Introduction
Immunotherapy has become one of the most rapidly
developed treatment methods in the field of cancer, which
essentially depends on activating the patient's own immune system
against tumors (1). A tumor
vaccine is used to introduce various forms of tumor antigen, which
can induce cellular and humoral immunity, as well as generating
long-term immune memory, thereby controlling or preventing tumor
progression (2).
Melanoma is a highly malignant tumor type, prone to
invasion and metastasis, with a poor prognosis (3). Melanoma also has strong
immunogenicity (4). Immunotherapy
has significant advantages in improving the curative effect of
melanoma, especially after the immune system is stimulated, and
often result in a long-term curative effect and lasting immune
memory (5). Therefore, tumor
vaccine-induced immunotherapy has a favorable application prospect
for the prevention and treatment of melanoma.
In eukaryotes, the ubiquitin-proteasome system (UPS)
is critical to maintaining the intracellular protein balance and
mediating specific irreversible protein degradation of short-lived
intracellular proteins, as well as some non-functional proteins
(6,7). When certain proteins closely
associated with apoptosis and cell cycle control are ubiquitin
labelled, they can be identified and degraded by the 26S proteasome
(8-10).
The effects of the UPS on cancer treatment have become a research
hotspot. In 2004, scientists Aaron Ciechanover, Avram Hershko and
Irwin Rose were awarded the Nobel Prize in Chemistry for the
discovery of ubiquitin-mediated protein degradation (11). In 2009, articles on the
antineoplastic effects of the UPS were published in Nature, where
the prospects of such a system in cancer treatment were described
from multiple points of view (12-17).
Bortezomib, as the first drug targeting the UPS, passed clinical
trials in 2003. After being approved by the Food and Drug
Administration, it was selected as a therapeutic medicine for
multiple malignant hematologic tumors, including relapsed or
refractory multiple myeloma (18,19).
However, simply inhibiting the proteasome may cut off the entire
UPS pathway. Consequently, not only is the transformation of a
great number intracellular proteins inhibited, but significant
adverse reactions are also observed (20). To reduce the influence on the other
biological functions of the UPS, numerous regulatory enzymes that
act on this pathway have become hot topics of research (20-22).
At present, MLN4924, as a compound that targets an NEDD4 E3
ubiquitin protein ligase (NEDD4) activating enzyme, is at phase I
of a clinical trial, and has shown promising antineoplastic
activity (23,24). Furthermore, melanoma treatment has
been revolutionized by immuno- and targeted therapy (25-27).
NEDD4 is a type of ubiquitin-like protein, and its
covalent bonding to a substrate is known as neddylation (28). An important physiological function
of neddylation is to regulate E3 ubiquitin-ligating enzymes and
promote the progression of the ubiquitination pathway (29,30).
Furthermore, the family of cullin proteins includes the substrates
on which NEDD4 primarily acts. This family is one of the major core
structures of E3 ubiquitin-ligating enzymes (31). As catalyzed by a series of enzymes,
NEDD4 is covalently bound to cullin proteins, thereby forming a
NEDD4 chain structure (31).
Consequently, the confirmational changes to E3 ubiquitin-ligating
enzymes, and E2 (which carries ubiquitin), promote entry to the
E3-substrate complex, such that the substrate can be ubiquitinated
(32). Numerous studies have
reported that neddylation not only enables a significant increase
in the ubiquitination efficiency of proteins to be degraded, but
also promotes the degradation of these proteins by the 26S
proteasome (33,34).
Previous studies have reported that NEDD4 serves an
important role in cancer development (29,30).
NEDD4 is highly expressed in patients with breast (35) and lung cancer (36), where it was closely associated with
clinical pathology (35,36). Nevertheless, NEDD4 expression in
melanoma, and its underlying mechanisms in melanoma occurrence and
development, remain unknown. In the present study, NEDD4 expression
in melanoma tissues and benign moles was detected via
immunohistochemistry (IHC). Moreover, cellular experiments were
conducted following NEDD4-knockout to determine its influence on
the biological activity of melanoma cells, and to identify the
associated underlying mechanisms.
Materials and methods
Patient samples, cell lines and cell
culture
Specimens of 30 cases with cutaneous melanoma (CMM;
stages I-II, 20 cases; and stages III-IV, 10 cases) and 10 cases
with benign moles, surgically removed between March 2017 and March
2019, were selected from The Affiliated Xingtai People's Hospital
of Hebei Medical University. The patients were not treated with
radiotherapy, chemotherapy or immunotherapy prior to surgery. All
patients were pathologically diagnosed with CMM after surgery, and
the collected specimens were divided into two parts. Some specimens
were frozen by diethyl pyrocarbonate (DEPC) water treatment,
immersed in RNA Latter and stored at -80˚C for further use; the
other specimens underwent paraffin embedding and slicing to perform
hematoxylin and eosin (H&E) staining and IHC. The present study
was approved by The Affiliated Xingtai People's Hospital Ethics
Committee (approval no. 2017012205). All patients signed informed
consent.
All cell lines used in the study were purchased from
the American Type Culture Collection. Normal skin cells (HaCaT) and
melanoma cell lines (SK-MEL-2, Malme-3M, MV3, A375 and MUM-2B
cells) were all cultured in DMEM (Invitrogen; Thermo Fisher
Scientific, Inc.) supplemented with 1% double-antibiotic (100 U/ml
penicillin and 100 mg/l streptomycin) and 10% FBS (Gibco; Thermo
Fisher Scientific, Inc.). The cells were maintained in an incubator
at 37˚C (5% CO2). After 3 days, the cell confluence had
reached 80-90%, and the cells were passaged.
