
(5'-GCUCUGACUUUAUUGCACUACU-3') or negative control
(5'-UCACAACCUCCUAGAAAGAGUAGA-3') (Beijing SyngenTech Co., Ltd.)
using Lipofectamine® 3000 (Invitrogen; Thermo Fisher
Scientific, Inc.), according to the manufacturer's protocol. After
transfection for 48 h, relative luciferase activity was measured
using a Dual-Luciferase Reporter assay system (Promega
Corporation). Firefly luciferase activity was normalized to
Renilla luciferase activity.
Statistical analysis
Statistical analysis was performed using GraphPad
Prism software (version 8.0; GraphPad Software, Inc.) and data are
presented as the mean ± SEM. Statistical differences between two
groups were analyzed using unpaired Student's t-test, while
statistical differences between multiple groups were analyzed using
one-way ANOVA followed by Tukey's post hoc test. P<0.05 was
considered to indicate a statistically significant difference.
Results
miR-301a-3p increases the viability of
MDA-MB-231 cells
The expression levels of miR-301a-3p were
upregulated in breast cancer tissues in datasets from the TCGA
database (Fig. 1A). In addition,
miR-301a-3p expression levels were found to be upregulated in TNBC
tissues compared with normal breast tissue and luminal type breast
cancer tissue according to the TCGA database (Fig. 1B). No statistically significant
differences were observed in miR-301a-3p expression levels between
TNBC and HER+ breast cancer samples (Fig. 1B). The expression levels of
miR-301a-3p in samples from patients with and without TNBC were
subsequently analyzed. RT-qPCR analysis revealed that the
expression levels of miR-301a-3p were upregulated in TNBC tissues
compared with adjacent non-TNBC tissues (Fig. 1C). Subsequently, the expression
levels of miR-301a-3p in normal MCF-10A breast epithelial cells and
MDA-MB-231 TNBC cells were determined. miR-301a-3p expression
levels were found to be upregulated in MDA-MB-231 cells compared
with those in MCF-10A cells (Fig.
1D). Thus, MDA-MB-231 cells were selected to further
investigate the role of miR-301a-3p in TNBC. The cells were divided
into five experimental groups: i) Untreated; ii) miR-301a-3p mimic;
iii) mimic NC; iv) miR-301a-3p inhibitor; and v) inhibitor NC. The
transfection of cells with the miR-301a-3p mimic or inhibitor led
to significantly increased and decreased miR-301a-3p expression
levels, respectively, compared with the respective NCs (Fig. 2A). No statistically significant
differences in cell viability were observed among the untreated, NC
mimic-transfected and NC inhibitor-transfected MDA-MB-231 cells
(Fig. 2B). However, miR-301a-3p
overexpression increased MDA-MB-231 cell viability, whereas
miR-301a-3p knockdown exerted the opposite effect on MDA-MB-231
cells (Fig. 2B). These findings
suggested that miR-301a-3p may serve as an oncogenic miRNA in
TNBC.
 | Figure 1miR-301a-3p expression is upregulated
in breast cancer tissues and cells. (A) miR-301a-3p expression was
analyzed in breast cancer tissues from TCGA database. P<0.01.
(B) miR-301a-3p expression in breast cancer subclasses was analyzed
from TCGA database. Normal vs. TNBC, luminal and HER2-positive, all
***P<0.001. Luminal vs. TNBC, **P<0.01.
HER2-positive vs. TNBC, not significant. Luminal vs. HER2-positive,
*P<0.05. (C) RT-qPCR analysis of miR-301a-3p in
non-TBNC and TBNC tissues. ***P<0.001. (D) RT-qPCR
analysis of miR-301a-3p expression in MCF-10A and MDA-MB-231 cells.
