Introduction
Spinal cord injury (SCI) is a serious neurological
disorder that involves damage to the spinal cord caused by trauma
or disease and its most typical manifestation is paralysis at the
injured site (1). SCI not only
causes physical and psychological trauma for patients, but is also
associated with an important economic burden for the entire society
(2,3). The annual SCI incidence worldwide is
15-40 cases per million people (2),
and the treatment of patients with SCI is currently limited
(4). Several factors can cause SCI,
including oxygen-free radicals, inflammation, ion disturbances,
neurotransmitter accumulation, excitatory amino acid accumulation,
axon loss, glial scar formation and apoptosis (5-10).
Therefore, there is an urgent requirement for novel effective
treatment methods for SCI.
Initial stage SCI involves a series of vascular,
biochemical and cellular events, and it is the most complicated and
the least understood stage of SCI (11,12).
The vascular alterations that occur during this stage lead to
serious complications, including oxidative stress, blood flow
reduction, edema, tissue disorders, depolarization, metabolic
dysfunction, loss of cell function, tissue dissolution and neuronal
cell death (13,14). Subsequent cellular changes include
increases in the numbers of macrophages and neutrophils, apoptosis,
Wallerian degeneration and severe inflammatory response (15,16).
This secondary SCI is a delayed progressive tissue injury after the
initial injury (17,18), during which inflammatory cells such
as macrophages, microglia, T cells and neutrophils cross the
blood-brain barrier and enter the injured area. These cells expand
and release inflammatory factors, such as TNF-α and IL-1β, whose
levels peak at 6-12 h and continue to increase within 4 days after
injury.
Biochemical and cellular alterations in the local
environment that occur after SCI gradually affect the neurons,
oligodendrocytes and astrocytes (19). For example, the center and areas
distal to the site of injury contain oligodendrocytes undergoing
apoptosis. Apoptosis leads to demyelination of the axons of the
preserved oligodendrocytes. Moreover, the phagocytic inflammatory
cells attracted to the injured area release reactive oxygen
species, thereby causing a number of reactions, such as DNA
oxidative damage, protein oxidation and lipid peroxidation
(19).
More than three quarters of the human genome are
transcribed, but <2% of the RNA is translated into proteins
(20). RNAs that are not translated
into proteins but exhibit other cellular functions are called
non-coding RNAs (ncRNAs) (21). In
the past two decades, different types of ncRNAs have been
discovered alongside rRNAs and tRNAs, including microRNAs
(miRNAs/miRs), long non-coding RNAs (lncRNAs) and circular RNAs
(circRNAs) (22). Whereas miRNAs
and lncRNAs are linear, circRNAs are characterized by a covalently
closed continuous loop without the 5' cap and 3' poly(A) tail
(22). In contrast to lncRNAs,
circRNAs are usually derived from protein-coding genes and complete
exons (23). Various types of
circRNAs exist according to their biogenesis, as follows: circRNAs
formed by reverse splicing and exon circularization, circovirus RNA
genomes, circRNA intermediates, intronic circRNAs and exonic
circRNAs (24). Due to the lack of
free ends, circRNAs are resistant to exonucleases; moreover, they
exhibit the potential for rolling circle amplification (25).
Research on the role of circRNAs in nerve damage has
rapidly evolved. Numerous studies have revealed the expression
pattern of circRNAs in traumatic brain injury and neuropathic pain
models through DNA microarrays and RNA-sequencing (26,27).
For instance, circRNA_0006928 may regulate neuronal apoptosis by
binding to miR-184(26). In chronic
inflammatory pain, circRNA-Filip1l, which is negatively regulated
by miR-1224 through binding and splicing, increases chronic
inflammatory pain and can regulate nociception by targeting
ubiquitin protein ligase E3 component n-recognin 5(28). circRNA expression increased in the
rat spinal cord after traumatic SCI, which indicated that there was
a relationship between circRNA and SCI (29). In addition, differentially expressed
circRNAs were identified in the rat sciatic nerve compression
model, and downregulated circRNAs were revealed compared with
control rats. Among them, circRNA_2837 was demonstrated to regulate
neuronal autophagy by acting as a binding sponge for the miR-34
family (30). Furthermore,
silencing circRNA_2837 can induce autophagy in primary spinal cord
neurons by targeting miR-34a (30).
Although these findings suggested that circRNAs exhibit regulatory
functions in nerve injury, their precise role is still unknown.
A previous study using a rat SCI model revealed that
a total of 150 circRNAs were significantly differentially expressed
in the rat spinal cord after SCI (fold-change ≥2; P≤0.05). Of
these, 99 circRNAs were upregulated, and 51 were downregulated
(31). Among them, circRNA_014301
was highly expressed at the injury site and is therefore of
interest (29,31). Although these studies revealed that
circRNA_014301 was significantly induced following SCI in the rat
model, whether it exhibits a specific regulatory function in SCI
remains unknown (32). The present
study aimed to analyze the effect of circRNA_014301 on the
inflammation and apoptosis of PC12 cells to assess the possible
regulatory role of circRNA_014301 in a cellular model of SCI and
identify a potential therapeutic target for SCI.
Materials and methods
Cell culture and treatments
The rat adrenal pheochromocytoma cell line PC12 was
obtained from The World Cell Factory (CyberKang (Shanghai)
Biotechnology Co., Ltd.). The cells were maintained in a complete
medium containing RPMI-1640 (Sigma-Aldrich; Merck KGaA) plus 2 mM
glutamine, 1.5 g/l sodium bicarbonate, 4.5 g/l glucose, 10 mM HEPES
and 1 mM sodium pyruvate, supplemented with 10% heat-inactivated
horse serum (Invitrogen; Thermo Fisher Scientific, Inc.) and 5% FBS
(Biological Industries), in a 5% CO2 incubator at 37˚C
with no antibiotics. When cell confluence reached >90%, the
cells were passaged at a ratio of 1:2 or 1:3 before use.