Reagents and instruments
A reverse transcription (RT) kit and PCR kit were
purchased from Takara Biotechnology Co., Ltd., while DEPC was from
Sangon Biotech Co., Ltd. TRIzol® reagent was purchased
from Invitrogen (Thermo Fisher Scientific, Inc.). Richard-Allan
Scientific™ Wright-Giemsa compound stain was acquired from Thermo
Fisher Scientific, Inc., and the Annexin V-FITC/PI apoptosis assay
kit was from Nanjing KeyGen Biotech Co., Ltd. The Transwell assay
was obtained from Corning, Inc., and crystal violet from
Sigma-Aldrich; Merck KGaA. NEDD4 (cat. no. ab46521), PTEN (cat. no.
ab170941), Notch1 (cat. no. ab52627) and GAPDH (cat. no. ab181602)
antibodies were acquired from Abcam. FBS were all purchased from
Gibco; Thermo Fisher Scientific, Inc. Small interfering RNA
(siRNA/si)-NEDD4 was purchased from Nanjing KeyGen Biotech Co.,
Ltd. Lipofectamine® 3000 (Invitrogen; Thermo Fisher
Scientific, Inc.) was used for transfection, and all PCR primers
were purchased from Sangon Biotech Co., Ltd.
H&E staining
Deparaffination and rehydration were performed using
the following conventional methods. The sections (5-µm) were
immersed in xylene for 5 min at room temperature, which was then
replaced for immersion for another 5 min. The sections were then
rehydrated with a descending alcohol series (absolute, 95%, 85% and
70% ethyl alcohol for 5 min each), following by washing in PBS
thrice for 3 min each. Next, the sections were immersed in nuclear
staining solution (Reagent I from the H&E kit; cat. no. KGA224;
Nanjing KeyGen Biotech Co., Ltd.) for 3-5 min at room temperature,
and then in color separation fluid I of Reagent II for ~20 sec,
followed by washing for 30-60 sec. Subsequently, the sections were
immersed in color separation fluid II of Reagent III for ~40 sec,
and then washed with water for 30-60 sec at room temperature, prior
to staining for 2 min in Reagent IV. Reagent V was used to wash the
sections twice, and excess solution was removed. After the sections
were dried via absorption using filter paper, they were mounted,
underwent light-microscopic examination and were imaged at x400
magnification.
IHC staining
Tissue sections (thickness, 3-µm) underwent
deparaffination (60˚C; 2 h), dehydration using a gradient ethanol
series (absolute ethanol, 95% ethanol, 85% ethanol and 70% ethanol
all for 5 min) and antigen retrieval at room temperature, which
involved thermally induced epitope repair in citrate buffer (pH
6.0) for 30 min. The sections were sealed with
H2O2 at room temperature for 2 h, and then
incubated with primary (cat. no. ab46521; Abcam) and secondary
(cat. no. KGAA35; Nanjing KeyGen Biotech Co., Ltd.) antibodies at
4˚C for 12 h, and at 37˚C for 2 h, respectively. Next, DAB staining
was conducted for 5 min at room temperature and nuclear staining
(hematoxylin) at room temperature for 5 min, followed by mounting
for subsequent light-microscopic examination (magnification, x400);
five visual fields were randomly select to determine the
corresponding relative optical density. The data were analyzed
using Quantity One software v.4.66 (Bio-Rad Laboratories,
Inc.).
Reverse transcription-quantitative
(RT-q) PCR
Total RNA was extracted from clinical specimens and
cells using TRIzol® reagent (Invitrogen; Thermo Fisher
Scientific, Inc.) which were centrifuged for 5 min at 12,000 x g at
4˚C. The corresponding supernatant was harvested. Next, 1 ml 75%
ethyl alcohol was added to 1 ml TRIzol® (Invitrogen;
Thermo Fisher Scientific, Inc.), and a NanoDrop 2000
spectrophotometer (NanoDrop Technologies; Thermo Fisher Scientific,
Inc.) was used to calculate the RNA concentration after 5-10 min of
drying at room temperature (or vacuum drying). The real-time PCR
Master Mix (SYBR-Green; Takara Bio, Inc.) and the ABI StepOne plus
real-time PCR system (Applied Biosystems; Thermo Fisher Scientific,
Inc) were used for qPCR. The following thermocycling conditions
were used: Initial denaturation at 95˚C for 10 min; followed by 40
cycles of denaturation at 95˚C for 15 sec and annealing at 60˚C for
1 min; and a final extension of 10 min at 72˚C and amplification
was monitored in a real-time manner. GAPDH was used as the
reference gene. For each sample, the test was repeated three times,
and sample gene expression was quantified using the
2-∆∆Cq (37) method.
