Introduction
Glioma remains to be an aggressive type of brain
tumor with limited improvement to its diagnosis and treatment
methoods over the past decades (1,2). The
molecular biology underlying glioma is complex, which obstructs the
development of specific diagnostic and treatment strategies
(3,4). The dysregulation of a number of
signaling pathways, including the Epidermal growth factor
receptor/AKT/PTEN, receptor tyrosine kinase (RTK)/PI3K,
Wnt/β-catenin and NF-κB pathways has been shown to occur during the
development of glioma (4-6).
Dysregulated RTK, RB and tumor protein p53 (TP53) pathways in
glioma have been highlighted by previous genomic analyses (7,8).
However, the mechanism promoting the aggressive pathophysiology of
glioma remain unclear.
BCL6 is an important regulator of the germinal
center response and protooncogene expression in diffuse large
B-cell lymphoma (9). This protein
has been reported to serve an important role in the pathology of
multiple tumors and diseases, including breast cancer, ovarian
cancer and glioma (10-12).
It has been previously shown that BCL6 exprssion is regulated by
transcription factors such as STAT3 and STAT5B and epigenetic
modifications, including DNA methylation and histone modifications
(13). Furthermore, BCL6 has been
proposed to be a prognostic marker and a targetable
glioma-promoting factor (12)
whereby knocking down he expression of this protein can inhibit the
pathophysiology of glioma cells and trigger cell apoptosis
(12). However, the precise role of
the BCL6 regulatory network within glioma requires additional
studies.
microRNA (miR or miRNA)-mediated regulation of BCL6
expression has been previously documented to serve a role in tumor
progression of breast cancer, by targeting the 3'-untranslated
region (3'UTR) sequence of BCL6 mRNA (14). miRNA-187-3p, miR-519d-3p and miR-554
can also suppress cell proliferation and invasion by downregulating
BCL6 expression in gastric, non-small cell lung and breast cancers
(14-16).
miRNAs are a class of small non-coding RNAs that can bind to target
mRNAs to regulate gene expression on a post-transcriptional level
(17,18). Previous expression studies on
specific miRNAs have reported their association with the pathology
and prognosis of glioma (19-21).
The different expression profile of miRNAs in glioma compared with
healthy brain tissues implicate their role in the regulation of
tumorigenic progression, possibly in processes including glioma
proliferation, metastasis, invasion, angiogenesis and drug
resistance (20). The development
of glioma is associated with a significant reduction in the levels
of antitumor miRNAs, such as miR-7 and miR-137(22). By contrast, the opposite trend is
noted with regards to the expression levels of the protooncogenic
miRNAs, including miR-10b, miR-21, miR-182, miR-222 and miR-423-5p,
all of which have been previously demonstrated to promote
tumorigenesis and development of glioma (20,21).
In the present study, the ability of miR-144-3p to
act as a suppressor of glioma development was investigated by using
reverse transcription-quantitative PCR (RT-qPCR), western blotting
and luciferase reporter assays and a tumor xenograft model. These
findings may provide a new and potential treatment strategy for
glioma.
Materials and methods
Tissue collection
A total of 25 glioma tissues were obtained from who
underwent surgical resection and 10 normal human brain tissues
(mean age, 54.74±6.52 years; 3 female and 7 male) were obtained
from patients with brain injury who underwent partial resections of
brain tissues to reduce intracranial pressure were collected from
individuals at the Peking University People's Hospital (Beijing,
China) between February 2017 and January 2018. The mean age of
patients was 60.36±8.81 years (≥60 years, n=16; <60, n=9),
including 11 female and 14 male. The inclusion criteria were that
all glioma samples had confirmed pathological diagnosis and
patients who did not receive any other therapies before surgery.
Patients who with complications, such as other malignant tumors,
serious systemic infections and other severe systemic diseases,
were excluded. The tissues were rapidly placed in liquid nitrogen
and subsequently stored at -80˚C. An aliquot of the tissue was
pathologically examined prior to further examination. All
participants met the following inclusion criteria that no
preoperative radiotherapy or chemotherapy was performed. The
clinical samples were collected after written informed consent was
obtained from each participant and after the study was authorized
by the Ethics Committee of Peking University People's Hospital
(Beijing, China).
Cell culture
The glioma U251 cell line was purchased from the
Cell Bank of Type Culture Collection of the Chinese Academy of
Sciences. U251 cells were cultured in DMEM (Invitrogen; Thermo
Fisher Scientific, Inc.) with 10% FBS (Invitrogen; Thermo Fisher
Scientific, Inc.) and 1% penicillin-streptomycin (Gibco; Thermo
Fisher Scientific, Inc.) at 37˚C in a humidified atmosphere in the
presence of 5% CO2.
Transfection assay
BCL6-overexpressing (pcDNA3.1, Cyagen Biosciences,
Inc.; NCBI Accession Gene ID: 604; https://www.ncbi.nlm.nih.gov/nuccore/NM_001130845.1).