Different concentrations of lipopolysaccharide (LPS;
0, 1, 2.5, 5 and 10 µg/ml; Sigma-Aldrich; Merck KGaA) were used to
treat PC12 cells for 24 h and to construct a PC12 cell inflammatory
model (33-35).
The small interfering (si)RNA for circRNA_014301
(si-circRNA_014301; 5'-CAGACAGGAGCTACTCGGATA TGAT-3') and the
si-negative control (si-NC; 5'-CATCTCCCA GCAGTGACACTGACTT-3') were
purchased from Shanghai GenePharma Co., Ltd. Cell transfection was
performed using Lipofectamine® 2000 (Thermo Fisher
Scientific, Inc.), as per the manufacturer's instructions. PC12
cells were seeded into 6-well plates at 2x104
cells/well. Cells were transfected with 100 nM of siRNA. The
culture plate was placed in a CO2 incubator at 37˚C for
12 h. Cells were subsequently stimulated with 5 µg/ml LPS for 24 h
to construct an SCI inflammation model. After the intervention is
over, cells are collected and tested. After transfection, RNA was
extracted, and the silencing efficiency was determined via reverse
transcription-quantitative (RT-q) PCR.
The experimental protocol was divided into four
treatments: Control (normally cultured cells without LPS), LPS (5
µg/ml) treatment, si-circRNA_014301 + LPS treatment and si-NC + LPS
treatment. Following transfection with si-circRNA_014301 or si-NC
for 24 h, LPS treatment was performed for 24 h to establish the
inflammatory model.
Total RNA extraction and
purification
Total RNA was extracted from PC12 cells using the
RNeasy Protect Mini kit (Qiagen, Inc.) following the manufacturer's
manual. RNA concentration was quantified using NanoDrop ND-1000
spectrophotometer (NanoDrop Technologies; Thermo Fisher Scientific,
Inc.), and its integrity was evaluated on an agarose gel. The RNA
was then purified with RNase-Free DNase Set (Qiagen, Inc.) and
digested for 30 min at 37˚C with RNase R (20 mg/ml; cat. no.
RNR07250; Epicenter Biotechnologies; Lucigen Corporation) to remove
linear RNA according to the manufacturer's instructions.
RNase R is known to degrade linear RNAs leaving
circRNAs intact, and is used to validate the depletion of linear
RNAs and the resistance of circRNAs to RNase R treatment (36). miRDB database (http://mirdb.org/) was utilized to predict miRNA-mRNA
interactions.
Isolation of nuclear and cytoplasmic
fractions
The nuclear and cytoplasmic fractions were isolated
using a PARIS™ kit (Thermo Fisher Scientific, Inc.) according to
the manufacturer's instructions. The expression of circRNA_014301
in the nuclear and cytoplasmic was tested via qPCR. U6 snRNA and
18S rRNA were employed as positive controls for nuclear and
cytoplasmic fractions, respectively.
RT-qPCR
The purified, RNase R-digested RNA was
reverse-transcribed to cDNA using PrimeScript RT reagent Kit
(Takara Biotechnology Co., Ltd.) for 15 min at 37˚C. cDNA
amplification was performed using SYBR® Green Realtime
PCR Master Mix (Toyobo Life Science). The settings of the
LightCycler 96 Real-Time PCR system (Roche Diagnostics) were as
follows: 94˚C for 30 sec; followed by 40 cycles of 94˚C for 5 sec
and 61˚C for 35 sec; followed by 97˚C for 10 sec, 65˚C for 1 min,
and 97˚C for 1 sec. Each reaction included three replicates. GAPDH
(forward, 5'-GCTCTC TGCTCCTCCCTGTTCTA-3'; and reverse, 5'-TGGTAACCA
GGCGTCCGATA-3') and U6 (forward, 5'-AAAGCAAAT CATCGGACGACC-3'; and
reverse, 5'-GTACAACACATT GTTTCCTCGGA-3') were used as the internal
reference genes (37,38). The primer sequences of 18S RNA
(forward, 5'-TGTGCCGCTAGAGGTGAAATT-3', and reverse 5'-TGG
CAAATGCTTTCGCTTT-3') were based on Tao et al (39). The primer sequences of
circRNA_014301 (forward, 5'-GCT GCTCTAGTGGTGACTCATG-3'; and reverse
5'-TTCTCC ATTCATCCAATCAACTTCG-3'), IL-1β (forward, 5'-GAC
CTTCCAGGATGAGGACA-3'; and reverse 5'-AGCTCA TATGGGTCCGACAG-3'),
IL-6 (forward, 5'-AGTTGCCTT CTTGGGACTGA-3'; and reverse
5'-CAGAATTGCCAT TGCACAAC-3') and TNF-α (forward, 5'-ACGGCATGGATC
TCAAAGAC-3'; and reverse 5'-GTGGGTGAGGAGCAC GTAGT-3') were based on
Gonzales et al (40). The
primer sequences of MTY1L (forward, 5'-GTCATGGTGTGAGGG GTCCC-3',
and reverse 5'-CACACTCTGTGATTCTTCAG-3') were based on Bruno et
al (41). Gene expression was
calculated using the 2-ΔΔCq method (31,42).