The primer sequences are presented in Table I.
 | Table IqPCR primer sequences. |
Table I
qPCR primer sequences.
| Gene name | Primer sequence
(5'-3') |
|---|
| NEDD4 E3 | F:
TTGCACTTTGCAGCCAGAAG |
| ubiquitin protein
ligase | R:
CTGTCTGGGAGTCAGCTTCA |
| Notch1 | F:
GCAGAGGCGTGGCAGACTAT |
| | R:
GGTTGGTGAGGCAGGCATTGT |
| PTEN | F:
GGTCTGAGTCGCCTGTCACCAT |
| | R:
CCGTGTTGGAGGCAGTAGAAGG |
| GAPDH | F:
CAAATTCCATGGCACCGTCA |
| | R:
AGCATCGCCCCACTTGATTT |
Transfection
A day before transfection, cells were seeded into a
culture plate. Culture medium without antibiotics was added into
each well, such that the confluence had reached 70-80% at the time
of transfection. Then, 2.5 µg siRNA was diluted using 250 µl
Opti-MEM (Thermo Fisher Scientific, Inc) without serum, followed by
gentle mixing and incubation at room temperature for 5 min. Then, 5
µl Lipofectamine® 3000 was diluted in 250 µl Opti-MEM
(without serum), mixed and incubated at room temperature for 5 min.
Next, the siRNA-Lipofectamine mixture was added to each well
(containing 500 µl culture media and cells), and cultured for 4-6 h
in DMEM at 37˚C. The culture medium was replaced with a fresh
medium (10% FBS), and the cells were placed in a 5% CO2
incubator (37˚C) for 48 h. The siRNA sequences were as follows:
si-NEDD4-1 forward, 5'-GGGAAGAGA GGCAGGAUAUTT-3' and reverse,
5'-AUAUCCUGCCUC UCUUCCCTT-3'; si-NEDD4-2 forward, 5'-CCUAACAGAU
GCUGAGAAUTT-3' and reverse, 5'-AUUCUCAGCAUC UGUUAGGTT-3';
si-NEDD4-3 forward, 5'-GUGAAAAGG GAUUGGAUUATT-3' and reverse,
5'-UAAUCCAAUCCC UUUUCACTT-3'; and si-negative control (NC) forward,
5'-UGACCUACAACUUCUAUGGTT-3' and reverse, 5'-UUC
UCCGAACGUGUCACGUTT-3'. Subsequent experimentation was performed 48
h after transfection.
Plate and soft agar colony formation
experiments
Cells in the logarithmic phase were harvested and
counted, and the concentration was adjusted to 1x103/ml.
In total, 3x103 cells were collected and placed in
culture medium. Distilled water was used to prepare 1.3 and 0.8%
agarose solutions with low melting points. After the agarose
solution was mixed with RPMI-1640 (MilliporeSigma) at a 1:1 ratio,
the mixture was evenly distributed on a plate where it was
naturally cooled and solidified into a double agar layer. Next, 1
ml cell suspension was added to the solidified upper agar layer,
and the plate was placed in an incubator (37˚C) for ~2 weeks. Both
colony morphology observation and counting were performed. For both
the experimental and control groups, the test was repeated
thrice.
Flow cytometry
Once the supernatant of the transfected cells was
removed, double staining was performed using the Annexin V-FITC/PI
apoptosis assay kit (BD Biosciences) per the manufacturer's
protocol. Apoptosis was analyzed via flow cytometry. Moreover, the
cells were divided into live cells, non-viable non-apoptotic cells,
viable apoptotic cells and non-viable apoptotic cells. The number
of apoptotic cells was determined using the Muse Cell Analyzer (EMD
Millipore). The Muse software v.1.1.2 (EMD Millipore) evaluated the
numbers of apoptotic rate.
Transwell assay
The transfected cells (1x104 cells/well)
were seeded into the upper chamber of a Transwell insert pre-coated
with Matrigel® (Corning, Inc.) in serum-free medium.
DMEM containing 15% FBS was added into the 96-well plate, and 100
µl (1x106/ml) cell suspension was added to the upper
chambers. After continuous cultivation for 24 h in an incubator,
the inserts were removed and washed with PBS, and the redundant
cells were removed using cotton swabs. Then, the membranes were
fixed for 15 min with 4% paraformaldehyde at room temperature,
dried and stained for 15 min with 0.5% crystal violet at 37˚C.
After washing three times with PBS, the plate was assessed under a
light microscope, images were captured, and the number of invasive
cells was calculated.
Wound-healing assay
The cells were inoculated into a 6-well plate
(2x105/ml; 2 ml/well). When reaching 80-90% confluency,
the supernatant was removed, which was followed by washing once
with PBS. Then, the monolayers were perpendicularly scratched along
the median line using a 10-µl pipette tip. After washing twice with
PBS, serum-free culture medium was added to the wells, and images
were captured at 0, 24 and 48 h under a light microscope (x100).
The corresponding wound-closure distance was calculated as wound
healing rate, using the following calculation: (0 h width - 24 h or
48 h width)/0 h width x 100%.
Western blotting
After the proteins were extracted from different
groups of cells, which were treated with different molecules using
RIPA lysis buffer (Beyotime Institute of Biotechnology), their
concentration was determined using a BCA assay. 10% SDS-PAGE was
performed to separate the proteins (50 µg per lane), which were
then transferred onto a PVDF membrane. After washing with
TBS-Tween-20 (TBST; 0.1%), skimmed milk was used to block the
membrane for 2 h at room temperature. Then, primary antibodies
against NEDD4, PTEN, Notch1 and GAPDH (Abcam) were added to the
membrane, which was incubated at 4˚C overnight. Then, the membrane
was rewarmed for ~30 min at room temperature, washed with TBST, and
incubated with the horseradish peroxidase conjugated goat anti
rabbit secondary antibody (1:5,000; cat. no. abs20002; Absin
Bioscience, Inc.) at 37˚C for 2 h on a shaker at room temperature
with secondary antibody. DAB was used for color development, which
was exposed using an imager. The bands were detected using an ECL
reagent (EMD Millipore). The grayscale values of the membranes were
semi quantified using ImageJ software v.1.52r (National Institutes
of Health) and the relative expression differences were determined
using GAPDH as the reference. The antibody dilutions were as
follows: NEDD4, 1:100; Notch1, 1:100; PTEN, 1:200; and GAPDH,
1:500.