***P<0.001. TCGA, The Cancer Genome Atlas; miR,
microRNA; BC, breast cancer; TNBC, triple-negative breast cancer;
HER2, human epidermal growth factor receptor 2; RT-qPCR, reverse
transcription-quantitative PCR. |
 | Figure 2miR-301a-3p regulates the
proliferation and apoptosis of MDA-MB-231 cells. (A) MDA-MB-231
cells, untreated (blank) or transfected with mimics NC, mimics
miR-301a-3p, inhibitor NC and inhibitor miR-301a-3p were subjected
to reverse transcription-quantitative PCR analysis of miR-301a-3p
expression. *P<0.05 vs. NC. (B) MDA-MB-231 cells,
untreated (blank) or transfected with mimics NC, mimics
miR-301a-3p, inhibitor NC and inhibitor miR-301a-3p were subjected
to MTT analysis of cell proliferation. *P<0.05 vs.
mimics NC; #P<0.05 vs. inhibitor NC. (C) MDA-MB-231
cells of untreated or transfected with mimics NC, mimics
miR-301a-3p, inhibitor NC and inhibitor miR-301a-3p were subjected
to flow cytometry analysis of apoptosis. (D) Quantification of
apoptosis as shown in C. *P<0.05 vs. NC. miR,
microRNA; NC, negative control. |
miR-301a-3p suppresses cell
apoptosis
The regulatory role of miR-301a-3p on apoptosis was
subsequently determined in MDA-MB-231 cells using PI/Annexin V-FITC
double staining and flow cytometric analysis. Compared with the
cells transfected with the mimic NC, transfection with the
miR-301a-3p mimic significantly inhibited the apoptosis of
MDA-MB-231 cells. Conversely, miR-301a-3p knockdown induced
MDA-MB-231 cell apoptosis (Fig. 2C
and D). These results suggested
that miR-301a-3p may suppress apoptosis in TNBC cells.
miR-301a-3p induces the migration and
invasion of MDA-MB-231 cells
TNBC is a highly metastatic cancer. Thus, transwell
plates with or without Matrigel precoating and wound healing assays
were performed to analyze cell invasion and migration,
respectively. No statistically significant differences were
observed among the untreated, mimic NC-transfected and inhibitor
NC-transfected MDA-MB-241 cells. The transfection of MDA-MB-231
cells with the miR-301a-3p mimic increased the migratory and
invasive abilities. By contrast, transfection of cells with the
miR-301a-3p inhibitor inhibited cell migration and invasion
(Fig. 3). These data indicated that
miR-301a-3p may function as an oncomiR in TNBC.
 | Figure 3miR-301a-3p promotes the migration and
invasion of MDA-MB-231 cells. (A) MDA-MB-231 cells, untreated
(blank) or transfected with mimics NC, mimics miR-301a-3p,
inhibitor NC and inhibitor miR-301a-3p were subjected to Transwell
migration (upper images) and invasion (lower images) assays
(magnification, x200). (B) MDA-MB-231 cells, untreated (blank) or
transfected with mimics NC, mimics miR-301a-3p, inhibitor NC and
inhibitor miR-301a-3p were subjected to wound healing assay. Images
were captured at 0 h (upper panels) and 24 h (lower panels)
(magnification, x200). (C and D) Quantification of cell migration
and invasion as shown in A and B. **P<0.01. miR,
microRNA; NC, negative control. |
miR-301a-3p downregulates the
expression levels of MEOX2
Downstream target genes of miR-301a-3p in MDA-MB-231
cells were subsequently identified. RT-qPCR analysis revealed that
MEOX2 mRNA expression levels were downregulated in MDA-MB-231 cells
following transfection with the miR-301a-3p mimic. Conversely,
transfection with the miR-301a-3p inhibitor upregulated the
expression levels of MEOX2 in MDA-MB-231 cells (Fig. 4A). The results of western blotting
demonstrated that MEOX2 protein expression levels were also
decreased and increased following the overexpression and knockdown
of miR-301a-3p, respectively (Fig.
4B). Analysis of data from TCGA database revealed that
miR-301a-3p expression levels were inversely associated with MEOX2
expression levels in breast cancer tissues (Fig. 4C). To investigate whether
miR-301a-3p bound to the 3'-UTR sequence of MEOX2, WT and MUT
3'-UTR sequences of MEOX2 were synthesized and cloned into
luciferase reporter vectors. The relative luciferase activity was
subsequently measured following the overexpression of miR-301a-3p.
The results demonstrated that miR-301a-3p overexpression reduced
the relative luciferase activity of MDA-MB-231 cells when
co-transfected with the WT, but not the MUT, 3'-UTR sequence of
MEOX2 (Fig. 4D and E). Collectively, these results suggested
that miR-301a-3p may directly target MEOX2 by binding to its 3'-UTR
sequence.