Cell viability assay
The viability of PC12 cells was measured using Cell
Counting Kit-8 (CCK-8) assay (Beyotime). Briefly, LPS treatment
after transfection was performed as aforementioned, and then the
cells were seeded onto 96-well plates at 2x104
cells/well and incubated in a humidified incubator at 37˚C for 6 h.
Subsequently, 10 µl of CCK-8 solution was added to each well, and
the plate was incubated for 1 h at 37˚C. The absorbance of each
well at 450 nm was recorded using a microplate reader (BioTek
Instruments, Inc.). The higher the OD 450 value, the faster the
cell proliferation.
ELISA
The contents of IL-1β (cat. no. RLB00), IL-6 (cat.
no. R6000B) and TNF-α (cat. no. RTA00) in the PC12 cells were
measured using corresponding Quantikine ELISA kits (R&D
Systems, Inc.) according to the manufacturer's instructions. Each
experiment was performed in three biological replicates.
Flow cytometry analysis of
apoptosis
PC12 cells in the various groups were harvested and
centrifuged at room temperature at 10,000 x g for 5 min, followed
by treatment with Annexin V-FITC binding buffer and 1 mg/ml of PI
solution (cat. no. G003-1-2; Nanjing Jiancheng Bioengineering
Institute) at room temperature for 15 min. Apoptotic cells were
detected using flow cytometry (CytoFLEX; Becton, Dickinson and
Company) and analyzed using CytExpert software 2.0 (Becton,
Dickinson and Company).
Western blotting
Total protein from the PC12 cell line was acquired
using RIPA buffer (cat. no. P0013B; Beyotime Institute of
Biotechnology) and centrifuged at 4˚C for 15 min, protein
concentrations were determined using BCA assays (cat. no. PC0020;
Beijing Solarbio Science & Technology Co., Ltd), and samples
(20 µg) were separated via 10% SDS-PAGE and transferred to PVDF
membranes. The membranes were incubated at room temperature for 2 h
in 5% nonfat dried milk. The target protein antibodies (1:1,000)
and anti-GAPDH (1:3,000) were incubated with the membranes at 4˚C
overnight. The primary antibodies used were as follows: Bax (cat.
no. ab32503; Abcam), Bcl-2 (cat. no. 3498; Abcam), cleaved
caspase-3 (cat. no. 9661; Abcam), phosphorylated (p)-NF-κB p65
(cat. no. 3033; Abcam), NF-κB p65 (cat. no. 8242; Abcam), with and
GAPDH (cat. no. M1000110; Beijing Solarbio Science & Technology
Co., Ltd.) as the reference antibody. The secondary antibodies
[goat anti- rabbit IgG-HRP (1:5,000; cat. no. SE134; Beijing
Solarbio Science & Technology Co., Ltd.) and goat anti-mouse
IgG-HRP (cat. no. GB23301; Wuhan Servicebio Technology Co., Ltd.)]
were subsequently incubated with membranes at room temperature for
1 h. ECL color developing solution (cat. no. PE0010; Beijing
Solarbio Science & Technology Co., Ltd.) was added to the PVDF
film for exposure and photographing. Quantity One software 4.0
(Bio-Rad Laboratories, Inc.) was used to perform grayscale analysis
of protein bands.
Statistical analysis
The results of three biological replicates are
presented as the mean ± standard deviation. SPSS 21.0 software (IBM
Corp.) was used for statistical analyses, and the data were
analyzed using one-way ANOVA followed by Tukey's post hoc test.
P<0.05 was used to indicate a statistically significant
difference.
Results
LPS treatment promotes inflammatory
response in PC12 cells
As presented in Fig.
1A, PC12 cell viability was reduced with increasing LPS
concentration. Cell viability was low with 5 µg/ml LPS treatment,
but this was significantly higher than that of the 10 µg/ml LPS
treatment.
As demonstrated in Fig.
1B, while 93.3% of the cells were unaffected after the control
treatment (LPS-0), cells underwent apoptosis under increasing LPS
concentrations. Moreover, higher LPS concentrations enhanced the
apoptotic effect. The proportion of early cell apoptosis was 4.41,
6.63, 7.41 and 13.4% with LPS-1, LPS-2.5, LPS-5 and LPS-10
treatment, respectively, whereas the proportion of late cell
apoptosis was 3.10, 4.38, 8.55 and 8.02% with LPS-1, LPS-2.5, LPS-5
and LPS-10 treatment, respectively. Furthermore, apoptosis was most
pronounced with LPS-5 and LPS-10 treatments (Fig. 1B). Thus, 5 μg/ml LPS evoked a
sufficiently high inflammatory response while mildly affecting cell
viability.
The expression and concentration of inflammatory
factors (IL-1β, IL-6 and TNF-α) both increased with increasing LPS
concentration (Fig. 1C and D). The expression levels of IL-1β, IL-6
and TNF-α increased slightly with 1-2.5 µg/ml LPS, increased
notably with 5 µg/ml LPS, and peaked with 10 µg/ml LPS. However,
there was no significant difference in the IL-1β concentration in
the 5-10 µg/ml LPS range.
