Introduction
Periodontal ligaments (PDL) connect the cementum to
the alveolar bone and provide mechanical support to the
periodontium (1). Human periodontal
ligament cells (hPDLCs), largely composed of fibroblasts, are
involved in PDL homeostasis and regeneration (2). The proliferation of hPDLCs may be
required for the ability of the PDL to maintain a normal cell
population and space under physiological conditions (3,4). Cell
proliferation is known to be activated by injury to the PDL, such
as infection or overload force (5,6).
Therefore, the proliferation capacity of hPDLCs may be needed for
the renewal and repair of the PDL.
Proliferation is controlled by cell cycle
progression, which is a complex and stepwise process. Cyclins and
cyclin-dependent kinases (CDKs) are the predominant proteins that
regulate the progression of the cell cycle. Cyclin D1 plays a
notable role in G1 phase progression of the cell cycle
via CDK4 activation (7). Cyclin D2
and cyclin A bind and activate CDK2, appearing in the
G1/S and S phases (8,9).
Cyclin B1, in partnership with CDK1, is associated with the
transition from the G2 phase to mitosis (10).
Porphyromonas gingivalis (Pg) is a
major periodontal pathogen whose lipopolysaccharide (LPS) induces
the production of inflammatory cytokines, such as IL-1β, -6, -8 and
TNF-α (11-14).
Pg-LPS has been identified as an important contributor to
periodontal inflammation, destruction and alveolar bone resorption
(11,15-17).
Numerous studies that investigated the interactions between
Pg-LPS and hPDLCs have demonstrated that Pg-LPS can
promote the immuno-inflammatory response of hPDLCs (16-18).
Cell proliferation and inflammatory cytokine release are the two
major inflammatory responses observed in this setting (11,16,17).
Given that pathological alterations caused by inflammatory insults
can impact the regenerative capacities of hPDLCs, the mechanism of
how LPS affects the proliferation of hPDLCs needs to be
investigated. However, the impact of Pg-LPS on hPDLC
proliferation has not been clearly resolved and remains a debated
subject. Yu et al (19)
reported that LPS-induced inflammation inhibits cell proliferation.
By contrast, studies by Kato et al (11) and Takemura et al (20) have suggested that Pg-LPS
promotes cell proliferation and induces pro-inflammatory cytokines.
At present, to the best of our knowledge, there are few reports on
the role of CDKs and cyclins in Pg-LPS-induced inflammation,
and the effects of Pg-LPS on the cell cycle in the process
of cell proliferation remain unclear. Therefore, the present study
aimed to investigate the effects of Pg-LPS on the
proliferation of hPDLCs and regulation of the cell cycle.
Materials and methods
Culture of hPDLCs
Ethical approval for the present study was granted
by the Institutional Review Board of the University of Hong
Kong/Hospital Authority Hong Kong West Cluster (approval no. IRB
UW13-120; Hong Kong, China). hPDLCs were isolated from the
non-decayed, healthy teeth of three donors (two females aged 13 and
15 years old separately; one male aged 14 years old) who had
undergone premolar extraction for orthodontic treatment at the
Division of Paediatric Dentistry and Orthodontics in The Faculty of
Dentistry, The University of Hong Kong from 10th January 2017 to
31st December 2018. Written consent to use the samples in
scientific research was signed by the parents or legal guardians of
the donors. The PDL tissues of the root were scraped and collected
from the middle third of the premolar root surfaces to avoid
contamination by cells derived from the gingiva and dental germ.
The primary hPDLCs were cultured in modified Eagle's medium-α
(α-MEM) containing 10% foetal bovine serum (HyClone; Cytiva) and 1%
antibiotic solution (100 U/ml penicillin and 100 µg/ml
streptomycin) at 37˚C in a humidified 5% CO2 atmosphere.
After achieving 80% confluence, cells were detached by treatment
with 0.25% trypsin (Thermo Fisher Scientific, Inc.) and
sub-cultured in fresh α-MEM. The hPDLCs were characterised by
immunocytochemical staining for vimentin and cytokeratin. The 3rd
to 5th passages of the hPDLCs were used as the test (treated with
Pg-LPS) and control groups (without Pg-LPS). The
hPDLCs were synchronised in serum-free α-MEM culture medium at 37˚C
for 24 h. After serum starvation, the cells in the test and control
groups were synchronised to the same stages of the cell cycle. This
was based on previous studies demonstrating that serum starvation
can arrest cells at the G0/G1 phase with high
efficiency and no toxic effects (5,21-24).
Cell viability and proliferation
detection using cell counting kit-8 (CCK-8) assay
The hPDLCs were seeded into 96-well plates at
5x103 cells/well. After serum starvation, the cells were
treated with Pg-LPS (InvivoGen) at various concentrations
(0.0001, 0.001, 0.01, 0.1, 1 and 10 µg/ml) in the medium of α-MEM
containing 10% FBS for different durations (0, 6, 12, 18, 24, 36
and 48 h, respectively) at 37˚C in vitro. Afterwards, 10 µl
CCK-8 solution (APExBIO Technology LLC) was added to each well of
the plates. After incubation for 4 h at 37˚C, the plates were
measured for absorbance at 450 nm using a microplate reader
(Molecular Devices, LLC).
Flow cytometry for cell proliferation
and cell cycle analysis
The dose- and time-dependent effects of
Pg-LPS were assessed separately by flow cytometry to monitor
cell proliferation and cell cycle. To test the dose-dependent
effects, hPDLCs were seeded into six-well plates at a density of
1.2x105 cells/well. After serum starvation, the cells
were treated with Pg-LPS at various concentrations (0.0001,
0.001, 0.01, 0.1, 1 and 10 µg/ml) for 24 h at 37˚C in vitro.
To investigate the time-dependent effects, hPDLCs were seeded into
six-well plates at a density of 0.6x105 cells/well.