Statistical analysis
Statistical analysis was performed using SPSS 22.0
software (IBM Corp.), and the data are expressed as the mean ± SD.
Statistical analysis was conducted using one-way ANOVA and RMANOVA
followed by Tukey's post hoc test. All experiments were repeated 3
times, and P<0.05 was considered to indicate a statistically
significant difference.
Results
Pathological changes and NEDD4 mRNA
and protein expression in CMM clinical samples
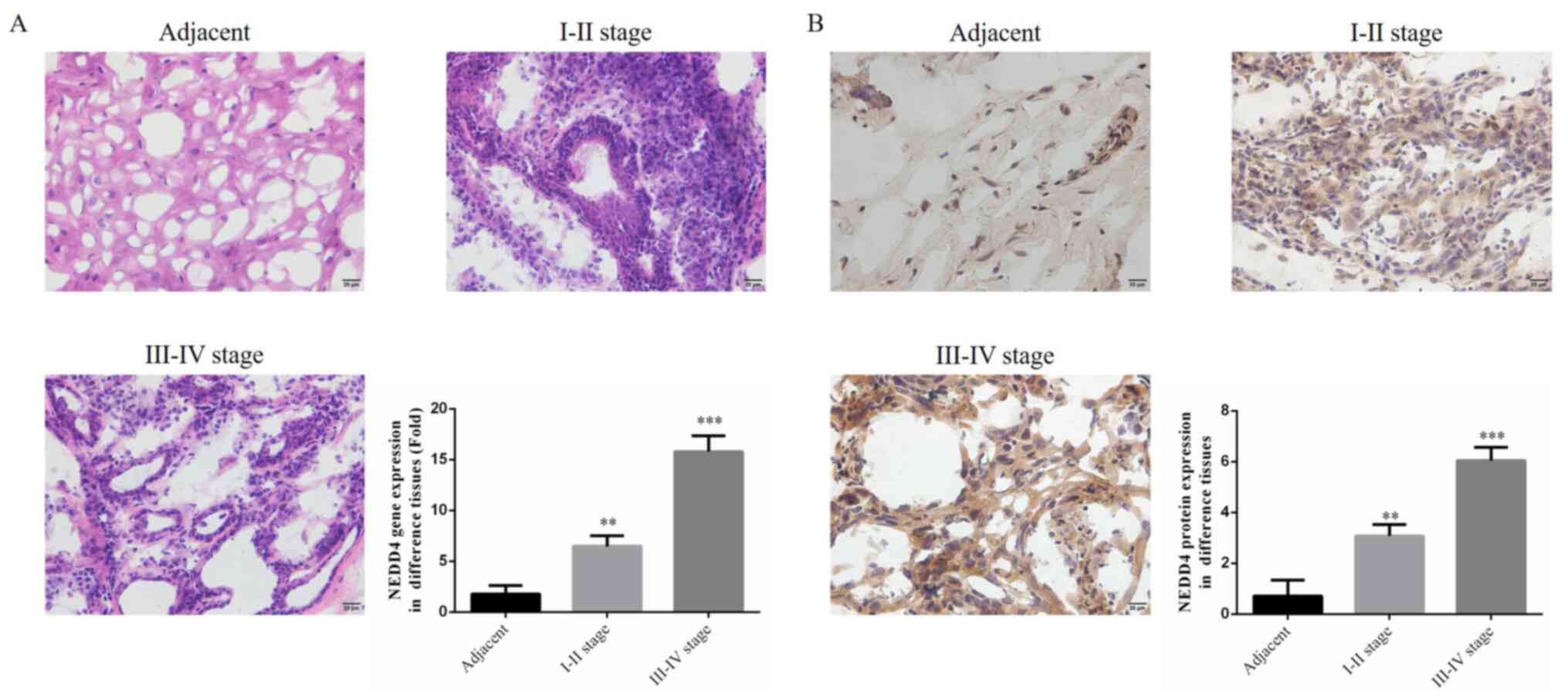
Using H&E staining, it was identified that
immune cell infiltration were increased in cancer tissues as the
stage increased, and there was no infiltration of the benign nevus.
With clear interstitial boundaries, NEDD4 mRNA expression was
significantly upregulated compared with that of the adjacent normal
tissues, as measured by RT-qPCR (P<0.01; Fig. 1A). Using IHC, it was found that,
compared with adjacent tissues, NEDD4 protein expression was
significantly enhanced in tumor tissues (P<0.01; Fig. 1B), and NEDD4 protein expression at
stages III-IV tissues was higher than that at stages I-II. The
aforementioned results indicated that NEDD4 was associated with
melanoma. NEDD4 might be associated with CMM.