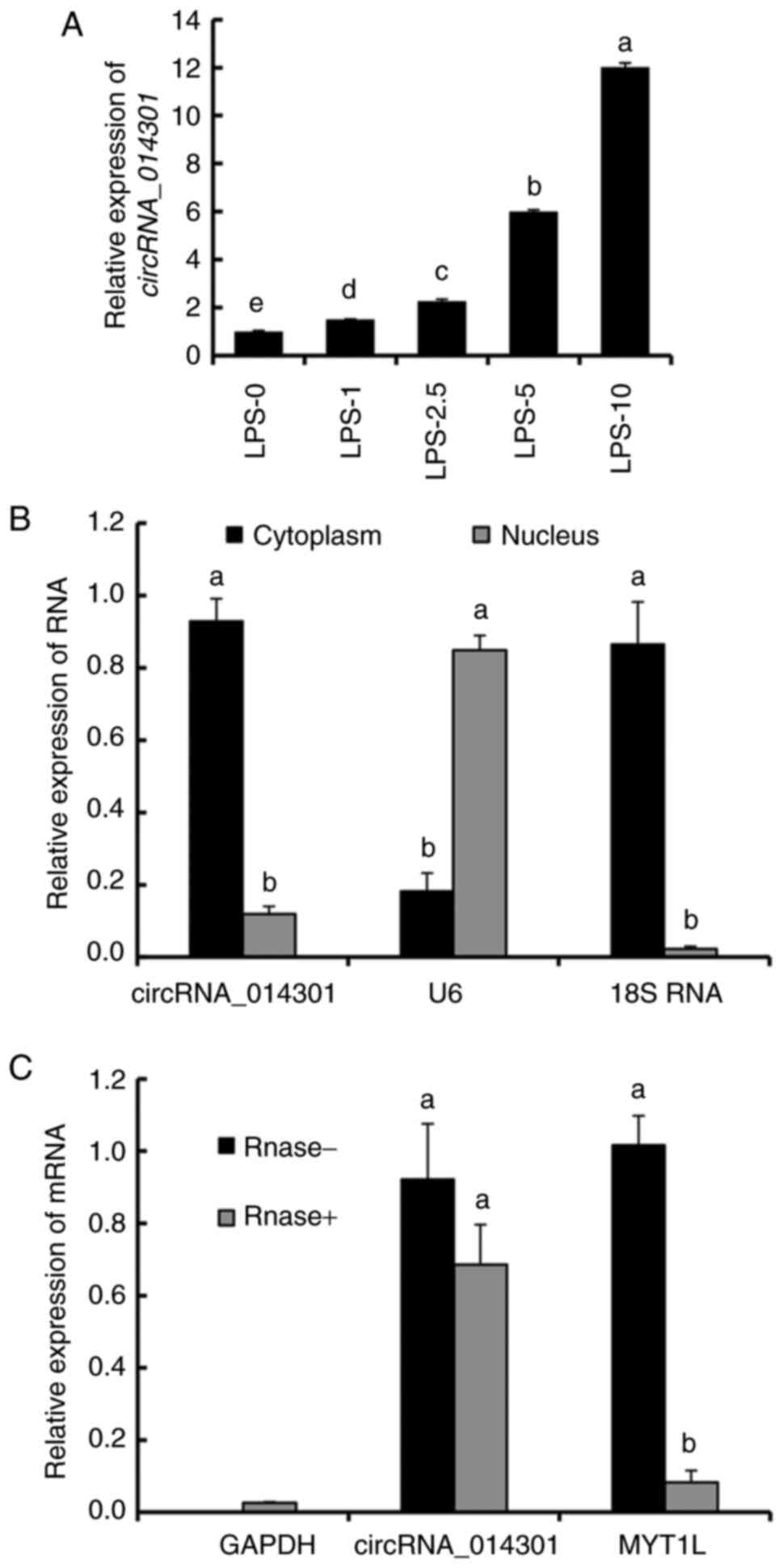
LPS treatment promotes circRNA_014301
expression in PC12 cells
The relative expression of circRNA_014301 increased
with increasing LPS concentration (Fig.
2A). It was slightly upregulated with 1-2.5 µg/ml LPS but was 6
and 12 times higher than that of the control with 5 µg/ml and 10
µg/ml LPS treatments, respectively. These results suggested that
LPS induced circRNA_014301 expression in PC12 cells.
Referring to the miRDB database, it was therefore
predicted that the following miRNAs could bind to circRNA_014301:
miR-200a-3p, miR-141-3p, miR-3120, miR-16-5p, miR-15a-5p,
miR-15b-5p, miR-195-5p, miR-497-5p, miR-322-5p, miR-344b-1-3p,
miR-410-3p, miR-331-5p and miR-501-5p (Table SI).
Whereas circRNA_014301 and 18S RNA were highly
expressed in the cytoplasm, U6 showed high enrichment in the
nucleus (Fig. 2B). There were no
significant changes in the expression of circRNA_014301 with
RNase- and RNase+ treatments. However, the
expression of MYT1L was significantly lower with the
RNase+ treatment compared with the RNase- treatment,
implying that MYT1L was cleaved by the RNase (Fig. 2C). These results suggested that
circRNA_014301 was predominantly expressed in the cytoplasm.
circRNA_014301 silencing inhibits
inflammation in PC12 cells
As indicated in Fig.
S1 siRNA eliminated circRNA_014301 expression in PC12 cells,
thus confirming the efficiency of the siRNA-mediated knockdown.
Compared with the control, PC12 cell viability was reduced with
LPS, si-circRNA_014301 + LPS and si-NC + LPS treatments. It was
also more enhanced with si-circRNA_014301 + LPS treatment than with
LPS and si-NC + LPS treatments (Fig.