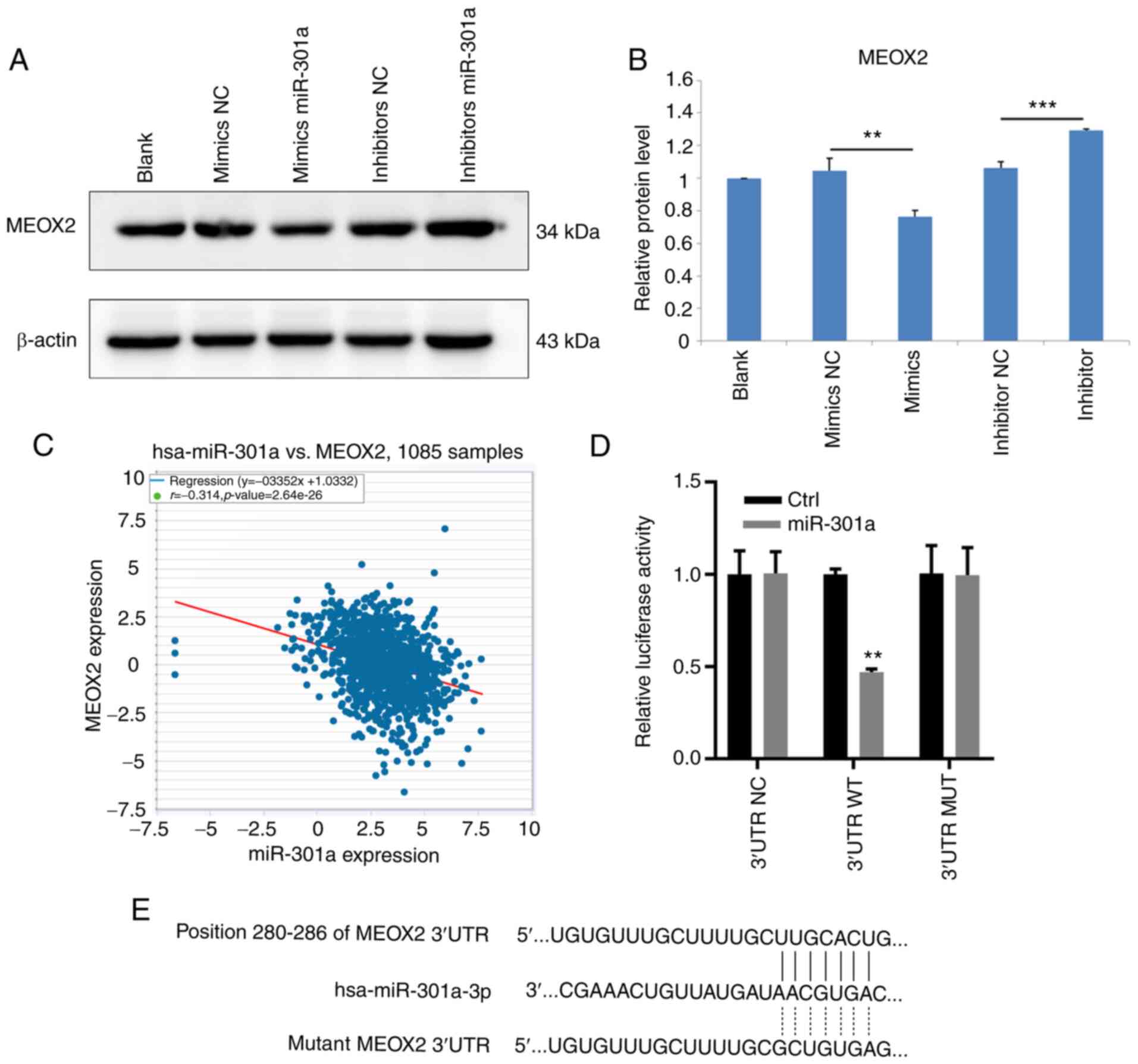 | Figure 4MEOX2 is a downstream target of
miR-301a-3p in MDA-MB-231 cells. (A) Western blot result of MEOX2
in MDA-MB-231 cells, untreated (blank) or transfected with mimics
NC, mimics miR-301a-3p, inhibitor NC and inhibitor miR-301a-3p. (B)
Relative protein level of MEOX2 in MDA-MB-231 cells, untreated or
transfected with mimics NC, mimics miR-301a-3p, inhibitor NC and
inhibitor miR-301a-3p. **P<0.01 vs. mimics NC; and
***P<0.001 vs. inhibitor NC. (C) Analysis of the
correlation between miR-301a-3p and MEOX2 in breast cancer tissues
from The Cancer Genome Atlas database. (D) WT or MUT sequence of
MEOX2 3'-UTR was inserted into luciferase reporter vectors. At 48 h
after transfecting MEOX2 overexpression vectors, luciferase
activity was examined as indicated. **P<0.01 vs.