Total RNA was extracted from U251 cells and reversed transcribed
into cDNA as a template for PCR amplification (forward,
5'-CCGGAATTCATGGCCTCGCCGGCTGACAGCTGT-3' and reverse,
5'-CCCAAGCTTTCAGCAGGCTTTGGGGAGCTC-3'; Ultra HiFidelity PCR Kit,
cat. no. KP203, Tiangen Biotech Co., Ltd.; thermocycling
conditions, 94˚C for 3 min, followed by 30 cycles of 94˚C for 30
sec, 60˚C for 30 sec and 72˚C for 5 min). After the BCL6 target
gene was obtained, it was ligated and recombined with pcDNA3.1
vector. BCL6 short hairpin (sh) RNA
(5'-tcgagtgctgttgacagtgagcgaGCCTGTTCTATAGCATCTTTAtagtgaagccacagatgtaTAAAGATGCTATAGAACAGGCgtgcctactgcctcggaa-3')
plasmid (pcDNA3.1), the corresponding negative control (NC) plasmid
(NC-shRNA, 5'-TTCTCCGAACGTGTCACGT-3'), miR-144-3p mimic
(5'-UCAUGUAGUAGAUAUGACAU-3'), miR-144-3p NC
(5'-UUCUCGAACGUGUCACGUUUU-3'), miR-144-3p inhibitor
(5'-AUGUCAUAUCUACUACAUGA-3'), and inhibitor NC
(5'-CAGUACUUUUGUGUAGUACAA-3') were obtained from Cyagen Bioscience,
Inc. A mixed solution containing the plasmid (2 µg) and miRs (20
nM) and Lipofectamine® 3000 (Invitrogen; Thermo Fisher
Scientific, Inc.) was diluted with serum-free DMEM. According to
the manufacturer's protocols, the transfection mixture was added to
the cell suspension and replaced with complete medium following
incubation for 8 h at 37˚C. Subsequent experiments were conducted
following 48 h of cell culture.
Luciferase reporter assay
Bioinformatic prediction were conducted by starBase
v2.0 (http://starbase.sysu.edu.cn/agoClipRNA.php?source=mRNA)
and TargetScanHuman v7.2 (http://www.targetscan.org/vert_72/). The pMIR-BCL6
Luciferase vector (Promega Corporation) was prepared using the
3'-UTR sequence of BCL6. To generate the reporter vectors
containing the potential binding sites of miR-144-3p, the DNA
fragments containing the 3'UTR of target the gene BCL6 (NCBI
Accession Gene ID: 604; https://www.ncbi.nlm.nih.gov/nuccore/NM_001130845.1)
were amplified (3'-UTR BCL6 forward, 5'-GTTGATGCTTTCGTCTCCAGC-3'
and reverse, 5'-ATCCCATGATGTAGTGCCTCT-3'). Ultra Super High
Fidelity PCR Kit (cat. no. KP203, Tiangen Biotech Co., Ltd.) was
conducted from the cDNA extracted from U251 cells by following
conditions: 94˚C for 3 min, followed by 30 cycles of 94˚C for 30
sec, 60˚C for 30 sec and 72˚C for 5 min. The fragments were then
cloned into the pmirGLO luciferase reporter plasmid (Promega
Corporation). Quick-Change Site-Directed Mutagenesis kit (cat. no.
210518; Stratagene; Agilent Technologies, Inc.) was used for gene
mutation for the production of the pMIR-BCL6-Mut plasmid. Following
the seeding of 5x103 U251 cells into 24-well plates, the
cells were co-transfected with 200 ng pMIR-BCL6 or pMIR-BCL6-Mut
and 50 nM miR-144-3p mimic or miR-control using the
Lipofectamine® 3000 reagent (Invitrogen; Thermo Fisher
Scientific, Inc.). Subsequently, the cells were harvested and lysed
following 36 h of cell culture after transfection. The
Dual-Luciferase® Reporter Assay System (Promega
Corporation) was used for measuring Renilla luciferase and
firefly luciferase activity using a Glomax® Luminometer
(Promega Corporation). Normalized Renilla-luciferase values
were represented relative to the control according to the
manufacturer's protocol.
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA was extracted from tissues and U251 cells
using TRIzol® Reagent (Invitrogen; Thermo Fisher
Scientific, Inc.) according to manufacturer's protocol.
PrimeScript™ RT reagent Kit (cat. no. RR037A; Takara Bio, Inc.) was
used to reverse transcribe the cDNA starting from total RNA using
the protocol of 37˚C for 15 min, 85˚C for 5 sec, and 4˚C
preservation. For mRNA and miR detection, qPCR experiments were
conducted using the SuperReal PreMix Plus (SYBR Green; cat. no.
FP205; Tiangen Biotech Co., Ltd.). qPCR was carried out under the
following conditions: 95˚C for 30 sec, followed by 35 cycles of
95˚C for 5 sec, 60˚C for 30 sec and 68˚C for 5 min. Each sample was
analyzed in triplicate, and quantified using the 2-ΔΔCq
method (23). The sequences of
primers were as follows: BCL6 forward, 5'-GTGATGGCCACGGCTATGTA-3'
and reverse, 5'-CATCCGGCTGTTGAGGAACT-3' and GAPDH (internal
reference) forward, 5'-ACCATCTTCCAGGAGCGAGA-3' and reverse,
5'-GACTCCACGACGTACTCAGC-3'.
miRNA qPCR
The miRcute Plus miRNA qPCR Kit (cat. no. FP411;
Tiangen Biotech Co., Ltd.) was used to measure miR-144-3p levels.
Following reverse transcription (42˚C for 60 min, 95˚C for 3 min
and 4˚C preservation) by the miRcute Plus miRNA First-Strand cDNA
Kit (cat. no. KR211; Tiangen Biotech Co., Ltd.), RT-qPCR was
conducted by a miR-144-3p forward primer and a universal reverse
primer. PCR reaction conditions were: Pre-denaturation at 95˚C for
15 min, followed by 40 cycles of denaturation at 94˚C for 20 sec
and annealing and extension at 60˚C for 34 sec. Each sample was
analyzed in triplicate, and quantified using the 2-ΔΔCq
method (23). The primers were as
follows: hsa-miR-144-3p, 5'-CCCTACAGTATAGATGATGTA-3'; and U6 small
nuclear RNA (internal reference), 5'-CAAATT CGTGAAGCGTTCCATAT-3'.