After serum starvation, the cells were stimulated with the
appropriate concentration (based on the previous result) of
Pg-LPS for different durations (0, 6, 12, 18, 24, 36 and 48
h) at 37˚C in vitro. Cells were harvested by trypsinisation,
washed twice with ice-cold phosphate-buffered saline (PBS) and
fixed in 70% ethanol on ice for 15 min. After the ethanol was
washed out with PBS, the cells were re-suspended in 500 µl DNA
staining solution containing 50 µg/ml propidium iodide (Invitrogen;
Thermo Fisher Scientific, Inc.), 0.05% Triton X-100 and 0.1% mg/ml
RNase A in PBS. The samples were kept in the dark at 37˚C for 40
min. A FACSVerse flow cytometer (BD Biosciences) acquired 10,000
events for each sample, and the percentage of cells in the
G0/G1, S and G2/M phases of the
cell cycle were determined using FlowJo software (version 10.0.7;
Tree Star, Inc.). The proliferation index and S-phase fraction
(SPF) were analysed using flow cytometry. The formulae used to
calculate the above were as follows (25): Proliferation index (%)=[(S +
G2/M)/(G0/G1 + S +
G2/M)] x100%; SPF (%)=[S/(G0/G1 +
S + G2/M)] x100%; G0/G1-phase
fraction
(%)=[(G0/G1)/(G0/G1 + S
+ G2/M)] x100%; G2/M-phase fraction
(%)=[(G2/M)/(G0/G1 + S +
G2/M)] x100%.
Detection of genes via reverse
transcription-quantitative PCR (RT-qPCR)
A RT-qPCR assay was carried out to analyse the mRNA
expression of genes associated with different stages of
proliferation. After hPDLCs were stimulated with 0.01 µg/ml
Pg-LPS for 18 or 24 h, the mRNA expression levels of cyclins
A, B1, D1, D2 and CDK1, 2 and 4 were detected by RT-qPCR. The
sequences of the primers used are listed in Table I. Total RNA was immediately
extracted from cells using TRIzol® reagent (Invitrogen;
Thermo Fisher Scientific, Inc.) and then reverse transcribed to
cDNA using SuperScript III Reverse Transcriptase (Invitrogen;
Thermo Fisher Scientific, Inc.) in accordance with the
manufacturer's instructions. qPCR was then performed using Power
SYBR®-Green PCR Master Mix (Applied Biosystems; Thermo
Fisher Scientific, Inc.). GAPDH was used as the housekeeping gene
for the internal control. The following thermocycling conditions
were used for qPCR: Initial denaturation at 95˚C for 10 min,
followed by 40 cycles of denaturation at 95˚C for 15 sec and
annealing and extension at 60˚C for 1 min. The relative mRNA
expression levels were quantified using the 2-∆∆Cq
method (26) and normalized to the
internal reference gene GAPDH.
 | Table IPrimer sequences used in reverse
transcription-quantitative PCR. |
Table I
Primer sequences used in reverse
transcription-quantitative PCR.
| Gene name | Forward sequence
(5'-3') | Reverse sequence
(5'-3') |
|---|
| Cyclin A |
GCCTTTCATTTAGCACTC |
TGAAGGTCCATGAGACAA |
| Cyclin B1 |
GGAAACATGAGAGCCATCCT |
TTCTGCATGAACCGATCAAT |
| Cyclin D1 |
CAAACAGATCATCCGCAAAC |
GCGTGTGAGGCGGTAGT |
| CDK1 |
TGAAACTGCTCGCACTTG |
ATGGTAGATCCCGGCTTATT |
| CDK2 |
CAGAAACAAGTTGACGGGAGA |
GACATCCAGCAGCTTGACAATA |
| CDK4 |
ACAGCTACCAGATGGCACTTACA |
CAAAGATACAGCCAACACTCCAC |
| Cyclin D2 |
GTGTGATGCCATATCAAGTCC |
TCGCATACACTGATCATGC |
| GAPDH |
TCCCTGAGCTGAACGGGAAG |
GGAGGAGTGGGTGTCGCTGT |
Detection of proteins by western
blotting (WB)
After the hPDLCs were incubated with 0.01 µg/ml
Pg-LPS for 24 h, the cells were washed with cold PBS and
lysed using a RIPA buffer (50 mM Tris; pH, 8.0; 150 mM NaCl; 5 mM
EDTA, pH 8.0; 0.8% Triton X-100) containing 1% protease inhibitor.
Protein concentrations were determined using the bicinchoninic acid
protein assay. Equal quantities of protein extracts (30 µg/lane)
were separated via 10% SDS-PAGE. After transferring the proteins to
PVDF membranes. The membranes were blocked for 1 h at room
temperature with 5% non-fat milk in TBST (10 mM Tris, 100 mM NaCl
and 0.1% Tween-20), after which they were probed overnight at 4˚C
with primary antibodies against cyclin A (1:500; cat. no. sc596;
Santa Cruz Biotechnology, Inc.), cyclin B1 (1:500; cat. no. sc245;
Santa Cruz Biotechnology, Inc.), cyclin D1 (1:1,000; cat. no.
ab134175; Abcam) and β-actin (1:1,000; cat. no. 8457; Cell
Signaling Technology, Inc.). The membranes were washed with TBST
and incubated with HRP-conjugated anti-IgG secondary antibodies
(anti-rabbit IgG-HRP-linked antibody, 1:3,000; cat. no. cst7074;
Cell Signaling Technology, Inc.; anti-mouse IgG-HRP-linked
antibody, 1:3,000; cat. no. sc2055; Santa Cruz Biotechnology, Inc.)
at room temperature for 2 h. After washing, the blots were
developed with enhanced chemiluminescence
(WesternBright® ECL HRP substrate; Advansta, Inc.) and
exposed to X-ray film (Kodak). The densities of western blotting
bands were measured by Quantity One 4.6.9 software (Bio-Rad
Laboratories, Inc.).
Statistical analysis
Statistical analysis was carried out using SPSS 19.0
software (IBM Corp.). A one-way analysis of variance (ANOVA) and
Tukey's post hoc test were conducted for the data of flow cytometry
and WB. A two-way ANOVA followed by Bonferroni test was performed
for the CCK-8 and RT-qPCR data. All data were expressed as the mean
± standard deviation from four independent experiments. P<0.05
was considered to indicate a statistically significant
difference.
Results
Cell viability and proliferation
detection by CCK-8 assay
As presented in Fig.