control. (E) Binding sequence of miR-301a-3p with the 3'-UTR of
MEOX2. MEOX2, mesenchyme homeobox 2; miR, microRNA; NC, negative
control; WT, wild-type; MUT, mutant; UTR, untranslated region. |
MEOX2 acts as a tumor suppressor gene
in TNBC
Finally, the role of MEOX2 in TNBC cells was
investigated. MEOX2 expression was knocked down by transfection
with siRNA and overexpressed by the pcDNA3.1 vector RT-qPCR and
western blotting confirmed successful MEOX2 knockdown and
overexpression in MDA-MB-231 cells (Fig. 5A and B). The results of the MTT assay revealed
that MEOX2 knockdown enhanced, while MEOX2 overexpression reduced
MDA-MB-231 cell viability (Fig.
5C). Furthermore, MEOX2 was overexpressed in cells also
overexpressing miR-301a-3p (Fig.
5D). The overexpression of MEOX2 further suppressed the
viability of cells overexpressing miR-301a-3p (Fig. 5E). MEOX2 overexpression also
increased the levels of apoptosis in MDA-MB-231 cells (Fig. 5F). These findings suggested that
MEOX2 may serve as a tumor suppressor in TNBC.
 | Figure 5MEOX2 suppresses MDA-MB-231 cell
viability. (A) RT-qPCR and (B) western blot analyses of MEOX2
expression in MEOX2-knockdown and -overexpressing MDA-MB-231 cells.
**P<0.01 vs. siCtrl and ***P<0.001 vs.
Ctrl. (C) MTT assay was used to determine the viability of
MEOX2-knockdown and -overexpressing MDA-MB-231 cells.
*P<0.05 vs. mimics, **P<0.01 vs. siCtrl
or vs. Ctrl, respectively. (D) RT-qPCR and western blot analyses of
MEOX2 expression following transfection with mimics NC, mimics
miR-301a-3p and mimics miR-301a-3p + MEOX2 in MDA-MB-231 cells.
**P<0.01 and ***P<0.001 vs. mimics NC.
(E) MTT assay was used to determine cell viability following
transfection with mimics NC, mimics miR-301a-3p and mimics
miR-301a-3p + MEOX2 in MDA-MB-231 cells. **P<0.01 vs.
mimics NC; and #P<0.05 vs. mimics miR301a. (F)
MDA-MB-231 cell apoptosis was analyzed following transfection with
mimics NC, mimics miR-301a-3p and mimics miR-301a-3p + MEOX2.
**P<0.01. MEOX2, mesenchyme homeobox 2; miR,
microRNA; NC, negative control; si, small interfering RNA; Ctrl,
control; RT-qPCR, reverse transcription-quantitative PCR. |
Discussion
Breast cancer is the most common malignancy among
women, and poses a major threat to women's health in both developed
and developing countries. Over the past few decades, researchers
have made significant efforts to determine the molecular mechanisms
underlying breast cancer tumorigenesis. The expression of the ER
has been reported as a major risk factor for breast cancer
development. Tamoxifen, which interrupts the interaction between
estrogen and the ER, is a drug approved for the treatment of
ER+ breast cancer. The discovery of HER-targeted therapy
also represented a significant milestone in breast cancer research.
Trastuzumab, which was developed in 1990, inhibits the
extracellular segment of HER2 and, combined with chemotherapy, can
effectively prolong the overall and progression-free survival of
patients with HER+ breast cancer (15-17).
However, 15-20% of patients with breast cancer are diagnosed with
TNBC, which remains difficult to treat as the currently available
targeted therapies are not applicable, as this type of cancer is
negative for ER, PR and HER2 expression. Therefore, there are
currently no effective treatment options for patients with TNBC,
except for traditional methods, such as surgical resection,
radiotherapy and chemotherapy.