NEDD4 gene expression in different
cell lines and siRNA treatment groups
Compared with normal epidermal HaCaT cells, NEDD4
gene expression was significantly upregulated in melanoma cell
lines (SK-MEL-2, Malme-3M, MV3, A375 and MUM-2B) (P<0.01), and
was highest in SK-MEL-2 and Malme-3M cells (Fig. 2A). accordingly, SK-MEL-2 and
Malme-3M cells were selected for subsequent experimentation. In
total, three types of siRNA that had knockdown effects on NEDD4
expression were used. NEDD4 mRNA expression was significantly
suppressed in the siRNA-transfected SK-MEL-2 and Malme-3M cells
compared with the si-NC groups (P<0.01; Fig. 2B). Moreover, NEDD4 mRNA expression
in the siRNA-2 group was the lowest in both the SK-MEL-2 and
Malme-3M cell lines. After siRNA-2 transfection (compared with the
NC group), NEDD4 mRNA expression was significantly decreased in
SK-MEL-2 and Malme-3M cell lines (P<0.001, Fig. S1). From the results, the SK-MEL-2
and Malme-3M cell lines were selected for the further experiments,
and si-RNA-2 could decrease the expression level of NEDD4 in these
cell lines.
NEDD4-knockdown suppresses cellular
proliferation
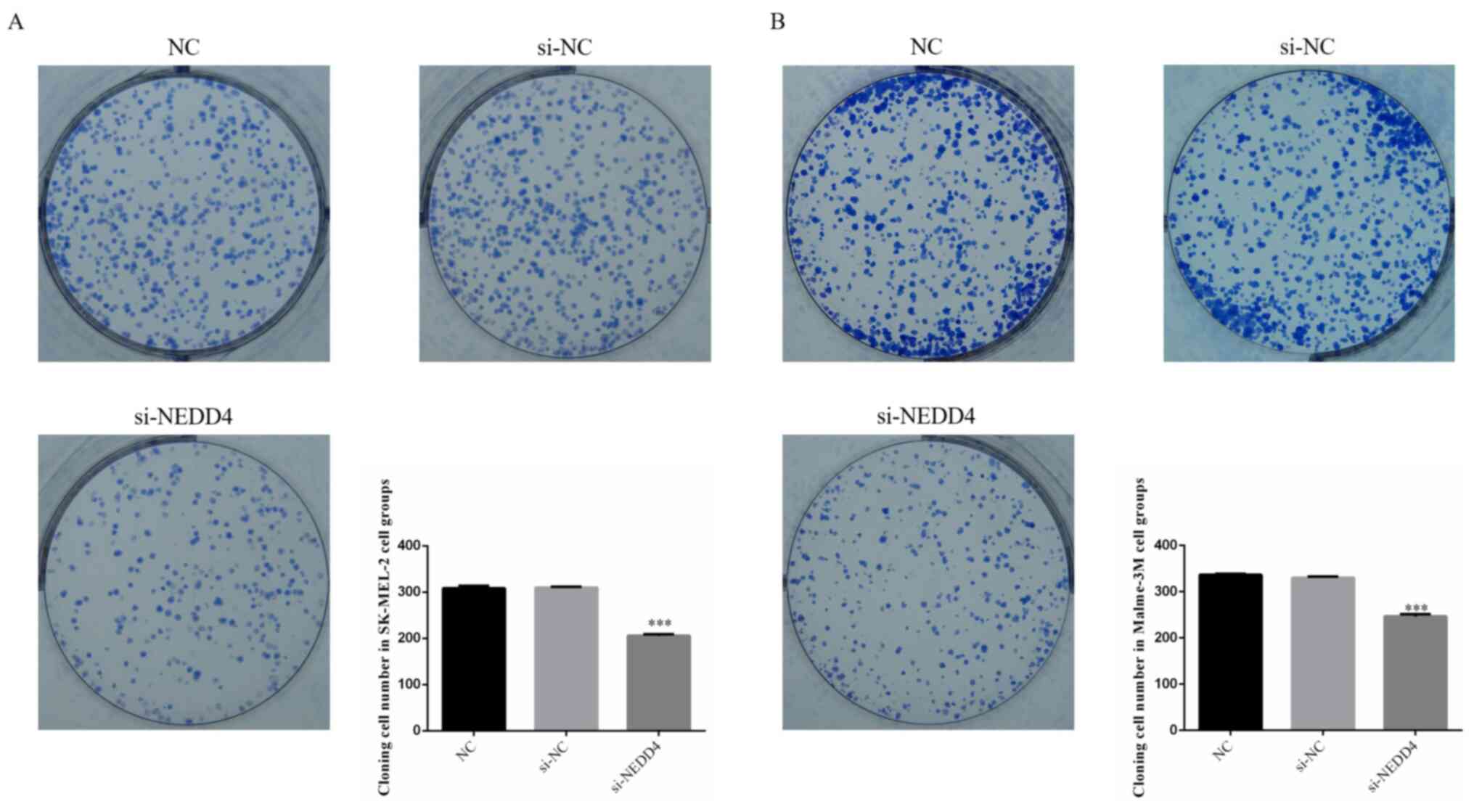
Following si-NEDD4 transfection, the colony cell
number was significantly decreased compared with that of the NC
group (P<0.001; Fig. 3A and
B) in SK-MEL-2 and Malme-3M cell
lines. NEDD4 knockdown could suppress cell proliferation in the
SK-MEL-2 and Malme-3M cell lines.
NEDD4-knockdown increases
apoptosis
Using flow cytometry, it was found that the cell
apoptotic rate of the si-NEDD4 groups was significantly increased
(P<0.001; Fig. 4A and B) compared with that of the NC groups in
SK-MEL-2 and Malme-3M cells. NEDD4 knockdown could increase cell
apoptosis in the SK-MEL-2 and Malme-3M cell lines.