1A, the hPDLCs were stimulated with Pg-LPS at different
concentrations (0.0001, 0.001, 0.01, 0.1, 1 and 10 µg/ml) for
various durations (6, 12, 18, 24, 36 and 48 h) in vitro. The
results showed that Pg-LPS could not inhibit the cell
viability of hPDLCs. The proliferation of hPDLCs was significantly
enhanced in the culture medium at concentrations of 1 and 10 µg/ml
Pg-LPS for 6 (P<0.05) and 18 h (P<0.01) compared with
the cells cultured without LPS. Pg-LPS at the concentrations
of 0.001, 0.01, 0.1, 1 and 10 µg/ml for 24, 36 and 48 h
significantly promoted the proliferation of hPDLCs compared with
controls (P<0.01). The proliferation of hPDLCs was also
significantly increased with Pg-LPS at the concentration of
0.0001 µg/ml for 48 h compared with the cells cultured without LPS
(P<0.01). These results suggested that Pg-LPS could not
inhibit the cell viability of hPDLCs. Instead, Pg-LPS can
increase hPDLC proliferation.
 | Figure 1Effects of Pg-LPS on the
proliferation and cell cycle of hPDLCs. (A) Cell viability and
proliferation detected using Cell Counting Kit-8 assay.
Proliferation of hPDLCs was significantly enhanced in culture
medium at the concentrations of 1 and 10 µg/ml Pg-LPS
compared with the cells cultured without LPS at 6 and 18 h.
Pg-LPS at the concentrations of 0.001, 0.01, 0.1, 1 and 10
µg/ml for 24, 36 and 48 h significantly promoted the proliferation
of hPDLCs compared with controls. The proliferation of hPDLCs was
also significantly increased with Pg-LPS at the
concentration of 0.0001 µg/ml for 48 h compared with the cells
cultured without LPS. (B) Cell proliferation and cell cycle
analysis using flow cytometry. After treatment with different
concentrations of Pg-LPS (0.0001, 0.001, 0.01, 0.1, 1 and 10
µg/ml) for 24 h, the proliferation index of hPDLCs was
significantly enhanced at concentrations ranging from 0.0001-10
µg/ml, but the SPF only increased significantly at 0.01 µg/ml
compared with that of cells without LPS. *P<0.05,
**P<0.01 vs. control group (0 µg/ml Pg-LPS) at
each time point. Pg, Porphyromonas gingivalis; LPS,
lipopolysaccharide; hPDLCs, human periodontal ligament cells; SPF,
S-phase fraction. |
Cell proliferation and cell cycle
analysis by flow cytometry
After serum starvation for 24 h, the majority of
cells were at the G0/G1 stage and few were in
the S or G2/M phases of the cell cycle (Fig. S1). To assess the dose-dependent
effect of Pg-LPS, hPDLCs were cultured with different
concentrations of LPS (0.0001, 0.001, 0.01, 0.1, 1 and 10 µg/ml);
the proliferation index of these treated cells was significantly
enhanced compared with cells cultured without LPS (P<0.01),
while there were no significant differences among the groups
stimulated with Pg-LPS at all concentrations tested
(Fig. 1B). However, the SPF was
only significantly increased with Pg-LPS at a concentration
of 0.01 µg/ml (P<0.05), compared with cells cultured without LPS
(Figs. 1B and S2). As presented in Figs. 2 and S3, when hPDLCs were incubated with 0.01
µg/ml Pg-LPS for 24 h, the G2/M-phase fraction of
the hPDLCs was higher compared with that in the control group
(P<0.01), whereas the G0/G1-phase fraction
was lower (P<0.01). Figs. 3 and
S4 present the data for
time-dependent effects. Both the proliferation index and SPF of the
hPDLCs increased, reached a peak at 24 h and then decreased from 24
to 48 h in both the Pg-LPS-stimulated and control groups.
The proliferation index and SPF of hPDLCs were significantly
increased after treatment with Pg-LPS at 24 h compared with
the cells cultured without LPS (P<0.01 and P<0.05,
respectively), while they were significantly decreased after
treatment with Pg-LPS for 36 h compared with the cells
cultured without LPS (P<0.01 and P<0.05, respectively). There
were no significant differences between Pg-LPS-stimulated
and control groups at 0, 6, 12, 18 and 48 h. These results suggest
that changes in PI and SPF were not dose-dependent, but were
time-dependent.
 | Figure 2Effects of 0.01 µg/ml Pg-LPS
on the proliferation index, the G0/G1-phase
fraction, the G2/M-phase fraction and SPF of hPDLCs.
When hPDLCs were incubated with 0.01 µg/ml Pg-LPS for 24 h,
the proliferation index, SPF and G2/M-phase fraction of
hPDLCs were higher compared with the control group, whereas the
G0/G1-phase fraction was lower.
*P<0.05, **P<0.01. Pg, Porphyromonas
gingivalis; LPS, lipopolysaccharide; hPDLCs, human periodontal
ligament cells; SPF, S-phase fraction. |
mRNA expression levels of cyclins and
CDKs in hPDLCs treated with Pg-LPS
The mRNA expression levels of cyclin A and cyclin D1
were significantly increased in the LPS-treated hPDLCs at 18 and 24
h compared with their control groups at the same time point
(P<0.01), and the levels of cyclin B1 were also increased at 24
h (P<0.01; Fig. 4). There were
no significant differences in CDK1, CDK2 and CDK4 mRNA expression
levels at 18 and 24 h between the LPS-treated and control groups at
the same time points (Fig. 4). By
tracking the trends of change, the levels of cyclin A, cyclin B1,
CDK1 and cyclin D2 in the LPS-treated group were revealed to have
risen sharply at 24 h compared with those at 18 h, whereas that of
cyclin D1 decreased (P<0.01; Fig.
4). The expression levels of cyclin A, cyclin B1, CDK1 and CDK2
in the control group were higher at 24 h compared with those at 18
h (P<0.05; Fig. 4). These
findings suggest that Pg-LPS can mainly promote the mRNA
expression levels of cyclins D1, A and B1.