The universal reverse primers of hsa-miR-144-3p and U6 were
purchased from Tiangen Biotech Co., Ltd (cat. no. FP411).
Western blotting analysis
The U251 cells were first lysed using RIPA lysis
buffer and protein concentrations were quantified by using a BCA
kit (Thermo Fisher Scientific, Inc.), before being subsequently
boiled at 100˚C for 5 min and stored at -20˚C. In total, 30 µg
proteins were separated by 10% SDS-PAGE gels and transferred onto
PVDF membranes, which were blocked with 5% milk in TBST (0.05%
Tween 20) for 1 h at room temperature. The membranes were incubated
with the following primary antibodies at 4˚C overnight: Anti-BCL6
(cat. no. ab33901; 1:1,000; Abcam) and anti-GAPDH (cat. no.
ab181602; 1:2,000; Abcam). The following morning, the membranes
were incubated with HRP-linked secondary antibody (cat. no.
ASR1089; 1:20,000; Abcepta Biotech Co., Ltd.) for 1 h at room
temperature. The bands were visualized using an enhanced
chemiluminescence detection system (cat. no. 1705060; Bio-Rad
Laboratories, Inc.). Optical densities of the bands were calculated
using a MiVnt image analysis system (Version 5.2.1; Bio-Rad
Laboratories, Inc.).
Cell Counting Kit-8 (CCK-8) assay
Cell viability was assessed using the CCK8 assay.
U251 cells were seeded at a density of 3x103 cells in
96-well plates before being transfected with the miR-144-3p mimic
(or NC) or inhibitor (or inhibitor NC) and cultured for 12, 24, 48
or 72 h at 37˚C. A total of 10 µl CCK-8 solution (Dojindo Molecular
Technologies, Inc.) was added into the culture medium in each well
and incubated for 1 h at 37˚C. The microplate reader (Thermo Fisher
Scientific Inc.) was then used for measuring the optical density in
each well at 450 nm.
Cell invasion assay
Transwell assay was conducted using a Transwell
chamber (8 µm; cat. no. 3422; Corning, Inc.) pre-coated with
Matrigel Matrix (Matrigel 1:8 dilution with DMEM put onto the upper
compartment surface of the membrane at the bottom of the Transwell
and incubated at 37˚C for 2 h; BD Biosciences) at 37˚C. The bottom
chamber of the wells contained 750 µl DMEM with 10% FBS whereas the
top chamber contained 250 µl cell suspension (1x105/ml
with 2% FBS). Following culture of the cells at 24 h at 37˚C, the
cells that failed to invade were removed from the top chamber and
subsequently the cells on the membrane were stained with 0.5%
crystal violet for 15 min at room temperature. The images were
obtained from three independents fields (magnification, x10)/well
using an inverted light microscope (Olympus Corporation). Following
culture of the cells at 24 h at 37˚C, the cells that failed to
invade were removed from the top chamber and subsequently the
number of cells on membrane were measured by the CCK-8 assay. The
added CCK-8 solution was used to determine the relative cell
invasive capacity by measuring the OD at 450 nm.
Tumor xenograft model
The in vivo animal assay was authorized by
the Institutional Animal Care and Use Committee and the Local
Ethics Board of the Peking University People's Hospital (Beijing,
China). In total, 6-8 week old male nude mice (n=24) were purchased
from Beijing Vital River Laboratory Animal Technology Co., Ltd and
fed under specific pathogen free conditions, in the mouse-feeding
facility with a 12-h light/dark cycle at 22±2˚C and 50-60%
humidity, with free foraging and activity and free access to food
and water. Following transfection for 48 h with a final
transfection concentration of 20 nM miR-144-3p mimic, miR-NC,
BCL6-shRNA or NC-shRNA (n=6/group), U251 cells (2x106 in
serum-free DMEM, 200 µl) were subcutaneously injected into the
flanks of the mice. Following implantation, tumor nodules appeared
at the initial site of injection after ~7 days. The tumor volume
was measured, including length and width. The tumor volume (TV) was
calculated very week, according to the following formula: TV
(mm3)=1/2 x width2 x length. The
tumor-burdened nude mice were sacrificed 28 days following tumor
implantation and the tumor tissues were obtained. The health of the
mice was monitored daily and all mice were sacrificed by
dislocation of the cervical vertebrae when symptoms of terminal
tumor burden were observed, including tumor volume >1,000
mm3, >20% weight loss, hunched posture, cranial
protrusion and significant neurologic symptoms (seizures and
abnormal gait or posture), which developed dyspnea. No animal
reached the criteria before these endpoints.
Statistical analysis
All data are presented as mean ± SEM from four
indepdent experiments. The SPSS version 22.0 (IBM Corp.) was used
for all statistical analyses, which was conducted using two-tailed
Student's t-test or one-way ANOVA with least significant difference
post hoc test. P<0.05 was considered to indicate a statistically
significant difference.