NEDD4-knockdown inhibits cellular
invasiveness
Using a Transwell assay, it was demonstrated that
the invasive cell number in the si-NEDD4 groups was significantly
decreased (P<0.001; Fig. 5A and
B) compared with that in the NC
groups in both the SK-MEL-2 and Malme-3M cell lines. Following
NEDD4 knockdown, the invasion abilities of the SK-MEL-2 and
Malme-3M cell lines were significantly decreased
NEDD4-knockdown decreases
wound-healing capacity
The wound-healing assay results revealed that the
closure rate of the si-NEDD4 groups, in which NEDD4 was knocked
down by si-NEDD4 transfection, was significantly suppressed at 24
and 48 h (P<0.01; Fig. 6A and
B) compared with that in the NC
group in both the SK-MEL-2 and Malme-3M cell lines. Following NEDD4
knockdown, the cell migration abilities of the SK-MEL-2 and
Malme-3M cell lines were significantly decreased.
NEDD4-knockdown affects the relative
gene expression of Notch 1 and PTEN
From RT-qPCR analysis, the results demonstrated that
NEDD4 and Notch1 gene expression was significantly decreased
(P<0.001; Fig. 7A and B), while PTEN gene expression was
significantly increased in the si-NEDD4 groups (P<0.001;
Fig. 7A and B) compared with those of the NC SK-MEL-2
and Malme-3M cell groups. NEDD4 knockdown might be regulated to
Notch1 and PTEN gene expression in the SK-MEL-2 and Malme-3M cell
lines.
NEDD4-knockdown affects the relative
protein expression of Notch 1 and PTEN
The western blotting results indicated that NEDD4
and Notch-1 protein expression was significantly downregulated
(P<0.001; Fig. 8), and PTEN
protein expression was significantly increased in the si-NEDD4
groups (P<0.001; Fig. 8),
compared with those of the NC group, in both SK-MEL-2 and Malme-3M
cells. NEDD4 knockdown might be regulated to Notch1 and PTEN
protein expression in the SK-MEL-2 and Malme-3M cell lines.
Discussion
Over the past few years, studies have reported that
NEDD4 serves an essential role in tumor development (33-36).
For example, relevant research findings have shown that abnormal
expression of NEDD4 is closely associated with the bioactivity and
prognosis of tumors (38-41).
However, to date, neither the expression nor relevant underlying
mechanisms of NEDD4 in melanoma have been extensively investigated.
In the present study, IHC and RT-qPCR analyses were initially used
to quantify NEDD4 expression in normal pigment nevus and CMM
tissues. After NEDD4-knockdown, not only was the bioactivity of
melanoma observed, but the corresponding mechanism was investigated
based on molecular biological experiments.
The present results demonstrated that the gene and
protein expression levels of NEDD4 in CMM tissues were
significantly increased compared with the normal pigment nevus
tissues. Moreover, as the stage increased, NEDD4 expression
consistently increased. In the cellular experiments, the
bioactivity (e.g., proliferation, invasion and migration ability)
of CMM cells (SK-MEL-2 and Malme-3M; with highly expressed NEDD4)
was decreased following NEDD4-knockdown. As shown by western
blotting and RT-qPCR, the mRNA and protein expression levels of
NEDD4 and Notch1 were significantly downregulated, while those of
PTEN were significantly increased.
At present, the Notch signal transduction pathway is
considered a research hotspot in the life sciences field. As one of
the major signaling pathways that mediates cell-to-cell contact, it
is capable of regulating cellular proliferation, differentiation
and apoptosis (42,43). Moreover, NEDD4-silencing was
reported to effectively suppress Notch1 expression (44). Additionally, Notch1 has a reverse
regulatory effect on downstream PTEN genes (42,45),
and PTEN deleted on chromosome 10 was the first tumor suppressor
gene found to possess a dual phosphatase activity. PTEN mutation or
expression loss is closely associated with the occurrence and
development of multiple malignant tumors (46). Furthermore, PTEN expression loss is
an important promoter of tumor occurrence. In the present study, it
was found that PTEN expression was increased, along with a
reduction in Notch1 expression, following NEDD4-knockdown. However,
there were some study limitations. Using siRNA to knockdown NEDD4
expression in SK-MEL-2 and Malme-3M cells, proliferation
significantly decreased, while apoptosis significantly increased.
However, invasion and migration ability were significantly
downregulated. Thus, the effects of apoptosis on invasion and
migration cannot be ruled out. Furthermore, the study only
discussed the effects of the NEDD4/Notch1/PTEN pathway in melanoma
development, and other pathways may also be implicated, which will
be investigated in future research.
In conclusion, the present study demonstrated that
interference with NEDD4 expression has the potential to effectively
inhibit the proliferative abilities of CMM cells, as well as
inhibit invasive and migratory capacity. Moreover, the underlying
mechanism of NEDD4 may be closely associated with the regulation of
the downstream Notch1/PTEN signaling pathway.
Supplementary Material
NEDD4 mRNA expression in different
cells lines, as determined by reverse transcription-quantitative
PCR. ***P<0.001 vs. the NC group. NC, cells treated
with culture medium; si-NC, cells transfected with si-NC; si-NEDD4,
cells transfected with si-NEDD4-NC, negative control; siRNA/si,
small interfering RNA; NEDD4, NEDD4 E3 ubiquitin protein
ligase.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
All authors contributed to the study conception and
design. Material preparation, data collection and analysis were
performed by FC, YC, XZ, LA, LY, ZL and LZ. The first draft of the
manuscript was written by FC and all authors commented on previous
versions of the manuscript. FC and RH confirm the authenticity of
all the raw data. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
The study was approved by The Affiliated Xingtai
People's Hospital Ethics Committee (Xingtai, China) and informed
consent was signed by all participants (approval no.