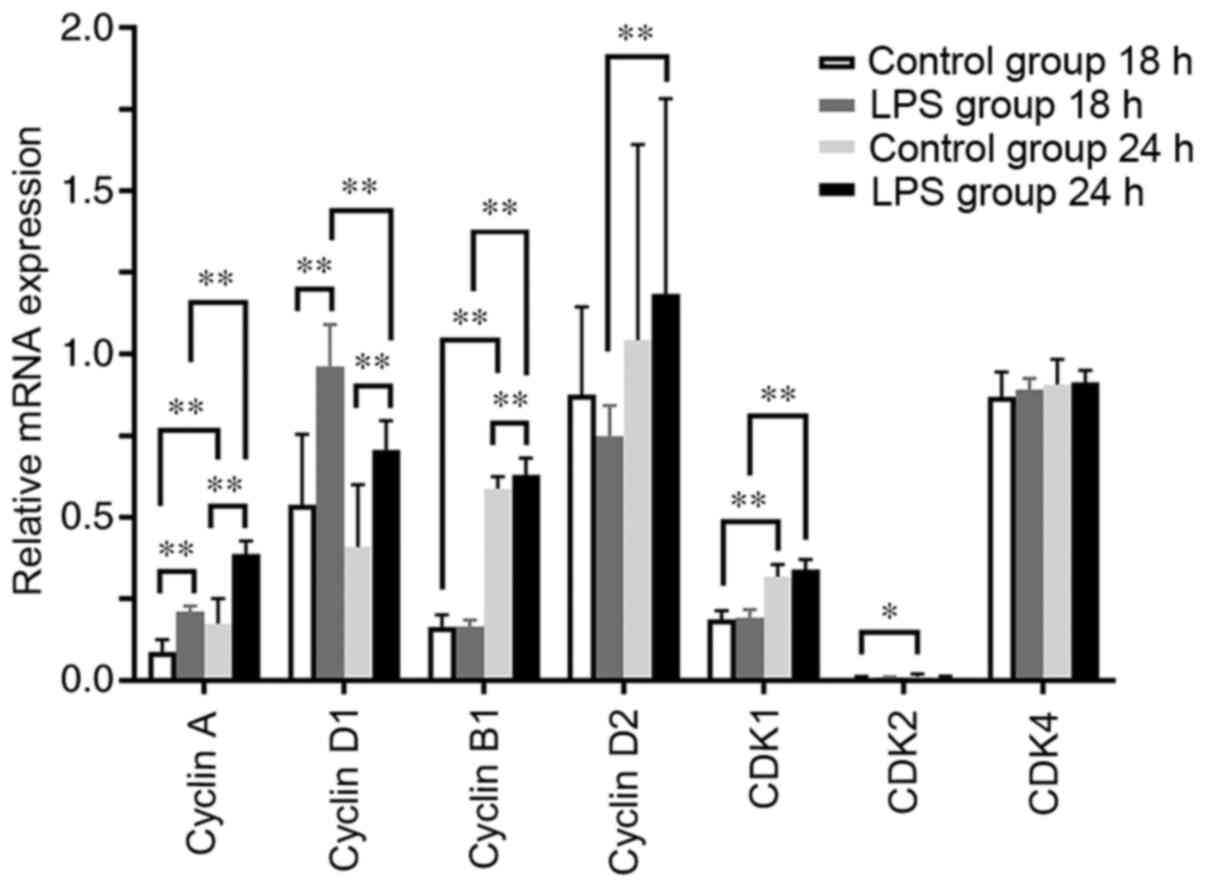 | Figure 4mRNA expression levels of cyclins A,
B1, D1 and D2, and CDK1, 2 and 4 in hPDLCs treated with or without
0.01 µg/ml Pg-LPS at 18 and 24 h. mRNA levels of cyclin A
and cyclin D1 were significantly increased in the LPS-treated group
at 18 h compared with those in the control group. mRNA levels of
cyclins A, D1 and B1 were significantly increased in the
LPS-treated group at 24 h compared with those in the control group.
mRNA levels of cyclin A, cyclin B1, CDK1 and CDK2 in the control
group were higher at 24 h compared with at 18 h. mRNA levels of
cyclin A, cyclin B1, cyclin D2 and CDK1 in the LPS-treated group
rose sharply, whereas that of cyclin D1 fell from 18 h to 24 h.
*P<0.05, **P<0.01. CDK,
cyclin-dependent kinase; hPDLCs, human periodontal ligament cells;
Pg, Porphyromonas gingivalis; LPS, lipopolysaccharide. |
Cyclin protein expression in hPDLCs
treated with Pg-LPS
hPDLCs were incubated in the presence or absence of
0.01 µg/ml Pg-LPS. After 24 h of treatment, the protein
expression levels of cyclins A, B1 and D1 were analysed using
western blotting. Total cell lysates were normalised to β-actin.
The levels of cyclin B1 and cyclin D1 in the LPS-treated groups
significantly increased after 24 h compared with control groups at
the same time point (P<0.05 and P<0.01, respectively;
Fig. 5). These results were in
concordance with the mRNA expression levels. There were no
significant differences in the expression levels of cyclin A
between the control and LPS group. These results revealed that
Pg-LPS can upregulate the protein expression levels of
cyclins D1and B1.
Discussion
The results of the present study indicated that high
concentrations of Pg-LPS (1 and 10 µg/ml) stimulated the
proliferation of hPDLCs at early stages (6 and 18 h), and that low
concentrations of Pg-LPS (0.001, 0.01, 0.1, 1 and 10 µg/ml)
stimulated the proliferation of hPDLCs at later stages (24, 36 and
48 h). Moreover, Pg-LPS at a concentration of 0.01 µg/ml
prominently increased the proliferation of hPDLCs by affecting the
G1, S and G2/M phases. A number of studies
have reported different results concerning the effects of LPS on
the proliferation of hPDLCs. Jönsson et al (27) demonstrated that PDL cell
proliferation is unaffected by stimulation for 72 h with a high
concentration (10 µg/ml) of Escherichia coli LPS. Huang
et al (28) reported that
stimulation with Pg-LPS (0.01, 0.1 µg/ml) for either 3 or 5
days has no effect on PDL cell proliferation. Jung et al
(29) also demonstrated similar
results when treating cells with Pg-LPS at different
concentrations (0.1, 0.5, 1, 5, 10 and 20 µg/ml) for 7 days. By
contrast, other studies have indicated that treatment with
Pg-LPS at lower concentrations such as 0.01 and 0.1 µg/ml
stimulates the proliferation of hPDLCs on day 3(30), whereas high concentrations of LPS,
such as 1(28), 10 and 100 µg/ml
(30), have been demonstrated
inhibit the proliferation of hPDLCs.