Results
Downregulation of miR-144-3p
expression and upregulation of BCL6 expression in glioma
tissues
To assess miR-144-3p expression on glioma, its
levels were measured in 25 glioma tissues and 10 normal brain
tissues. RT-qPCR data suggested that miR-144-3p expression was
significantly reduced, whilst BCL6 expression was significantly
increased in glioma tissues compared with those in the normal brain
tissues (P<0.001; Fig. 1). These
results suggest that miR-144-3p and BCL6 are involved in glioma
development.
miR-144-3p suppresses BCL6 expression
by targeting its 3'-UTR sequence in U251 cells
Bioinformatic prediction and targetscan demonstrated
that BCL6 could be a potential target gene of miR-144-3p (Fig. 2A). To verify the association between
miR-144-3p and BCL6, the pMIR vector was packaged with WT or MUT
BCL6 3'-UTR sequences before luciferase reporter assay was
performed in U251 cells. The data indicated that luciferase
activity was significantly downregulated in glioma U251 cells
co-transfected with miR-144-3p mimic and WT BCL6 3'-UTR compared
with that in the WT control group (P<0.001). However, luciferase
activity levels were unaltered in U251 cells transfected with MUT
BCL6 3'-UTR (Fig. 2B). Therefore,
the data confirmed that BCL6 is a target gene for miR-144-3p in
glioma cells.
The role of miR-144-3p on BCL6 expression was next
investigated following transfection with the miR-144-3p mimic or
inhibitor into U251 cells. Transfection with the miR-144-3p mimic
significantly upregulated miR-144-3p levels whereas transfection
with the miR-144-3p inhibitor significantly decreased miR-144-3p
expression (P<0.001; Fig. 3A).
Western blotting analysis of BCL6 protein levels was next
performed. The protein expression levels of BCL6 were significantly
downregulated following transfection of the cells with the
miR-144-3p mimic, whilst they were enhanced following miR-144-3p
knockdown in U251 cells compared with those in the control group
(P<0.001; Fig. 3B).
miR-144-3p reduces U251 cell
proliferation and invasion in vitro
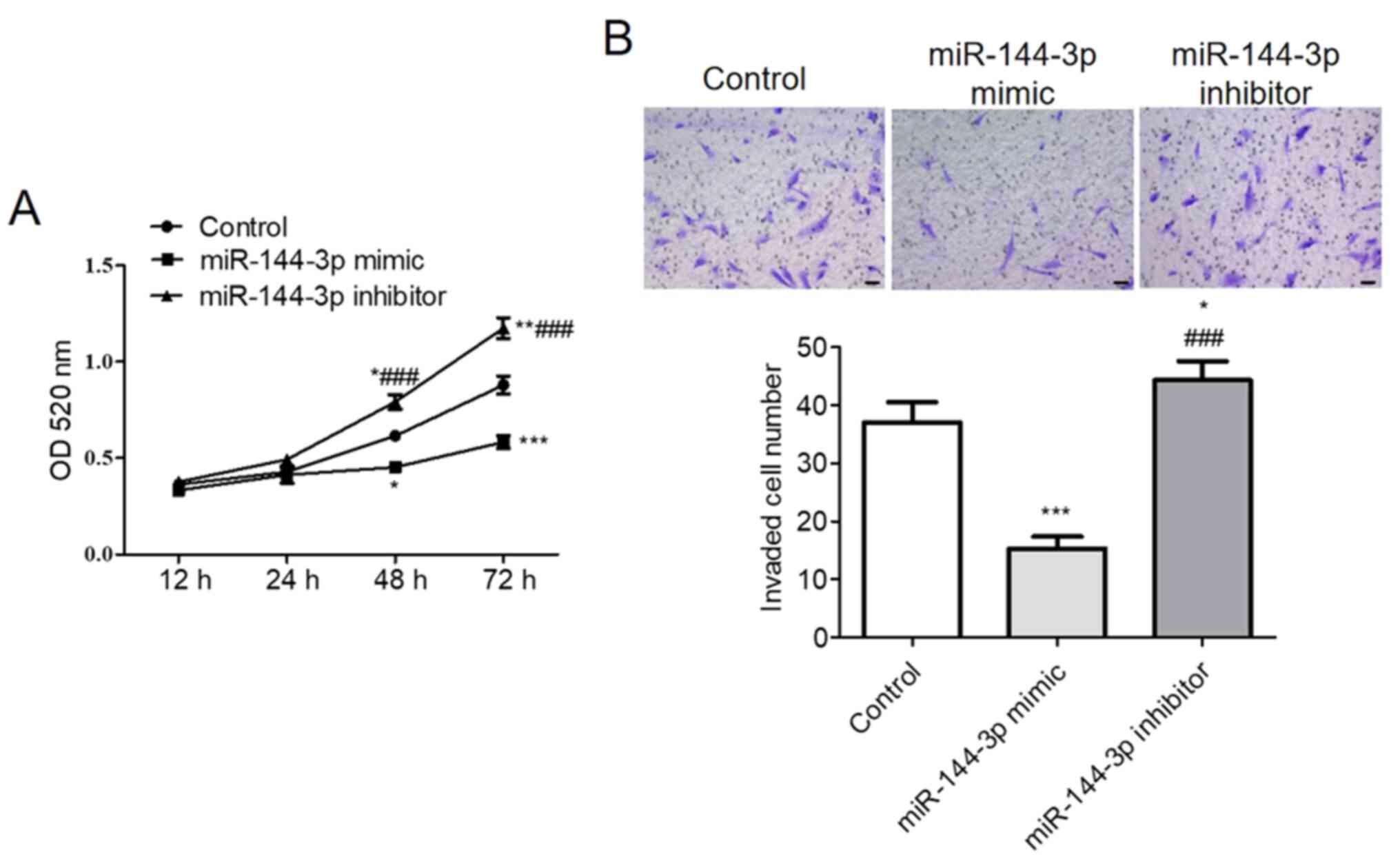
CCK-8 assay demonstrated that upregulation of
miR-144-3p significantly reduced U251 cell viability whereas
miR-144-3p knockdown significantly increased U251 cell viability
compared with that in control (P<0.001; Fig. 4A). The Transwell assay results
suggested that increasing miR-144-3p expression levels
significantly suppressed U251 cell invasion compared with that in
control, whilst miR-144-3p inhibition increased U251 cell invasion
compared with that in the control (P<0.001; Fig. 4B).