The present study aimed to determine the role of
miR-301a-3p in TNBC. miR-301a-3p expression levels were upregulated
in breast cancer tissues. Notably, the expression levels of
miR-301a-3p were upregulated in TNBC samples compared with adjacent
non-TNBC samples. The overexpression of miR-301a-3p increased the
viability and migratory ability of MDA-MB-231 cells. By contrast,
miR-301a-3p knockdown suppressed the viability and migration of
MDA-MB-231 cells. Mechanistically, miR-301a-3p downregulated the
expression levels of MEOX2 by directly binding to the 3'-UTR
sequence of MEOX2. Further experiments demonstrated that MEOX2
acted as a tumor suppressor gene in TNBC.
miR-301a, which comprises miR-301a-3p and
miR-301a-5p, was found to play a crucial role in cancer
development, including pancreatic, colorectal and gastric cancer.
The expression levels of miR-301a were reported to be upregulated
in pancreatic cancer and promoted pancreatic cancer cell
proliferation by targeting Bim (18). The knockdown of miR-301a-3p
expression in pancreatic cancer cells was also found to increase
the sensitivity of cells to gemcitabine treatment (19). In addition, miR-301a promoted the
growth and invasion of colorectal cancer by targeting suppressor of
cytokine signaling 6 or transforming growth factor β receptor 2
(20,21). In gastric cancer, miR-301a was also
discovered to function as an oncogene. The overexpression of
miR-301a was associated with a poor prognosis in gastric cancer and
was found to downregulate RUNX family transcription factor 3
expression levels to promote disease progression (22,23).
The role of miR-301a in breast cancer has also been previously
investigated. For example, upregulated expression levels of
miR-301a were found to be inversely associated with the prognosis
of breast cancer (8,24). The overexpression of miR-301a also
promoted breast cancer cell migration and invasion by
downregulating the expression levels of PTEN, a tumor suppressor
gene (9). The results of the
present study revealed that the expression levels of miR-301a-3p
were upregulated in TNBC tissues. MDA-MB-231 cells transfected with
miR-301a-3p mimics exhibited increased viability compared with
MDA-MB-231 cells transfected with NC mimics. The opposite effects
were observed following miR-301a-3p knockdown. Furthermore,
miR-301a-3p overexpression promoted MDA-MB-231 cell migration and
invasion. These results suggested that miR-301a-3p may act as an
oncogene in TNBC.
MEOX2 has been reported to function both as an
oncogene and tumor suppressor, depending on the cancer type. For
example, single-nucleotide polymorphism-based sequencing studies
reported that MEOX2 served as a tumor suppressor gene in Wilms'
tumor (25). In another study,
MEOX2 expression was upregulated in laryngeal carcinoma and lung
cancer tissues (26,27), and the overexpression of MEOX2
promoted laryngeal cancer growth by activating the PI3K/AKT
signaling pathway. The upregulated expression levels of MEOX2 were
also found to contribute to chemoresistance in lung cancer. In
another study, MEOX2 was found to be downregulated by miR-301a in
hepatocellular carcinoma (HCC) (28); miR-301a promoted HCC cell
proliferation, migration and invasion by downregulating MEOX2
expression. However, to the best of our knowledge, the association
between miR-301a-3p and MEOX2 in TNBC remains to be determined. The
findings of the present study revealed that overexpression of
miR-301a-3p downregulated MEOX2 expression levels in MDA-MB-231
cells. The results of the dual luciferase reporter assay
demonstrated that miR-301a-3p directly bound to the 3'-UTR sequence
of MEOX2. In addition, the knockdown of MEOX2 expression promoted
TNBC cell viability. Further analysis also identified that the
expression levels of miR-301a-3p were inversely correlated with
those of MEOX2 in breast cancer samples. Thus, it was suggested
that the reduction in MEOX2 expression may promote TNBC cell
viability.
In conclusion, the results of the present study
indicated that miR-301a-3p may serve as an oncogene in TNBC. The
overexpression of miR-301a-3p promoted TNBC cell viability and
migration, while the knockdown of miR-301a-3p exerted the opposite
effects. Furthermore, miR-301a-3p was found to exert its effects in
TNBC by downregulating the expression levels of MEOX2. The findings
of the preset study highlight the importance of miR-301a-3p in TNBC
and may indicate a possible target for the treatment of this
disease.