These aforementioned findings highlight differences
in both the dose- and time-dependent effects of Pg-LPS on
the proliferation of hPDLCs. These contradictory results may be due
to several reasons. First, hPDLCs in culture consist of several
subpopulations (31). Second, there
is a large amount of inter-individual heterogeneity in the
responses of hPDLCs to Pg-LPS (32). Third, the properties of hPDLCs may
change with different passages (33). Fourth, the proliferation capability
of hPDLCs is a function of the patient's age and health status
(23). Fifth, these results depend
on certain characteristics of hPDLCs, the chemical structures of
LPS and a number of culture environmental factors, including
primary cell separation methods, culture medium composition, oxygen
concentration, culture time and other physical or chemical
stimulation (34). Finally, in the
majority of the aforementioned studies, cell proliferation was
detected using an MTT assay (28-30).
However, using quantitative measurements, such as the CCK-8 assay,
which is reported to be more stable and sensitive than MTT,
improves the quality of results (35,36).
In the present study, the 3rd to 5th passages of
hPDLCs from young patients with good oral health were selected to
eliminate the influence of ageing or diseases on proliferative
ability (5,37). Pg-LPS was used instead of
E. coli LPS as Pg is a major periodontal pathogen
that is capable of promoting cell proliferation and inflammatory
cytokine production (5,18). The present study detected both dose-
and time-dependent effects using CCK-8 assay and flow cytometry to
monitor cell proliferation and cell cycle.
As the cell cycle progresses, the preparatory
G0/G1 phase ensures that everything is ready
for DNA synthesis, after which DNA replication occurs during the S
phase (38). The G2/M
phase is the nuclear fission stage, which focuses on orderly
division of the cell into two daughter cells (39). Proliferation index and SPF are two
factors of flow cytometry indicative of cell proliferation
activity. They provide an approximation of the growth fraction and
usually represent the velocity of cell division (40). The present study revealed that the
proliferation index of hPDLCs increased significantly after
treatment with Pg-LPS (0.001, 0.01, 0.1, 1 and 10 µg/ml) for
24 h which was consistent with the results of CCK-8 assay. However,
there were no significant differences in the proliferation indices
of hPDLCs among the groups stimulated with Pg-LPS at
different concentrations (0.0001, 0.001, 0.01, 0.1, 1 and 10
µg/ml). The SPF of hPDLCs only significantly increased when treated
with Pg-LPS at a concentration of 0.01 µg/ml, whilst there
were no significant differences after treatment with Pg-LPS
at the other concentrations (0.0001, 0.001, 0.1, 1 and 10 µg/ml)
compared with cells cultured without LPS. These results indicated
that changes in the proliferation index and SPF were not
dose-dependent. After the hPDLCs were stimulated with 0.01 µg/ml
Pg-LPS for 24 h, both the SPF and G2/M-phase
fraction were increased compared with those in the control group,
whereas the G0/G1-phase fraction was
significantly decreased. This suggested that Pg-LPS enhanced
the proliferation of hPDLCs by affecting both the S and
G2/M phases, while simultaneously reducing the
proportion of cells in the G0/G1 phase.
The cell cycle is a complex process that is needed
for the proliferation of cells. CDKs and cyclins are central to
this process. Extensive work on gene knockout mouse models of cell
cycle regulators has revealed compensatory mechanisms that regulate
the interactions among cyclins and CDKs (41,42).
In addition, CDK2 has been revealed to be dispensable in the
regulation of the mitotic cell cycle, as both CDK4 and CDK1 can
cover for its functions (43). In
the present study, the results of western blotting demonstrated
that Pg-LPS treatment increased the expression levels of
cyclin D1 and cyclin B1, but not of cyclin A. This implied that the
CDK4/cyclin D1 and CDK1/cyclin B1 complexes, as potential master
regulators, may compensate for the function of the CDK2/cyclin A
complex in cell cycle control upon exposure to Pg-LPS. In
the current study, the mRNA expression levels were consistent with
those of protein expression for cyclin D1 and cyclin B1, but the
cyclin A mRNA and protein expression levels were not concomitant.
The inconsistency between cyclin A mRNA and protein expression
levels may arise from several factors. First, the relationship
between mRNA and protein expression levels is not strictly linear,
as different regulatory mechanisms, such as gene transcription,
post-transcriptional regulation, translation and protein
modification, act on the synthesized mRNA and protein (44). Second, transcription and translation
can be regulated in a spatiotemporal manner, and protein expression
in known to lag behind mRNA transcription (45). Therefore, protein expression levels
may not concur with mRNA transcription levels at a given time
point. Third, the detection sensitivities are different for RT-qPCR
and western blotting, with RT-qPCR having increased sensitivity.
Overall, the results of RT-qPCR and western blotting analyses
confirmed that Pg-LPS was required for proliferation and
acted by upregulating cyclin D1 and cyclin B1, which were involved
in the G1 and G2/M phases, respectively.
Upon further investigating the time dependency of
cell proliferation after exposure to Pg-LPS, the present
study revealed that Pg-LPS significantly augmented the
proliferation index and SPF of hPDLCs at the 24-h time point and
significantly reduced both at the 36-h time point but not at the
other time points (6, 12, 18 or 48 h). The proliferation of hPDLCs,
as indicated by SPF and proliferation index, indicated the same
trends in change for both the test and control groups, suggesting
that Pg-LPS did not change the overall time-dependency. Both
the proliferation index and SPF of hPDLCs gradually increased,
peaked at 24 h and then decreased from 24 to 48 h in both the
Pg-LPS-stimulated and control groups. Maintaining the proper
incubation period is therefore critical for in vitro studies
involving hPDLCs. The present study demonstrated that when control
hPDLCs (without Pg-LPS) were incubated for durations of 18
to 24 h, cyclin A, cyclin B1, CDK1 and CDK2 significantly
increased. When hPDLCs in the test group (treated with 0.01 µg/ml
Pg-LPS) were incubated for the same durations, cyclin D2
levels also rose significantly and cyclin D1 levels fell. These
observations may help explain why Pg-LPS significantly
augmented the proliferation index and SPF of hPDLCs at the 24 h
time point but not at the 18 h time point, and why, when
progressing from 18 to 24 h, cyclin D2 increased to accelerate the
G1/S transition and cyclin D1 decreased to slowed down
G1 progression. Changes that occur over longer durations
will require further investigations in the future.