BCL6 lie downstream of
miR-144-3p-mediated glioma cell proliferation and invasion in
vitro
To examine the downstream effects of miR-144-3p on
glioma cell viability and invasion, miR-144-3p-overexpressing U251
cells were co-transfected with a BCL6-activation plasmid. western
blotting analysis was first performed to measure the effects of
co-transfection on BCL6 expression. The transfection efficiency of
pcDNA3.1-BCL6 and BCL6-shRNA was assessed by RT-qPCR, where
pcDNA3.1-BCL6 upregulated BCL6 expression and BCL6-shRNA inhibited
BCL6 expression compared wth that in the control group (Fig. S1). BCL6 expression levels were
significantly higher in the miR-144-3p + BCL6 group compared with
those in the miR-144-3p group (Fig.
5A). In addition, cell viability and invasion were
significantly increased in the miR-144-3p + BCL6 group, compared
with those in the miR-144-3p group (Fig. 5B and C). Therefore, BCL6 overexpression could
partly attenuate the inhibitory effects of miR-144-3p on the
viability and invasion of U251 cells, suggesting that BCL6 serve an
important role in miR-144-3p-mediated regulation of glioma cell
proliferation and invasion.
Overexpression of miR-144-3p or BCL6
knockdown suppress tumor growth in vivo
The in vitro results suggested that
miR-144-3p suppressed glioma cell proliferation and invasion by
targeting BCL6. Therefore, the effects of the miR-144-3p/BCL-6 axis
on glioma tumorigenesis was assessed further in vivo using
xengraft nude mouse model transplanted with a subcutaneous glioma
tumors. Following transfection with miR-144-3p mimic, miR-NC,
BCL6-shRNA or NC-shRNA, U251 cells (2x106) were
subcutaneous injected into the flanks of the mice. The tumor
volumes were measured every week. The tumor size is shown in
Fig. 6A. A significantly decreased
average volume of tumors was observed in mice injected with cells
transfected with either miR-144-3p mimic or cells transfected with
BCL6-shRNA compared with those injected with either miR-NC or
NC-shRNA, respectively (Fig. 6B).
These results suggested that the miR-144-3p/BCL6 axis is involved
in malignant glioma progression in vivo.
Discussion
BCL6 has been reported to to be a possible
prognostic biomarker and potential therapeutic target that exhibits
abnormal expression levels and serves a regulatory role in breast
cancer, epithelial ovarian cancer and glioma (10-12).
Specifically, BCL6 can serve as a potential indicator of tumor
malignancy in breast cancer and glioma (10-12).
It has been demonstrated that miRNA is an epigenetic regulator that
targets the 3'-UTR sequence of target mRNAs and serves an important
role in the development of several diseases (13). Increased miR-10b expression inhibits
osteoblast differentiation by directly targeting the 3'-UTR
sequence of BCL6 to increase the expression levels of STAT1 and
block the nuclear translocation of Runx2, all of which were
reversed by miR-10b downregulation (24). By contrast, inhibition of miR-10b
induces osteoblast differentiation in a manner that was partially
reversed by downregulating BCL6 expression (24). Furthermore, BCL6 has been shown to
regulate miRNA levels and promotes lymphomagenesis by repressing
miRNA-155(25). In the present
study, high levels of BCL6 were noted in glioma tissues. These
findings are in agreement with those reported in a previous study
by Xu et al (12),
suggesting that BCL6 may be a tumor promoter for glioma
pathogenesis and a valuable prognostic indicator in patients with
glioma patients.
A number of studies have indicated that miR-144-3p
is involved in a diverse range of biological functions, including
cell proliferation, angiogenesis of lung vessels, oxidative stress
and osteoporosis (26-28).
The contribution of miR-144-3p in tumorigenesis is complex, where
its role as a tumor physiology remains controversial. Previous
studies have shown that miR-144-3p causes a decreased expression of
target genes in various tumor tissues. miRNA-144-3p has been shown
to interact with ETS-1 in laryngeal squamous cell carcinoma
(29) but downregulates mTOR levels
in salivary adenoid carcinoma (30). In addition, miR-144-3p can target
enhancer of zeste 2 polycomb repressive complex 2 subunit in lung
adenocarcinoma (31) whilst
downregulating centrosomal protein 55 in prostate cancer (32). miR-144-3p has also been shown to
regulates serum/glucocorticoid regulated kinase family member 3
function in hepatocellular carcinoma (33) but targets FOSB and proline rich
protein 11 in pancreatic cancer (34). Furthermore, miR-144-3p can regulate
c-MET in multiple myeloma (32) and
inhibit tumorigenesis of oral squamous cell carcinoma by targeting
the endoplasmic reticulum oxidoreductase 1α/STAT3 signaling pathway
(35). miR-144-3p inhibits human
neuroblastoma cell proliferation by regulating homeobox A7
expression (36). Jiang et
al (37) previously
demonstrated that miR-144-3p suppressed TGF-β1-induced A549 cell
malignancy by surpressing Src expression (37). Wu et al (38) reported that miR-144-3p expression
was downregulated in cervical cancer tissues and cell lines, where
miR-144-3p overexpression suppressed cancer cell malignancy.