2017012205).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Riley RS, June CH, Langer R and Mitchell
MJ: Delivery technologies for cancer immunotherapy. Nat Rev Drug
Discov. 18:175–196. 2019.PubMed/NCBI View Article : Google Scholar
|
|
2
|
National Cancer Institute (NCI): SEER Stat
Fact Sheet: Melanoma of the skin. http://seer.cancer.gov/statfacts/html/melan.html.
Accessed June 20, 2015.
|
|
3
|
Joshua AM, Evans A, Van der Kwast T,
Zielenska M, Meeker AK, Chinnaiyan A and Squire JA: Prostatic
preneoplasia and beyond. Biochim Biophys Acta. 1785:156–181.
2008.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Hershko A: The ubiquitin system for
protein degradation and some of its roles in the control of the
cell-division cycle (Nobel lecture). Angew Chem Int Ed.
44:5932–5943. 2005.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Lee AY and Brady MS: Neoadjuvant
immunotherapy for melanoma. J Surg Oncol. 123:782–788.
2021.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Hjerpe R and Rodríguez MS: Alternative UPS
drug targets upstream the 26S proteasome. Int J Biochem Cell Biol.
40:1126–1140. 2008.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Tsvetkov P, Reuven N and Shaul Y:
Ubiquitin-independent p53 proteasomal degradation. Cell Death
Differ. 17:103–108. 2010.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Allende-Vega N and Saville MK: Targeting
the ubiquitin-proteasome system to activate wild-type p53 for
cancer therapy. Semin Cancer Biol. 20:29–39. 2010.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Yu ZK, Gervais JL and Zhang H: Human CUL-1
associates with the SKP1/SKP2 complex and regulates p21(CIP1/WAF1)
and cyclin D proteins. Proc Natl Acad Sci USA. 95:11324–11329.
1998.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Ciechanover A: The 2008 Lindau Nobel
Laureate Meeting: Aaron Ciechanover, Chemistry 2004. J Vis Exp.
29(1559)2009.PubMed/NCBI View
Article : Google Scholar
|
|
11
|
Hoeller D and Dikic I: Targeting the
ubiquitin system in cancer therapy. Nature. 458:438–444.
2009.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Hochstrasser M: Origin and function of
ubiquitin-like proteins. Nature. 458:422–429. 2009.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Bhoj VG and Chen ZJ: Ubiquitylation in
innate and adaptive immunity. Nature. 458:430–437. 2009.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Hirsch C, Gauss R, Horn SC, Neuber O and
Sommer T: The ubiquitylation machinery of the endoplasmic
reticulum. Nature. 458:453–460. 2009.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Bergink S and Jentsch S: Principles of
ubiquitin and SUMO modifications in DNA repair. Nature.
458:461–467. 2009.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Raiborg C and Stenmark H: The ESCRT
machinery in endosomal sorting of ubiquitylated membrane proteins.
Nature. 458:445–452. 2009.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Paoluzzi L and O'Connor OA: Mechanistic
rationale and clinical evidence for the efficacy of proteasome
inhibitors against indolent and mantle cell lymphomas. BioDrugs.
20:13–23. 2006.PubMed/NCBI View Article : Google Scholar
|
|
18
|
O'Connor OA: Marked clinical activity of
the proteasome inhibitor bortezomib in patients with follicular and
mantle-cell lymphoma. Clin Lymphoma Myeloma. 6:191–199.
2005.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Yang Y, Kitagaki J, Dai RM, Tsai YC,
Lorick KL, Ludwig RL, Pierre SA, Jensen JP, Davydov IV, Oberoi P,
et al: Inhibitors of ubiquitin-activating enzyme (E1), a new class
of potential cancer therapeutics. Cancer Res. 67:9472–9481.
2007.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Soucy TA, Smith PG and Rolfe M: Targeting
NEDD8-activated cullin-RING ligases for the treatment of cancer.
Clin Cancer Res. 15:3912–3916. 2009.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Soucy TA, Smith PG, Milhollen MA, Berger
AJ, Gavin JM, Adhikari S, Brownell JE, Burke KE, Cardin DP,
Critchley S, et al: An inhibitor of NEDD8-activating enzyme as a
new approach to treat cancer. Nature. 458:732–736. 2009.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Brownell JE, Sintchak MD, Gavin JM, Liao
H, Bruzzese FJ, Bump NJ, Soucy TA, Milhollen MA, Yang X, Burkhardt
AL, et al: Substrate-assisted inhibition of ubiquitin-like
protein-activating enzymes: The NEDD8 E1 inhibitor MLN4924 forms a
NEDD8-AMP mimetic in situ. Mol Cell. 37:102–111. 2010.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Lin JJ, Milhollen MA, Smith PG, Narayanan
U and Dutta A: NEDD8-targeting drug MLN4924 elicits DNA
rereplication by stabilizing Cdt1 in S phase, triggering checkpoint
activation, apoptosis, and senescence in cancer cells. Cancer Res.
70:10310–10320. 2010.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Rabut G and Peter M: Function and
regulation of protein neddylation. ‘Protein modifications: Beyond
the usual suspects' review series. EMBO Rep. 9:969–976.