Currently, the intrinsic mechanisms by which
Pg-LPS affects cell proliferation and regulates the cell
cycle are unknown. Numerous studies have suggested that
Pg-LPS produces pro-inflammatory cytokines mainly through
the Toll-like receptor 4 pathway, which influences cell
proliferation (5,19). However, the effect of Pg-LPS
on the cell cycle and the concentration of Pg-LPS that
affects hPDLCs proliferation are still matters of debate
(18,46-48). The present study demonstrated that Pg-LPS
promoted the proliferation of hPDLCs and that Pg-LPS
prominently affected the G1, S and G2/M
phases at a concentration of 0.01 µg/ml. Pg-LPS was also
demonstrated to likely affect the cell cycle through the actions of
cyclin D1 and cyclin B1. However, the exact role of cyclin A in the
cell cycle control is not yet clear as the result of
inconsistencies between the RT-qPCR and WB data in the present
study.
In summary, Pg-LPS may significantly
stimulate the proliferation of hPDLCs through the upregulation of
cyclins D1, A and B1. The mechanism of Pg-LPS on the
proliferation and cell cycle regulation of hPDLCs remains unclear,
and further studies are required. The results from the current
study only address the short-term effects of Pg-LPS on the
proliferation of hPDLCs. However, periodontal disease is a chronic
inflammatory condition that can occur due to multiple and complex
factors. Hence, more investigations conducted over longer durations
are warranted to further understand periodontal pathogenesis and
devise more effective therapeutic strategies.
Supplementary Material
Flow cytometry dot plot and the
corresponding histogram of hPDLCs after serum starvation for 24 h.
(A) Dot plot of FSC vs. SSC gated hPDLCs after serum starvation for
24 h. (B) Histogram analysis of hPDLCs after serum starvation for
24 h. hPDLC, human periodontal ligament cells; FSC, forward
scatter; SSC, side scatter; Freq., frequency.
Effects of Pg-LPS on the
proliferation and cell cycle of hPDLCs. (a) Dot plots and (b)
corresponding histograms of hPDLCs cultured for 24 h as follows:
(A) Without Pg-LPS; (B) with 0.0001 μg/ml Pg-LPS; (C)
with 0.001 μg/ml Pg-LPS; (D) with 0.01 μg/ml Pg-LPS;
(E) with 0.1 μg/ml Pg-LPS; (F) with 1 μg/ml Pg-LPS;
and (G) with 10 μg/ml Pg-LPS. Proportion of cells in G0/G1
are represented in histograms by a green peak on the left and cells
in G2/M are represented by a blue peak on the right; cells in
S-phase are represented by the middle yellow area. hPDLCs, human
periodontal ligament cells; Pg, Porphyromonas
gingivalis; LPS, lipopolysaccharide; FSC, forward scatter; SSC,
side scatter.
Dot plots and the corresponding
histograms of hPDLCs cultured without or with Pg-LPS at 0.01
μg/ml for 24 h. (A) Dot plot and histogram of hPDLCs cultured with
Pg-LPS at 0.01 μg/ml. (B) Dot plot and histogram of hPDLCs
cultured without Pg-LPS. hPDLCs, human periodontal ligament
cells; Pg, Porphyromonas gingivalis; LPS,
lipopolysaccharide; Freq., frequency; FSC, forward scatter; SSC,
side scatter.
Dot plots and the corresponding
histograms of hPDLCs cultured without or with Pg-LPS at 0.01
μg/ml for different durations. (a) Dot plot and (b) histogram of
hPDLCs cultured as follows: (A) With Pg-LPS for 0 h; (B)
with Pg-LPS for 6 h; (C) with Pg-LPS for 12 h; (D)
with Pg-LPS for 18 h; (E) with Pg-LPS for 24 h; (F)
with Pg-LPS for 36 h; (G) with Pg-LPS for 48 h; (H)
without Pg-LPS for 0 h; (I) without Pg-LPS for 6 h;
(J) without Pg-LPS for 12 h; (K) without Pg-LPS for
18 h; (L) without Pg-LPS for 24 h; (M) without Pg-LPS
for 36 h; and (N) without Pg-LPS for 48 h. hPDLCs, human
periodontal ligament cells; Pg, Porphyromonas
gingivalis; LPS, lipopolysaccharide; FSC, forward scatter; SSC,
side scatter.
Acknowledgements
The authors would like to acknowledge Mr Tong Wai
Man (flow cytometry and reverse transcription-quantitative PCR
technical support), Ms Yu Ching Lam (cell culture technical
support) and Ms Tong Hoi Yee (western blotting technical support),
all from Centralized Research Laboratory, Faculty of Dentistry, The
University of Hong Kong (Hong Kong, China).
Funding
The project was supported by the General Research Fund from the
Research Grants Council of Hong Kong (grant no. 17106619).
Availability of data and materials
The datasets used and/or analysed during the
current study are available from the corresponding author on
reasonable request.
Authors' contributions
YY, CZ and LJ made substantial contributions to the
overall structural design of the study, methodology and data
analysis. JL, YH and ZT performed the experiments. JL detected the
cell proliferation and cell cycle by flow cytometry and analysed
mRNA expression levels of the cyclins and CDKs by RT-qPCR. YH
performed the western blotting of the cyclins. ZT and MG were major
contributors in the CCK-8 assay for cell proliferation. JL and YH
analyzed the data, drafted and revised the manuscript under the
guidance of CZ, LJ and YY. JL and YH confirmed the authenticity of
all the raw data. All authors have read and approved the final
manuscript.