However, previous studies have also shown that miR-144-3p is
involved in tumorigenesis. A previous study by Liu et al
(39) found that miR-144-3p
expression was increased in patients with papillary thyroid
carcinoma, such that its overexpression promoted cell survival and
cell cycle progression by targeting paired box gene 8(39). In addition, miR-144-3p has been
identified to be a novel plasma diagnostic indicator for clear cell
renal cell carcinoma (ccRCC) (40).
AT-Rich Interaction Domain 1A expression is suppressed by
miR-144-3p upregulation, which induces ccRCC malignancy and
mediates evasion of Sunitinib treatment (41). It has also been reported that
miR-144-3p levels are upregulated in both peripheral blood and bone
marrow samples of patients with acute myeloid leukemia compared
with those in healthy individuals, whereas miR-144-3p targets
nuclear factor, erythroid 2 like 2 in HL-60 cells (42). Song et al (43) reported that miR-144-3p facilitated
the progression of nasopharyngeal carcinoma by mediating PTEN via
crosstalk with the PI3K-AKT signaling pathway (43).
In addition, previous studies have demonstrated that
miR-144-3p levels were significantly downregulated in glioma
tissues compared with those in normal brain tissues, which were
also decreased with tumor grade (44-46).
miR-144-3p levels exhibited a positive correlation with overall
survival of glioma patients (44).
Following human cytomegalovirus (HCMV) infection, miR-144-3p levels
are decreased whereas the expression of DNA Topoisomerase IIα
(TOP2A) increases (44). Increased
miR-144-3p levels also inhibit clone formation, cell proliferation
and invasion in HCMV-infected glioma cells by targeting TOP2A in
vitro (44). In glioblastoma
cells, the overexpression of miR-144-3p reduces the viability of
cells and increases cell death, resulting in enhancement of the
therapeutic effects of radiation and temozolomide (45). Frizzle class receptor 7 is another
reported target of miR-144-3p-mediated suppression of glioma
metastasis (46). Song et al
(44) showed that the expression of
miR-144-3p in HCMV-positive samples was markedly lower compared
with that in HCMV-negative samples, where miR-144-3p expression
correlated negatively with TOP2A expression. In addition, Lan et
al (45) previously
demonstrated that miR-144-3p expression is decreased in glioma
tissues and associated with the OS of glioma patients. In this
previous study (45), miR-144-3p
exhibited a relatively high expression levels in non-neoplastic
brain tissues whereas its expression was distinctly decreased in
glioma tissues. In addition, there was also a significant
difference in miR-44-3p expression between high grade (III-IV) and
low-grade (I-II) glioma tissues. This decreased expression of
miR-144-3p was associated with higher pathological grades, lower
Karnofsky performance scores (45)
and poorer survival rates (45).
However, the latent role of miR-144-3p in glioma remains unclear.
The present study demonstrated lower levels of miR-144-3p
expression in 25 glioma patients, whilst overexpresseion of
miR-144-3p notably suppressed cell proliferation and invasion of
U251 cells. Upregulating BCL6 levels could reverse the inhibition
caused by miR-144-3p overexpression. This occurred due to the
direct targeting of the 3'-UTR sequence of BCL6 by miR-144-3p. The
results indicated that BCL6 acted as a downstream interacting
factor of miR-144-3p in glioma. Xu et al (12) revealed that BCL6 knockdown
suppressed the tumorigenic phenotype of glioblastoma cells and
induced cell degeneration (12).
Therefore, miR-144-3p-mediated BCL6 inhibition may serve to be a
potential strategy for treating glioma.
However, it should be noted that the lack of
co-transfected cell xenograft experiments is a potential limitation
of the present study. In addition, larger clinical samples sizes
will need to be collected and conducted in future studies.
To conclude, the present study revealed that
miR-144-3p and BCL6 were clinical predictors of patients with
glioma and that the therapeutic potential of miR-144-3p/BCL6 axis
should be exploited further for glioma treatment. Therefore,
additional analysis of a higher number of samples and multiple
grade tumors is required in the future to verify the diagnostic and
prognostic potential of the miR-144-3p/BCL6 axis in glioma.
Supplementary Material
BCL6 mRNA level in U251 cells after
transfection. U251 cells were transfected with BCL6 overexpressing
plasmid (pcDNA3.1), BCL6 shRNA plasmid (pcDNA3.1) or the
corresponding controls. After 48 h transfection, transfection
efficiency was assessed by reverse transcription quantitative PCR.
Data are presented as the mean ± SEM. n=3. ***P<0.001
vs. Control. NC, negative control; sh, short hairpin.
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
The data used and/or analyzed in the current work
are available from the corresponding author on reasonable
request.
Authors' Contributions
REL and JRZ conceived and designed the study. JRZ
carried out the experiments. REL and JRZ performed data analysis,
wrote and revised the manuscript. All authors read and approved the
manuscript.
Ethics approval and consent to
participate
Research involving human participants and animal
experiments was authorized by the Ethics Committee of Peking
University People's Hospital (Beijing, China). The NIH Guide for
the Care and Use of Laboratory Animals (the 8th edition) was
followed. All procedures performed in the studies involving human
participants were in accordance with the ethical standards of the
Medical Ethics Expert Committee of National Health and Family
Planning and adhered to the 1964 Helsinki declaration and its later
amendments or comparable ethical standards. The study protocol was
approved by the Ethical Committee of Peking University People's
Hospital (Beijing, China). Written informed consent was obtained
from all individual participants included in this work.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Zhang Q, Xiang W, Yi DY, Xue BZ, Wen WW,
Abdelmaksoud A, Xiong NX, Jiang XB, Zhao HY and Fu P: Current
status and potential challenges of mesenchymal stem cell-based
therapy for malignant gliomas. Stem Cell Res Ther.