Acknowledgements
Not applicable.
Funding
Funding: The present study was supported by the Beijing
Obstetrics and Gynecology Hospital, Capital Medical University
(grant no. FCYY201816).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
HL and GW designed the current study. HL performed
the experiments. GW supervised the present study. HL and GW confirm
the authenticity of all the raw data. HL and GW drafted, reviewed
and edited the manuscript. All authors read and approved the final
version of manuscript.
Ethics approval and consent to
participate
The protocol of the present study was approved by
the Clinical Research Ethics Committee of Beijing Obstetrics and
Gynecology Hospital, and written, informed consent was obtained
from each patient for the use of their tissue prior to
participation.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Harbeck N, Penault-Llorca F, Cortes J,
Gnant M, Houssami N, Poortmans P, Ruddy K, Tsang J and Cardoso F:
Breast cancer. Nat Rev Dis Primers. 5(66)2019.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Waks AG and Winer EP: Breast cancer
treatment: A review. JAMA. 321:288–300. 2019.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Huang J, Chan PS, Lok V, Chen X, Ding H,
Jin Y, Yuan J, Lao XQ, Zheng ZJ and Wong MC: Global incidence and
mortality of breast cancer: A trend analysis. Aging (Albany NY).
13:5748–5803. 2021.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Perou CM, Sørlie T, Eisen MB, van de Rijn
M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA,
et al: Molecular portraits of human breast tumours. Nature.
406:747–752. 2000.PubMed/NCBI View
Article : Google Scholar
|
|
5
|
Sørlie T, Perou CM, Tibshirani R, Aas T,
Geisler S, Johnsen H, Hastie T, Eisen MB, van de Rijn M, Jeffrey
SS, et al: Gene expression patterns of breast carcinomas
distinguish tumor subclasses with clinical implications. Proc Natl
Acad Sci USA. 98:10869–10874. 2001.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Gregory RI, Chendrimada TP, Cooch N and
Shiekhattar R: Human RISC couples microRNA biogenesis and
posttranscriptional gene silencing. Cell. 123:631–640.
2005.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Mycko MP, Cichalewska M, Machlanska A,
Cwiklinska H, Mariasiewicz M and Selmaj KW: MicroRNA-301a
regulation of a T-helper 17 immune response controls autoimmune
demyelination. Proc Natl Acad Sci USA. 109:E1248–E1257.
2012.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Yu H, Li H, Qian H, Jiao X, Zhu X, Jiang
X, Dai G and Huang J: Upregulation of miR-301a correlates with poor
prognosis in triple-negative breast cancer. Med Oncol.
31(283)2014.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Ma F, Zhang J, Zhong L, Wang L, Liu Y,
Wang Y, Peng L and Guo B: Upregulated microRNA-301a in breast
cancer promotes tumor metastasis by targeting PTEN and activating
Wnt/β-catenin signaling. Gene. 535:191–197. 2014.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Li X, Li J, Cai Y, Peng S, Wang J, Xiao Z,
Wang Y, Tao Y, Li J, Leng Q, et al: Hyperglycaemia-induced miR-301a
promotes cell proliferation by repressing p21 and Smad4 in prostate
cancer. Cancer Lett. 418:211–220. 2018.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Wang YG, Wang T, Shi M and Zhai B: Long
noncoding RNA EPB41L4A-AS2 inhibits hepatocellular carcinoma
development by sponging miR-301a-5p and targeting FOXL1. J Exp Clin
Cancer Res. 38(153)2019.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Li X, Zhong M, Wang J, Wang L, Lin Z, Cao
Z, Huang Z, Zhang F, Li Y, Liu M and Ma X: miR-301a promotes lung
tumorigenesis by suppressing Runx3. Mol Cancer.
18(99)2019.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Hu H, Zhang Q, Chen W, Wu T, Liu S, Li X,
Luo B, Zhang T, Yan G, Lu H and Lu Z: MicoRNA-301a promotes
pancreatic cancer invasion and metastasis through the JAK/STAT3
signaling pathway by targeting SOCS5. Carcinogenesis. 41:502–514.