Ethics approval and consent to
participate
Ethical approval for the present study was granted
by the Institutional Review Board of the University of Hong
Kong/Hospital Authority Hong Kong West Cluster (approval no. IRB
UW13-120; Hong Kong, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Garant PR: Collagen resorption by
fibroblasts. A theory of fibroblastic maintenance of the
periodontal ligament. J Periodontol. 47:380–390. 1976.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Hughes FJ, Ghuman M and Talal A:
Periodontal regeneration: A challenge for the tissue engineer? Proc
Inst Mech Eng H. 224:1345–1358. 2010.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Lekic P and McCulloch C: Periodontal
ligament cell populations: The central role of fibroblasts in
creating a unique tissue. Anat Rec. 245:327–341. 1996.PubMed/NCBI View Article : Google Scholar
|
|
4
|
McCulloch CA and Melcher AH: Continuous
labelling of the periodontal ligament of mice. J Periodontal Res.
18:231–241. 1983.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Liu J, Tang X, Li C, Pan C, Li Q, Geng F
and Pan Y: Porphyromonas gingivalis promotes the cell cycle
and inflammatory cytokine production in periodontal ligament
fibroblasts. Arch Oral Biol. 60:1153–1161. 2015.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Mabuchi R, Matsuzaka K and Shimono M: Cell
proliferation and cell death in periodontal ligaments during
orthodontic tooth movement. J Periodontal Res. 37:118–124.
2002.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Sherr CJ: G1 phase progression: Cycling on
cue. Cell. 79:551–555. 1994.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Girard F, Strausfeld U, Fernandez A and
Lamb NJ: Cyclin A is required for the onset of DNA replication in
mammalian fibroblasts. Cell. 67:1169–1179. 1991.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Sweeney KJ, Sarcevic B, Sutherland RL and
Musgrove EA: Cyclin D2 activates Cdk2 in preference to Cdk4 in
human breast epithelial cells. Oncogene. 14:1329–1340.
1997.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Takizawa CG and Morgan DO: Control of
mitosis by changes in the subcellular location of cyclin-B1-Cdk1
and Cdc25C. Curr Opin Cell Biol. 12:658–665. 2000.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Kato H, Taguchi Y, Tominaga K, Umeda M and
Tanaka A: Porphyromonas gingivalis LPS inhibits osteoblastic
differentiation and promotes pro-inflammatory cytokine production
in human periodontal ligament stem cells. Arch Oral Biol.
59:167–175. 2014.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Morandini AC, Sipert CR, Gasparoto TH,
Greghi SL, Passanezi E, Rezende ML, Sant'ana AP, Campanelli AP,
Garlet GP and Santos CF: Differential production of macrophage
inflammatory protein-1alpha, stromal-derived factor-1, and IL-6 by
human cultured periodontal ligament and gingival fibroblasts
challenged with lipopolysaccharide from P. gingivalis. J
Periodontol. 81:310–317. 2010.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Pathirana RD, O'Brien-Simpson NM and
Reynolds EC: Host immune responses to Porphyromonas
gingivalis antigens. Periodontol 2000. 52:218–237.
2010.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Wada N, Maeda H, Yoshimine Y and Akamine
A: Lipopolysaccharide stimulates expression of osteoprotegerin and
receptor activator of NF-kappa B ligand in periodontal ligament
fibroblasts through the induction of interleukin-1 beta and tumor
necrosis factor-alpha. Bone. 35:629–635. 2004.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Krajewski AC, Biessei J, Kunze M, Maersch
S, Perabo L and Noack MJ: Influence of lipopolysaccharide and
interleukin-6 on RANKL and OPG expression and release in human
periodontal ligament cells. APMIS. 117:746–754. 2009.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Park YD, Kim YS, Jung YM, Lee SI, Lee YM,
Bang JB and Kim EC: Porphyromonas gingivalis
lipopolysaccharide regulates interleukin (IL)-17 and IL-23
expression via SIRT1 modulation in human periodontal ligament
cells. Cytokine. 60:284–293. 2012.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Seo T, Cha S, Kim TI, Lee JS and Woo KM:
Porphyromonas gingivalis-derived lipopolysaccharide-mediated
activation of MAPK signaling regulates inflammatory response and
differentiation in human periodontal ligament fibroblasts. J
Microbiol. 50:311–319. 2012.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Yamaji Y, Kubota T, Sasaguri K, Sato S,
Suzuki Y, Kumada H and Umemoto T: Inflammatory cytokine gene
expression in human periodontal ligament fibroblasts stimulated
with bacterial lipopolysaccharides. Infect Immun. 63:3576–3581.
1995.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Yu B, Li Q and Zhou M: LPS-induced
upregulation of the TLR4 signaling pathway inhibits osteogenic
differentiation of human periodontal ligament stem cells under
inflammatory conditions. Int J Mol Med. 43:2341–2351.
2019.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Takemura A, Matsuda N, Kimura S, Fujiwara
T, Nakagawa I and Hamada S: Porphyromonas gingivalis
lipopolysaccharide modulates the responsiveness of human
periodontal ligament fibroblasts to platelet-derived growth factor.
J Periodontal Res. 33:400–407. 1998.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Tong J, Sun D, Yang C, Wang Y, Sun S, Li
Q, Bao J and Liu Y: Serum starvation and thymidine double blocking
achieved efficient cell cycle synchronization and altered the
expression of p27, p53, bcl-2 in canine breast cancer cells. Res
Vet Sci. 105:10–14. 2016.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Khammanit R, Chantakru S, Kitiyanant Y and
Saikhun J: Effect of serum starvation and chemical inhibitors on
cell cycle synchronization of canine dermal fibroblasts.
Theriogenology. 70:27–34. 2008.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Nishimura F, Terranova VP, Braithwaite M,
Orman R, Ohyama H, Mineshiba J, Chou HH, Takashiba S and Murayama
Y: Comparison of in vitro proliferative capacity of human
periodontal ligament cells in juvenile and aged donors. Oral Dis.
3:162–166. 1997.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Liu F, Wu BL, Gao J, Huang X, Ma DD and
Chen T: Effects of serum starvation on cell cycle synchronization
in human dental pulp cells. Chin J Conserv Dent. 21:67–71.