3A).
 | Figure 3circRNA_014301 promotes inflammation
in PC12 cells. (A) PC12 cell viability was decreased with LPS
treatment compared with the control, but it was increased in
LPC-treated cells after siRNA silencing of circRNA_014301. (B)
Compared with control treatment, the concentration of inflammatory
factors (IL-1β, IL-6 and TNF-α) was increased after LPS treatment,
but it was reduced after siRNA silencing of circRNA_014301. (C) LPS
promoted the expression of inflammatory factors (IL-1β, IL-6 and
TNF-α) compared with control treatment, which was suppressed by the
siRNA-mediated circRNA_014301 knockdown. (D) LPS increased the
ratio of p-NF-κB/NF-κB compared with control treatment, whereas
siRNA-mediated silencing of circRNA_014301 suppressed it. Groups
labelled with the same lower-case letter are not significantly
different to one another; groups with different letters are
significantly different (P<0.05). LPS, lipopolysaccharide;
circRNA, circular RNA; OD, optical density; p, phosphorylated; si,
small interfering; NC, negative control. |
As demonstrated in Fig.
3B and C, the expression levels
and concentration of inflammatory factors (IL-1β, IL-6 and TNF-α)
with LPS and si-NC + LPS treatments were significantly higher than
those of the control and si-circRNA_014301 + LPS treatments, as
analyzed using ELISA and RT-qPCR. The expression level and
concentration of TNF-α with si-circRNA_014301 + LPS treatment were
significantly higher than those of the control treatment. The
concentrations of IL-1β and IL-6 with si-circRNA_014301 + LPS
treatment were significantly higher than those of the control
treatment. However, there were no significant changes in the
expression levels of IL-1β and IL-6 between the control and
si-circRNA_014301 + LPS treatment.
The ratio of p-NF-κB/NF-κB was higher with LPS
treatment compared with that of the control, followed by the si-NC
+ LPS and si-circRNA_014301 + LPS treatments (Fig. 3D).
circRNA_014301 silencing inhibits
apoptosis in PC12 cells
As demonstrated in Fig.
4A, 2.60 and 2.02% of PC12 cells underwent early and late
apoptosis, respectively, with the control treatment. Compared with
the control, cell apoptosis increased with all other treatments.
With LPS treatment, 6.04 and 10.4% of cells underwent early and
late apoptosis, respectively. With si-circRNA_014301 + LPS
treatment, the apoptotic effect was milder, with 4.61 and 3.50% of
cells undergoing early and late apoptosis, respectively.
Remarkably, apoptosis was the highest with si-NC + LPS treatment,
where 6.63 and 12.2% of cells underwent early and late apoptosis,
respectively. These findings suggested that the siRNA-mediated
knockdown of circRNA_014301 expression alleviated apoptosis in PC12
cells.
As presented in Fig.
4B, the protein expression of apoptotic markers (Bax, Bcl-2 and
cleaved caspase-3) differed with the various treatments. The
relative protein expression of Bax and cleaved caspase-3 was
similar, being the highest with LPS treatment, followed by si-NC +
LPS, si-circRNA_014301 + LPS and control treatments (Fig. 4C). Conversely, the protein
expression levels of Bcl2 were the highest on the control
treatment, followed by si-circRNA_014301 + LPS, si-NC + LPS and LPS
treatments (Fig. 4C).
Discussion
SCI is invariably associated with spinal cord
inflammation that adversely affects the outcome of SCI (43). The PC12 cell line was derived from a
pheochromocytoma of rat adrenal medulla. Tischler (44) discovered that these cells
differentiate into neurons under treatment with nerve growth
factor. He also indicated that they can synthesize and store
several neurotransmitters, such as dopamine. PC12 cells can be used
as an in vitro cell model of neuronal cells for high-throughput
experiments (45). Compared with
primary cultured nerve cells, PC12 cell uniformity allows
experimental consistency, long-enough cell survival time for
observation and maintained nerve cell characteristics (46). The PC12 cell line has been widely
used to study nervous system diseases and pathological and
physiological characteristics of neurons and to create SCI cellular
models (47). In the preset study,
increasing concentration of LPS were used to stimulate apoptosis
and inflammatory injury in PC12 cells and create a cell model of
SCI (34,48). Jiang and Wang (33) reported that 5 µg/ml LPS treatment
could induce inflammatory injury in PC12 cells. Indeed, 5 µg/ml LPS
treatment evoked a strong inflammatory response while mildly
reducing the cell viability. Therefore, 5 µg/mL LPS treatment was
deemed suitable for generating a PC12 cell inflammatory model.
Ashwal-Fluss et al (49) demonstrated that circRNAs are
produced co-transcriptionally and compete with mRNAs for regular
splicing. Therefore, the biogenesis of circRNAs results in reduced
synthesis of mRNAs from the same locus. Thus, the production of
circRNA acts as an RNA trap for mRNA production (50). circRNA_014301 (51,52)
and MYT1L are both located in the cytoplasm. Whereas
MYT1L was digested under the presence of RNase,
circRNA_01430 was unaffected by the addition of the enzyme. These
results suggested that circRNA_014301 was indeed located in the
cytoplasm in accordance with Capel et al (51) and Patop et al (52), who revealed that this circRNA_014301
was predominantly cytoplasmic.
A representative function of circRNA is that it can
act as a miRNA sponge to regulate the stability or translational
efficiency of other RNAs (53).
circRNAs and miRNAs can also act as transcriptional regulators or
protein-bound RNAs and can even directly be translated into
proteins (54). As this type of
circRNAs regulates target gene expression, they are called
competitive endogenous RNAs (ceRNAs) (53). Further investigation of circRNA and
miRNA regulatory pathways will be the focus of follow-up
research.