2020.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
15
|
von Minckwitz G, Huang CS, Mano MS, Loibl
S, Mamounas EP, Untch M, Wolmark N, Rastogi P, Schneeweiss A,
Redondo A, et al: Trastuzumab emtansine for residual invasive
HER2-positive breast cancer. N Engl J Med. 380:617–628.
2019.PubMed/NCBI View Article : Google Scholar
|
|
16
|
von Minckwitz G, Procter M, de Azambuja E,
Zardavas D, Benyunes M, Viale G, Suter T, Arahmani A, Rouchet N,
Clark E, et al: Adjuvant pertuzumab and trastuzumab in early
HER2-positive breast cancer. N Engl J Med. 377:122–131.
2017.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Slamon D, Eiermann W, Robert N, Pienkowski
T, Martin M, Press M, Mackey J, Glaspy J, Chan A, Pawlicki M, et
al: Adjuvant trastuzumab in HER2-positive breast cancer. N Engl J
Med. 365:1273–1283. 2011.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Chen Z, Chen LY, Dai HY, Wang P, Gao S and
Wang K: miR-301a promotes pancreatic cancer cell proliferation by
directly inhibiting Bim expression. J Cell Biochem. 113:3229–3235.
2012.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Xia X, Zhang K, Luo G, Cen G, Cao J, Huang
K and Qiu Z: Downregulation of miR-301a-3p sensitizes pancreatic
cancer cells to gemcitabine treatment via PTEN. Am J Transl Res.
9:1886–1895. 2017.PubMed/NCBI
|
|
20
|
Fang Y, Sun B, Xiang J and Chen Z:
MiR-301a promotes colorectal cancer cell growth and invasion by
directly targeting SOCS6. Cell Physiol Biochem. 35:227–236.
2015.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Zhang W, Zhang T, Jin R, Zhao H, Hu J,
Feng B, Zang L, Zheng M and Wang M: MicroRNA-301a promotes
migration and invasion by targeting TGFBR2 in human colorectal
cancer. J Exp Clin Cancer Res. 33(113)2014.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Wang M, Li C, Yu B, Su L, Li J, Ju J, Yu
Y, Gu Q, Zhu Z and Liu B: Overexpressed miR-301a promotes cell
proliferation and invasion by targeting RUNX3 in gastric cancer. J
Gastroenterol. 48:1023–1033. 2013.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Xu XD, He XJ, Tao HQ, Zhang W, Wang YY, Ye
ZY and Zhao ZS: Abnormal expression of miR-301a in gastric cancer
associated with progression and poor prognosis. J Surg Oncol.
108:197–202. 2013.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Zheng JZ, Huang YN, Yao L, Liu YR, Liu S,
Hu X, Liu ZB and Shao ZM: Elevated miR-301a expression indicates a
poor prognosis for breast cancer patients. Sci Rep.
8(2225)2018.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Ohshima J, Haruta M, Arai Y, Kasai F,
Fujiwara Y, Ariga T, Okita H, Fukuzawa M, Hata J, Horie H and
Kaneko Y: Two candidate tumor suppressor genes, MEOX2 and SOSTDC1,
identified in a 7p21 homozygous deletion region in a Wilms tumor.
Genes Chromosomes Cancer. 48:1037–1050. 2009.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Tian L, Tao Z, Ye H, Li G, Zhan Z and Tuo
H: Over-expression of MEOX2 promotes apoptosis through inhibiting
the PI3K/Akt pathway in laryngeal cancer cells. Neoplasma.
65:745–752. 2018.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Ávila-Moreno F, Armas-López L,
Álvarez-Moran A, López-Bujanda Z, Ortiz-Quintero B, Hidalgo-Miranda
A, Urrea-Ramírez F, Rivera-Rosales RM, Vázquez-Manríquez E,
Peña-Mirabal E, et al: Overexpression of MEOX2 and TWIST1 is
associated with H3K27me3 levels and determines lung cancer
chemoresistance and prognosis. PLoS One. 9(e114104)2014.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Zhou P, Jiang W, Wu L, Chang R, Wu K and
Wang Z: miR-301a is a candidate oncogene that targets the homeobox
gene Gax in human hepatocellular carcinoma. Dig Dis Sci.
57:1171–1180. 2012.PubMed/NCBI View Article : Google Scholar
|



