2011.
|
|
25
|
Mikami K, Haseba T and Ohno Y: Ethanol
induces transient arrest of cell division (G2 + M block) followed
by G0/G1 block: Dose effects of short- and longer-term ethanol
exposure on cell cycle and cell functions. Alcohol Alcohol.
32:145–152. 1997.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Jönsson D, Nebel D, Bratthall G and
Nilsson BO: LPS-induced MCP-1 and IL-6 production is not reversed
by oestrogen in human periodontal ligament cells. Arch Oral Biol.
53:896–902. 2008.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Huang M, Yan F, Yao L, Li D, Zheng Y, Li Y
and Lin M: Effects of cyclosporine A and Porphyromonas
gingivalis-lipopolysaccharide on proliferation of human
periodontal ligament fibroblasts in vitro. Chin J Stomatol Res.
5:470–476. 2011.
|
|
29
|
Jung IH, Lee DE, Yun JH, Cho AR, Kim CS,
You YJ, Kim SJ and Choi SH: Anti-inflammatory effect of
(-)-epigallocatechin-3-gallate on Porphyromonas gingivalis
lipopolysaccharide-stimulated fibroblasts and stem cells derived
from human periodontal ligament. J Periodontal Implant Sci.
42:185–195. 2012.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Zhang F, Wu Z, Wan L and Yuan N: Effects
of lipopolysaccharides on proliferation and alkaline phosphatase
activity of periodontal ligament cells. Chin J Conserv Dent.
13:27–29. 2003.
|
|
31
|
Jönsson D, Nebel D, Bratthall G and
Nilsson BO: The human periodontal ligament cell: A fibroblast-like
cell acting as an immune cell. J Periodontal Res. 46:153–157.
2011.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Scheres N, Laine ML, de Vries TJ, Everts V
and van Winkelhoff AJ: Gingival and periodontal ligament
fibroblasts differ in their inflammatory response to viable
Porphyromonas gingivalis. J Periodontal Res. 45:262–270.
2010.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Itaya T, Kagami H, Okada K, Yamawaki A,
Narita Y, Inoue M, Sumita Y and Ueda M: Characteristic changes of
periodontal ligament-derived cells during passage. J Periodontal
Res. 44:425–433. 2009.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Jia L, Wen Y and Xu X: Effects of culture
conditions in vitro on the biological characteristics of
periodontal ligament stem cells. Int J Stomatol. 45:255–260.
2018.
|
|
35
|
Tominaga H, Ishiyama M, Ohseto F, Sasamoto
K, Hamamoto T, Suzuki K and Watanabe M: A water-soluble tetrazolium
salt useful for colorimetric cell viability assay. Anal Commun.
36:47–50. 1999.
|
|
36
|
Failli A, Legitimo A, Orsini G, Castagna
M, Spisni R, Miccoli P and Consolini R: Antiproliferative effects
of 5-fluorouracil and oxaliplatin in colon cancer cell lines:
Comparison of three different cytotoxicity assays. J Biol Regul
Homeost Agents. 27:275–284. 2013.PubMed/NCBI
|
|
37
|
Shiba H, Nakanishi K, Sakata M, Fujita T,
Uchida Y and Kurihara H: Effects of ageing on proliferative
ability, and the expressions of secreted protein, acidic and rich
in cysteine (SPARC) and osteoprotegerin (osteoclastogenesis
inhibitory factor) in cultures of human periodontal ligament cells.
Mech Ageing Dev. 117:69–77. 2000.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Masai H, Matsumoto S, You Z,
Yoshizawa-Sugata N and Oda M: Eukaryotic chromosome DNA
replication: Where, when, and how? Annu Rev Biochem. 79:89–130.
2010.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Schafer KA: The cell cycle: A review. Vet
Pathol. 35:461–478. 1998.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Gray JW, Dolbeare F, Pallavicini MG,
Beisker W and Waldman F: Cell cycle analysis using flow cytometry.
Int J Radiat Biol Relat Stud Phys Chem Med. 49:237–255.
1986.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Malumbres M, Sotillo R, Santamaria D,
Galan J, Cerezo A, Ortega S, Dubus P and Barbacid M: Mammalian
cells cycle without the D-type cyclin-dependent kinases Cdk4 and
Cdk6. Cell. 118:493–504. 2004.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Kozar K, Ciemerych MA, Rebel VI,
Shigematsu H, Zagozdzon A, Sicinska E, Geng Y, Yu Q, Bhattacharya
S, Bronson RT, et al: Mouse development and cell proliferation in
the absence of D-cyclins. Cell. 118:477–491. 2004.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Satyanarayana A and Kaldis P: Mammalian
cell-cycle regulation: Several Cdks, numerous cyclins and diverse
compensatory mechanisms. Oncogene. 28:2925–2939. 2009.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Mehra A, Lee KH and Hatzimanikatis V:
Insights into the relation between mRNA and protein expression
patterns: I. Theoretical considerations. Biotechnol Bioeng.
84:822–833. 2003.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Gedeon T and Bokes P: Delayed protein
synthesis reduces the correlation between mRNA and protein
fluctuations. Biophys J. 103:377–385. 2012.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Du A, Zhao S, Wan L, Liu T, Peng Z, Zhou
Z, Liao Z and Fang H: MicroRNA expression profile of human
periodontal ligament cells under the influence of Porphyromonas
gingivalis LPS. J Cell Mol Med. 20:1329–1338. 2016.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Han Y, Wang F, Shao L, Huang P and Xu Y:
LncRNA TUG1 mediates lipopolysaccharide-induced proliferative
inhibition and apoptosis of human periodontal ligament cells by
sponging miR-132. Acta Biochim Biophys Sin (Shanghai).
51:1208–1215. 2019.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Francis M, Pandya M, Gopinathan G, Lyu H,
Ma W, Foyle D, Nares S and Luan X: Histone methylation mechanisms
modulate the inflammatory response of periodontal ligament
progenitors. Stem Cells Dev. 28:1015–1025. 2019.PubMed/NCBI View Article : Google Scholar
|



