Inflammation serves a crucial role in the
pathogenesis of SCI (55). The
induced inflammation may result in a further decrease in functional
recovery due to the development of scar tissue and the necrosis or
apoptosis of neurons and oligodendrocytes (56). Nonetheless, potentially beneficial
effects of the inflammatory process have also been reported,
illustrating the dual nature of inflammation after SCI trauma
(57). Apoptosis is essential for
the clearance of potentially injurious inflammatory cells and the
subsequent efficient resolution of inflammation (58). The expression of inflammatory genes
(IL-1 and IL-1β) and the regulation of inflammatory agents (nitric
oxide synthase and cyclooxygenase 2) that serve a potential role in
the inflammatory pathways mediating damage of the central nervous
system are regulated by the NF-κB family of transcription factors
(59). It has been revealed that
inhibiting the activation of NF-κB regulates the secondary damage
in SCI to a large extent (60). The
present study demonstrated that NF-κB activation was suppressed
following siRNA-mediated knockdown of circRNA_014301, indicating
that circRNA_014301 is involved in the NF-κB pathway, and this
hypothesis requires further experimental verification.
Large circRNAs are associated with the biological
activities of endothelial cells (61). Li et al (35) indicated that circRNA
hsa_circ_0003575 silencing promoted cell proliferation and
angiogenesis in oxidized low-density lipoprotein-induced
endothelial cells. Furthermore, Dang et al (62) demonstrated that the knockdown of
circRNA hsa_circ_001079 suppressed proliferation and promoted
apoptosis in hypoxia-induced endothelial cells. circRNA_014301
silencing suppressed PC12 cell inflammation in the present study.
The protective regulatory effect of circRNA_014301 silencing may
inhibit the development of SCI. Subsequent in vivo experiments
should be performed for in-depth verification of circRNA_014301
function. Based on the aforementioned findings, it is hypothesized
that circRNA_014301 may constitute a potential biomarker for SCI
detection.
Supplementary Material
Relative expression of circRNA_014301
after siRNA knockdown in PC12 cells. aNo significant
difference (P<0.05) between control and si-NC;
bSignificant difference (P<0.05) between control and
si-cricRNA_014301. circRNA, circular RNA; si, small interfering;
NC, negative control.
miRNAs binding to circRNA_014301 as
detected using the miRDB database.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
XX performed the experiments, analyzed the results
and wrote the manuscript. YX and KX contributed to data analysis
and manuscript revision. All authors have read and approved the
final manuscript. XX and YX confirm the authenticity of all the raw
data.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Park S, Park K, Lee Y, Chang KT and Hong
Y: New prophylactic and therapeutic strategies for spinal cord
injury. J Lifestyle Med. 3:34–40. 2013.PubMed/NCBI
|
|
2
|
Sekhon LHS and Fehlings MG: Epidemiology,
demographics, and pathophysiology of acute spinal cord injury.
Spine (Phila Pa 1976). 26 (Suppl 24):S2–S12. 2001.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Bellon K, Kolakowsky-Hayner SA, Chen D,
McDowell S, Bitterman B and Klaas SJ: Evidence-based practice in
primary prevention of spinal cord injury. Top Spinal Cord Inj
Rehabil. 19:25–30. 2013.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Wouda EMN and Stienstra Y: vander WTS,
Kerstjens H, deLange WCM, Coppes M, Kuijlen J, Marga T, and
Akkerman OW: Neurological and functional recovery in tuberculosis
patients with spinal cord injury in The Netherlands. Neuro Rehab.
40:439–445. 2017.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Tator CH: Review of experimental spinal
cord injury with emphasis on the local and systemic circulatory
effects. Neurochirurgie. 37:291–302. 1991.PubMed/NCBI
|
|
6
|
Torres-Espín A, Forero J, Fenrich KK,
Lucas-Osma AM, Krajacic A, Schmidt E, Vavrek R, Raposo P, Bennett
DJ, Popovich PG, et al: Eliciting inflammation enables successful
rehabilitative training in chronic spinal cord injury. Brain.
141:1946–1962. 2018.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Agrawal SK and Fehlings MG: Mechanisms of
secondary injury to spinal cord axons in vitro: Role of Na+,
Na(+)-K(+)-ATPase, the Na(+)-H+ exchanger, and the Na(+)-Ca2+
exchanger. J Neurosci. 16:545–552. 1996.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Kiyatkin EA and Sharma HS: Not just the
brain: Methamphetamine disrupts blood-spinal cord barrier and
induces acute glial activation and structural damage of spinal cord
cells. CNS Neurol Disord Drug Targets. 14:282–294. 2015.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Mazzone GL, Veeraraghavan P,
Gonzalez-Inchauspe C, Nistri A and Uchitel OD: ASIC channel
inhibition enhances excitotoxic neuronal death in an in vitro model
of spinal cord injury. Neuroscience. 343:398–410. 2017.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Casha S, Yu WR and Fehlings MG:
Oligodendroglial apoptosis occurs along degenerating axons and is
associated with FAS and p75 expression following spinal cord injury
in the rat. Neuroscience. 103:203–218. 2001.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Darian-Smith C: Synaptic plasticity,
neurogenesis, and functional recovery after spinal cord injury.
Neuroscientist. 15:149–165. 2009.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Tator CH and Fehlings MG: Review of the
secondary injury theory of acute spinal cord trauma with emphasis
on vascular mechanisms. J Neurosurg. 75:15–26. 1991.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Liu D, Xu GY, Pan E and McAdoo DJ:
Neurotoxicity of glutamate at the concentration released upon
spinal cord injury. Neuroscience. 93:1383–1389. 1999.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Goodman JH, Bingham WG Jr and Hunt WE:
Platelet aggregation in experimental spinal cord injury.
Ultrastructural observations. Arch Neurol. 36:197–201.
1979.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Beattie MS, Farooqui AA and Bresnahan JC:
Review of current evidence for apoptosis after spinal cord injury.