9(228)2018.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Alphandery E: Glioblastoma treatments: An
account of recent industrial developments. Front Pharmacol.
9(879)2018.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Towner RA and Smith N: In vivo and in situ
detection of macromolecular free radicals using immuno-spin
trapping and molecular magnetic resonance imaging. Antioxid Redox
Signal. 28:1404–1415. 2018.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Feng E, Liang T, Wang X, Du J, Tang K,
Wang X, Wang F and You G: Correlation of alteration of HLA-F
expression and clinical characterization in 593 brain glioma
samples. J Neuroinflammation. 16(33)2019.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Tompa M, Kalovits F, Nagy A and Kalman B:
Contribution of the Wnt pathway to defining biology of
glioblastoma. Neuromolecular Med. 20:437–451. 2018.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Yamini B: NF-κB, mesenchymal
differentiation and glioblastoma. Cells. 7(125)2018.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Brennan CW, Verhaak RG, McKenna A, Campos
B, Noushmehr H, Salama SR, Zheng S, Chakravarty D, Sanborn JZ,
Berman SH, et al: The somatic genomic landscape of glioblastoma.
Cell. 155:462–477. 2013.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Cancer Genome Atlas Research Network.
Comprehensive genomic characterization defines human glioblastoma
genes and core pathways. Nature. 455:1061–1068. 2008.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Sato M, Kanemoto H, Kagawa Y, Kobayashi T,
Goto-Koshino Y, Mochizuki H, Takahashi M, Fujino Y, Ohno K and
Tsujimoto H: Evaluation of the prognostic significance of BCL6 gene
expression in canine high-grade B-cell lymphoma. Vet J.
191:108–114. 2012.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Ang L, Zheng L, Wang J, Huang J, Hu HG,
Zou Q, Zhao Y, Liu QM, Zhao M and Wu ZS: Expression of and
correlation between BCL6 and ZEB family members in patients with
breast cancer. Exp Ther Med. 14:3985–3992. 2017.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Zhu L, Feng H, Jin S, Tan M, Gao S, Zhuang
H, Hu Z, Wang H, Song Z and Lin B: High expressions of BCL6 and
Lewis y antigen are correlated with high tumor burden and poor
prognosis in epithelial ovarian cancer. Tumour Biol.
39(1010428317711655)2017.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Xu L, Chen Y, Dutra-Clarke M, Mayakonda A,
Hazawa M, Savinoff SE, Doan N, Said JW, Yong WH, Watkins A, et al:
BCL6 promotes glioma and serves as a therapeutic target. Proc Natl
Acad Sci USA. 114:3981–3986. 2017.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Wei Z, Gao W, Wu Y, Ni B and Tian Y:
Mutual interaction between BCL6 and miRNAs contributing to the
pathogenesis of various cancers. Clin Transl Oncol. 17:841–846.
2015.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Zhu Z, Wang S, Zhu J, Yang Q, Dong H and
Huang J: MicroRNA-544 down-regulates both Bcl6 and Stat3 to inhibit
tumor growth of human triple negative breast cancer. Biol Chem.
397:1087–1095. 2016.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Sun C, Li S, Yang C, Xi Y, Wang L, Zhang F
and Li D: MicroRNA-187-3p mitigates non-small cell lung cancer
(NSCLC) development through down-regulation of BCL6. Biochem
Biophys Res Commun. 471:82–88. 2016.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Li YY, Shao JP, Zhang SP, Xing GQ and Liu
HJ: miR-519d-3p inhibits cell proliferation and invasion of gastric
cancer by downregulating B-cell lymphoma 6. Cytogenet Genome Res.
154:12–19. 2018.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Dragomir M, Mafra ACP, Dias SMG, Vasilescu
C and Calin GA: Using microRNA networks to understand cancer. Int J
Mol Sci. 19(1871)2018.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Sun Z, Shi K, Yang S, Liu J, Zhou Q, Wang
G, Song J, Li Z, Zhang Z and Yuan W: Effect of exosomal miRNA on
cancer biology and clinical applications. Mol Cancer.
17(147)2018.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Zhou Q, Liu J, Quan J, Liu W, Tan H and Li
W: MicroRNAs as potential biomarkers for the diagnosis of glioma: A
systematic review and meta-analysis. Cancer Sci. 109:2651–2659.
2018.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Wang S, Yin Y and Liu S: Roles of
microRNAs during glioma tumorigenesis and progression. Histol
Histopathol. 34:213–222. 2019.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Huang SW, Ali ND, Zhong L and Shi J:
MicroRNAs as biomarkers for human glioblastoma: Progress and
potential. Acta Pharmacol Sin. 39:1405–1413. 2018.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Koshkin FA, Chistyakov DA, Nikitin AG,
Konovalov AN, Potapov AA, Usachyov DY, Pitskhelauri DI, Kobyakov
GL, Shishkina LV and Chekhonin VP: Profile of microRNA expression
in brain tumors of different malignancy. Bull Exp Biol Med.
157:794–797. 2014.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Yang J, Wang S, Wang F, Mu X, Qu Y, Zhao Z
and Yu X: Downregulation of miR-10b promotes osteoblast
differentiation through targeting Bcl6. Int J Mol Med.
39:1605–1612. 2017.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Basso K, Schneider C, Shen Q, Holmes AB,
Setty M, Leslie C and Dalla-Favera R: BCL6 positively regulates AID
and germinal center gene expression via repression of miR-155. J
Exp Med. 209:2455–2465. 2012.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Yang C, Lv K, Chen B, Yang Y, Ai X, Yu H,
Yang Y, Yi B and Lu K: miR144-3p inhibits PMVECs excessive
proliferation in angiogenesis of hepatopulmonary syndrome via Tie2.