2008.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Leonardi GC, Candido S, Falzone L,
Spandidos DA and Libra M: Cutaneous melanoma and the immunotherapy
revolution (Review). Int J Oncol. 57:609–618. 2020.PubMed/NCBI View Article : Google Scholar : (Review).
|
|
26
|
Kuryk L, Bertinato L, Staniszewska M,
Pancer K, Wieczorek M, Salmaso S, Caliceti P and Garofalo M: From
conventional therapies to immunotherapy: Melanoma treatment in
review. Cancers (Basel). 12(3057)2020.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Tanda ET, Vanni I, Boutros A, Andreotti V,
Bruno W, Ghiorzo P and Spagnolo F: Current state of target
treatment in BRAF mutated melanoma. Front Mol Biosci.
7(154)2020.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Qiu Y, Zheng Y, Wu KP and Schulman BA:
Insights into links between autophagy and the ubiquitin system from
the structure of LC3B bound to the LIR motif from the E3 ligase
NEDD4. Protein Sci. 26:1674–1680. 2017.PubMed/NCBI View
Article : Google Scholar
|
|
29
|
Huang DT, Ayrault O, Hunt HW, Taherbhoy
AM, Duda DM, Scott DC, Borg LA, Neale G, Murray PJ, Roussel MF, et
al: E2-RING expansion of the NEDD8 cascade confers specificity to
cullin modification. Mol Cell. 33:483–495. 2009.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Butt G, Yaylim I, Attar R, Aras A, Romero
MA, Qureshi MZ, Purenovic J and Farooqi AA: NEDD4 family of E3
ubiquitin ligases in breast cancer: Spotlight on SMURFs, WWPs and
NEDD4. Adv Exp Med Biol. 1152:365–375. 2019.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Wang ZW, Hu X, Ye M, Lin M, Chu M and Shen
X: NEDD4 E3 ligase: Functions and mechanism in human cancer. Semin
Cancer Biol. 67:92–101. 2020.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Zhang Y, Qian H, Wu B, You S, Wu S, Lu S,
Wang P, Cao L, Zhang N and Sun Y: E3 Ubiquitin ligase NEDD4
family-regulatory network in cardiovascular disease. Int J Biol
Sci. 16:2727–2740. 2020.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Soysouvanh F, Giuliano S, Habel N,
El-Hachem N, Pisibon C, Bertolotto C and Ballotti R: An Update on
the role of ubiquitination in melanoma development and therapies. J
Clin Med. 10(1133)2021.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Xie P, Peng Z, Chen Y, Li H, Du M, Tan Y,
Zhang X, Lu Z, Cui CP, Liu CH, et al: Neddylation of PTEN regulates
its nuclear import and promotes tumor development. Cell Res.
31:291–311. 2021.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Guo Y, Yang L, Lei S, Tan W and Long J:
NEDD4 negatively regulates GITR via ubiquitination in immune
microenvironment of melanoma. OncoTargets Ther. 12:10629–10637.
2019.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Huang X, Chen J, Cao W, Yang L, Chen Q, He
J, Yi Q, Huang H, Zhang E and Cai Z: The many substrates and
functions of NEDD4-1. Cell Death Dis. 10(904)2019.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) Method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Zou X, Levy-Cohen G and Blank M: Molecular
functions of NEDD4 E3 ubiquitin ligases in cancer. Biochim Biophys
Acta. 1856:91–106. 2015.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Zhang X, Zhang YL, Qiu G, Pian L, Guo L,
Cao H, Liu J, Zhao Y, Li X, Xu Z, et al: Hepatic neddylation
targets and stabilizes electron transfer flavoproteins to
facilitate fatty acid β-oxidation. Proc Natl Acad Sci USA.
117:2473–2483. 2020.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Luhtala S, Staff S, Kallioniemi A, Tanner
M and Isola J: Clinicopathological and prognostic correlations of
HER3 expression and its degradation regulators, NEDD4-1 and NRDP1,
in primary breast cancer. BMC Cancer. 18(1045)2018.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Shao G, Wang R, Sun A, Wei J, Peng K, Dai
Q, Yang W and Lin Q: The E3 ubiquitin ligase NEDD4 mediates cell
migration signaling of EGFR in lung cancer cells. Mol Cancer.
17(24)2018.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Wen W, Li J, Wang L, Xing Y, Li X, Ruan H,
Xi X, Xiong J and Kuang R: Inhibition of NEDD4 inhibits cell growth
and invasion and induces cell apoptosis in bladder cancer cells.
Cell Cycle. 16:1509–1514. 2017.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Meurette O: Shaping of the tumor
microenvironment by Notch signaling. Adv Exp Med Biol. 1223:1–16.
2020.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Rubey M, Chhabra NF, Gradinger D,
Sanz-Moreno A, Lickert H, Przemeck GKH and Hrabě de Angelis M:
DLL1- and DLL4-mediated Notch signaling is essential for adult
pancreatic islet homeostasis. Diabetes. 69:915–926. 2020.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Fleming T, Balderas-Márquez JE, Epardo D,
Ávila-Mendoza J, Carranza M, Luna M, Harvey S, Arámburo C and
Martínez-Moreno CG: Growth hormone neuroprotection against kainate
excitotoxicity in the retina is mediated by Notch/PTEN/Akt
signaling. Invest Ophthalmol Vis Sci. 60:4532–4547. 2019.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Zheng D, Tao M, Liang X, Li Y, Jin J and
He Q: p66Shc regulates podocyte autophagy in high glucose
environment through the Notch-PTEN-PI3K/Akt/mTOR pathway. Histol
Histopathol. 35:405–415. 2020.PubMed/NCBI View Article : Google Scholar
|