J Neurotrauma. 17:915–925. 2000.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Silva NA, Sousa N, Reis RL and Salgado AJ:
From basics to clinical: A comprehensive review on spinal cord
injury. Prog Neurobiol. 114:25–57. 2014.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Yip PK and Malaspina A: Spinal cord trauma
and the molecular point of no return. Mol Neurodegener.
7(6)2012.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Norenberg MD, Smith J and Marcillo A: The
pathology of human spinal cord injury: Defining the problems. J
Neurotrauma. 21:429–440. 2004.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Jain NB, Ayers GD, Peterson EN, Harris MB,
Morse L, O'Connor KC and Garshick E: Traumatic spinal cord injury
in the United States, 1993-2012. JAMA. 313:2236–2243.
2015.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Fatica A and Bozzoni I: Long non-coding
RNAs: New players in cell differentiation and development. Nat Rev
Genet. 15:7–21. 2014.PubMed/NCBI View
Article : Google Scholar
|
|
21
|
Cech TR and Steitz JA: The noncoding RNA
revolution-trashing old rules to forge new ones. Cell. 157:77–94.
2014.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Qu S, Yang X, Li X, Wang J, Gao Y, Shang
R, Sun W, Dou K and Li H: Circular RNA: A new star of noncoding
RNAs. Cancer Let. 365:141–148. 2015.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Pamudurti NR, Bartok O, Jens M,
Ashwal-Fluss R, Stottmeister C, Ruhe L, Hanan M, Wyler E,
Perez-Hernandez D, Ramberger E, et al: Translation of CircRNAs. Mol
Cell. 66:9–21.e7. 2017.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Chen LL: The biogenesis and emerging roles
of circular RNAs. Nat Rev Mol Cell Biol. 17:205–211.
2016.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Lasda E and Parker R: Circular RNAs:
Diversity of form and function. RNA. 20:1829–1842. 2014.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Zhou J, Xiong Q, Chen H, Yang C and Fan Y:
Identification of the spinal expression profile of non-coding RNAs
involved in neuropathic pain following spared nerve injury by
sequence analysis. Front Mol Neurosci. 10(91)2017.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Zhao RT, Zhou J, Dong XL, Bi CW, Jiang RC,
Dong JF, Tian Y, Yuan HJ and Zhang JN: Circular ribonucleic acid
expression alteration in exosomes from the brain extracellular
space after traumatic brain injury in mice. J Neurotrauma.
35:2056–2066. 2018.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Pan Z, Li GF, Sun ML, Xie L, Liu D, Zhang
Q, Yang XX, Xia S, Liu X, Zhou H, et al: MicroRNA-1224 splicing
circularRNA-Filip1l in an Ago2- dependent manner regulates chronic
inflammatory pain via targeting Ubr5. J Neurosci. 39:2125–2143.
2019.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Zhou ZB, Du D, Chen KZ, Deng FL, Niu YL
and Zhu L: Differential expression profiles and functional
predication of circRNA in traumatic spinal cord injury of rats. J
Neurother. 36:2287–2297. 2019.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Zhou ZB, Niu YL, Huang GX, Lu JJ, Chen A
and Zhu L: Silencing of circRNA.2837 Plays a Protective Role in
Sciatic Nerve Injury by Sponging the miR-34 Family via Regulating
Neuronal Autophagy. Mol Ther Nucleic Acids. 12:718–729.
2018.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Liu Y, Liu J and Liu B: Identification of
circular RNA expression profiles and their implication in spinal
cord injury rats at the immediate phase. J Mol Neurosci.
70:1894–1905. 2020.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Qin C, Liu CB, Yang DG, Gao F, Zhang X,
Zhang C, Du LJ, Yang ML and Li JJ: Circular RNA expression
alteration and bioinformatics analysis in rats after traumatic
spinal cord injury. Front Mol Neurosci. 11(497)2019.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Jiang J and Wang G: Matrine protects PC12
cells from lipopolysaccharide-evoked inflammatory injury via
upregulation of miR-9. Pharm Biol. 58:314–320. 2020.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Li R, Yin F, Guo Y, Ruan Q and Zhu Q:
Angelica polysaccharide protects PC-12 cells from
lipopolysaccharide-induced injury via down-regulating microRNA-223.
Biomed Pharmacother. 108:1320–1327. 2018.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Li CY, Ma L and Yu B: Circular RNA
hsa_circ_0003575 regulates oxLDL induced vascular endothelial cells
proliferation and angiogenesis. Biomed Pharmacother. 95:1514–1519.
2017.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Pandey PR, Rout PK, Das A, Gorospe M and
Panda AC: RPAD (RNase R treatment, polyadenylation, and poly(A)+
RNA depletion) method to isolate highly pure circular RNA. Methods.
155:41–48. 2019.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Wang C, Wang Q, Lou Y, Xu J, Feng Z, Chen
Y, Tang Q, Zheng G, Zhang Z, Wu Y, et al: Salidroside attenuates
neuroinflammation and improves functional recovery after spinal
cord injury through microglia polarization regulation. J Cell Mol
Med. 22:1148–1166. 2018.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Yanbin Z and Jing Z: CircSAMD4A
accelerates cell proliferation of osteosarcoma by sponging miR-1244
and regulating MDM2 mRNA expression. Biochem Biophys Res Commun.
516:102–111. 2019.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Tao L, Fan X, Sun J and Zhang Z: Metformin
prevented high glucose-induced endothelial reactive oxygen species
via OGG1 in an AMPKα-Lin-28 dependent pathway. Life Sci.