Exp Cell Res. 365:24–32. 2018.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Wang C, He H, Wang L, Jiang Y and Xu Y:
Reduced miR-144-3p expression in serum and bone mediates
osteoporosis pathogenesis by targeting RANK. Biochem Cell Biol.
96:627–635. 2018.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Li Y, Zhao Y, Cheng M, Qiao Y, Wang Y,
Xiong W and Yue W: Suppression of microRNA-144-3p attenuates
oxygen-glucose deprivation/reoxygenation-induced neuronal injury by
promoting Brg1/Nrf2/ARE signaling. J Biochem Mol Toxicol.
32(e22044)2018.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Zhang SY, Lu ZM, Lin YF, Chen LS, Luo XN,
Song XH, Chen SH and Wu YL: miR-144-3p, a tumor suppressive
microRNA targeting ETS-1 in laryngeal squamous cell carcinoma.
Oncotarget. 7:11637–11650. 2016.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Huo F, Zhang C, He H and Wang Y:
MicroRNA-144-3p inhibits proliferation and induces apoptosis of
human salivary adenoid carcinoma cells via targeting of mTOR.
Biotechnol Lett. 38:409–416. 2016.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Liu C, Yang Z, Deng Z, Zhou Y, Gong Q,
Zhao R and Chen T: Downregulated miR-144-3p contributes to
progression of lung adenocarcinoma through elevating the expression
of EZH2. Cancer Med. 7:5554–5566. 2018.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Zheng H, Guo Z, Zheng X, Cheng W and Huang
X: MicroRNA-144-3p inhibits cell proliferation and induces cell
apoptosis in prostate cancer by targeting CEP55. Am J Transl Res.
10:2457–2468. 2018.PubMed/NCBI
|
|
33
|
Wu M, Huang C, Huang X, Liang R, Feng Y
and Luo X: MicroRNA-144-3p suppresses tumor growth and angiogenesis
by targeting SGK3 in hepatocellular carcinoma. Oncol Rep.
38:2173–2181. 2017.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Liu S, Luan J and Ding Y: miR-144-3p
Targets FosB Proto-oncogene, AP-1 transcription factor subunit
(FOSB) to suppress proliferation, migration, and invasion of PANC-1
pancreatic cancer cells. Oncol Res. 26:683–690. 2018.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Li X, Li Y, Jiang C, Chen L and Gan N:
MicroRNA-144-3p inhibits tumorigenesis of oral squamous cell
carcinoma by downregulating ERO1L. J Cancer. 11:759–768.
2020.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Cao XY, Sun ZY, Zhang LJ, Chen MK and Yuan
B: MicroRNA-144-3p suppresses human neuroblastoma cell
proliferation by targeting HOXA7. Eur Rev Med Pharmacol Sci.
23:716–723. 2019.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Jiang W, Xu Z, Yu L, Che J, Zhang J and
Yang J: MicroRNA-144-3p suppressed TGF-β1-induced lung cancer cell
invasion and adhesion by regulating the Src-Akt-Erk pathway. Cell
Biol Int 2019 (Epub ahead of print).
|
|
38
|
Wu J, Zhao Y, Li F and Qiao B: miR-144-3p:
A novel tumor suppressor targeting MAPK6 in cervical cancer. J
Physiol Biochem. 75:143–152. 2019.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Liu C, Su C, Chen Y and Li G: miR-144-3p
promotes the tumor growth and metastasis of papillary thyroid
carcinoma by targeting paired box gene 8. Cancer Cell Int.
18(54)2018.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Lou N, Ruan AM, Qiu B, Bao L, Xu YC, Zhao
Y, Sun RL, Zhang ST, Xu GH, Ruan HL, et al: miR-144-3p as a novel
plasma diagnostic biomarker for clear cell renal cell carcinoma.
Urol Oncol. 35:36.e7–36.e14. 2017.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Xiao W, Lou N, Ruan H, Bao L, Xiong Z,
Yuan C, Tong J, Xu G, Zhou Y, Qu Y, et al: Mir-144-3p promotes cell
proliferation, metastasis, sunitinib resistance in clear cell renal
cell carcinoma by downregulating ARID1A. Cell Physiol Biochem.
43:2420–2433. 2017.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Sun X, Liu D, Xue Y and Hu X: Enforced
miR-144-3p expression as a non-invasive biomarker for the acute
myeloid leukemia patients mainly by targeting NRF2. Clin Lab.
63:679–687. 2017.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Song L, Chen L, Luan Q and Kong Q:
miR-144-3p facilitates nasopharyngeal carcinoma via crosstalk with
PTEN. J Cell Physiol. 234:17912–17924. 2019.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Song J, Ma Q, Hu M, Qian D, Wang B and He
N: The Inhibition of miR-144-3p on cell proliferation and
metastasis by targeting TOP2A in HCMV-positive glioblastoma cells.
Molecules. 23(3259)2018.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Lan F, Yu H, Hu M, Xia T and Yue X:
miR-144-3p exerts anti-tumor effects in glioblastoma by targeting
c-Met. J Neurochem. 135:274–286. 2015.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Cheng ZX, Song YX, Wang ZY, Wang Y and
Dong Y: miR-144-3p serves as a tumor suppressor by targeting FZD7
and predicts the prognosis of human glioblastoma. Eur Rev Med
Pharmacol Sci. 21:4079–4086. 2017.PubMed/NCBI
|