268(119015)2021.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Gonzales AM and Orlando RA: Curcumin and
resveratrol inhibit nuclear factor-kappaB-mediated cytokine
expression in adipocytes. Nutr Metab (Lond). 5(17)2008.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Bruno IG, Karam R, Huang L, Bhardwaj A,
Lou CH, Shum EY, Song HW, Corbett MA, Gifford WD, Gecz J, et al:
Identification of a microRNA that activates gene expression by
repressing nonsense-mediated RNA decay. Mol Cell. 42:500–510.
2011.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Anwar MA, Al Shehabi TS and Eid AH:
Inflammogenesis of secondary spinal cord injury. Front Cell
Neurosci. 10(98)2016.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Greene LA and Tischler AS: Establishment
of a noradrenergic clonal line of rat adrenal pheochromocytoma
cells which respond to nerve growth factor. Proc Natl Acad Sci USA.
73:2424–2428. 1976.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Dichter MA, Tischler AS and Greene LA:
Nerve growth factor-induced increase in electrical excitability and
acetylcholine sensitivity of a rat pheochromocytoma cell line.
Nature. 268:501–504. 1977.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Hillion JA, Takahashi K, Maric D, Ruetzler
C, Barker JL and Hallenbeck JM: Development of an ischemic
tolerance model in a PC12 cell line. J Cereb Blood Flow Metab.
25:154–162. 2005.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Zhang G, Liu Y, Xu L, Sha CH, Zhang HB and
Xu WB: Resveratrol alleviates lipopolysaccharide-induced
inflammation in PC12 cells and in rat model. BMC Biotechnol.
19(10)2019.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Xie Y, Zhang H, Zhang Y, Wang C, Duan D
and Wang Z: Chinese Angelica polysaccharide (CAP) alleviates
LPS-induced inflammation and apoptosis by down-regulating COX-1 in
PC12 cells. Cell Physiol Biochem. 49:1380–1388. 2018.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Ashwal-Fluss R, Meyer M, Pamudurti NR,
Ivanov A, Bartok O, Hanan M, Evantal N, Memczak S, Rajewsky N and
Kadener S: circRNA biogenesis competes with pre-mRNA splicing. Mol
Cell. 56:55–66. 2014.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Jens M and Rajewsky N: Competition between
target sites of regulators shapes post-transcriptional gene
regulation. Nat Rev Genet. 16:113–126. 2015.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Capel B, Swain A, Nicolis S, Hacker A,
Walter M, Koopman P, Goodfellow P and Lovell-Badge R: Circular
transcripts of the testis-determining gene Sry in adult mouse
testis. Cell. 73:1019–1030. 1993.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Patop IL, Wüst S and Kadener S: Past,
present, and future of circRNAs. EMBO J. 38(e100836)2019.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Bak RO and Mikkelsen JG: miRNA sponges:
Soaking up miRNAs for regulation of gene expression. Wiley
Interdiscip Rev RNA. 5:317–333. 2014.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Liu J, Li Z, Teng W and Ye X:
Identification of downregulated circRNAs from tissue and plasma of
patients with gastric cancer and construction of a
circRNA-miRNA-mRNA network. J Cell Biochem. 121:4590–4600.
2020.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Beattie MS: Inflammation and apoptosis:
Linked therapeutic targets in spinal cord injury. Trends Mol Med.
10:580–583. 2004.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Acarin L, González B and Castellano B:
Neuronal, astroglial and microglial cytokine expression after an
excitotoxic lesion in the immature rat brain. Eur J Neurosci.
12:3505–3520. 2000.PubMed/NCBI View Article : Google Scholar
|
|
57
|
Hausmann ON: Post-traumatic inflammation
following spinal cord injury. Spinal Cord. 41:369–378.
2003.PubMed/NCBI View Article : Google Scholar
|
|
58
|
Rossi AG, Sawatzky DA, Walker A, Ward C,
Sheldrake TA, Riley NA, Caldicott A, Martinez-Losa M, Walker TR,
Duffin R, et al: Cyclin-dependent kinase inhibitors enhance the
resolution of inflammation by promoting inflammatory cell
apoptosis. Nat Med. 12:1056–1064. 2006.PubMed/NCBI View
Article : Google Scholar : Erratum in: Nat
Med 12: 1434, 2006.
|
|
59
|
Hu X, Nesic-Taylor O, Qiu J, Rea HC,
Fabian R, Rassin DK and Perez-Polo JR: Activation of nuclear
factor-kappaB signaling pathway by interleukin-1 after
hypoxia/ischemia in neonatal rat hippocampus and cortex. J
Neurochem. 93:26–37. 2005.PubMed/NCBI View Article : Google Scholar
|
|
60
|
Brambilla R, Bracchi-Ricard V, Hu WH,
Frydel B, Bramwell A, Karmally S, Green EJ and Bethea JR:
Inhibition of astroglial nuclear factor kappaB reduces inflammation
and improves functional recovery after spinal cord injury. J Exp
Med. 202:145–156. 2005.PubMed/NCBI View Article : Google Scholar
|
|
61
|
Qin M, Wang W, Zhou H, Wang X, Wang F and
Wang H: Circular RNA circ_0003645 silencing alleviates inflammation
and apoptosis via the NF-κB pathway in endothelial cells induced by
oxLDL. Gene. 755(144900)2020.PubMed/NCBI View Article : Google Scholar
|
|
62
|
Dang RY, Liu FL and Li Y: Circular RNA
hsa_circ_0010729 regulates vascular endothelial cell proliferation
and apoptosis by targeting the miR-186/HIF-1α axis. Biochem Biophys
Res Commun. 490:104–110. 2017.PubMed/NCBI View Article : Google Scholar
|