Introduction
Cardiovascular disease, in particular coronary heart
disease (CHD), has become the leading cause of death worldwide
(1,2). Emerging evidence has demonstrated that
dysfunction of human vascular smooth muscle cells (hVSMCs) serves
an important role in the progression of angiocardiopathy, including
CHD (3,4). Numerous studies have indicated that
abnormal regulation of VSMCs is crucial in the progression of CHD
(3-5).
Furthermore, a number of reports have indicated that microRNAs
(miRNAs/miRs) are indispensable regulators of VSMC function
(6,7).
miRNAs, a family of small non-coding RNAs (~22
nucleotides), can regulate gene expression by binding to the
3'-untranslated region (3'-UTR) of target mRNAs (8). miRNA functions have been reported in
various biological behaviors, including cell proliferation,
invasion and apoptosis (9).
Previous reports have demonstrated that miRNAs are important
regulators in the intricate biological progression of various
cardiovascular diseases, including cardiac hypertrophy, heart
failure, myocardial ischemia and reperfusion (10-14).
However, to the best of our knowledge, little is known concerning
the function of miR-208a-3p in the development of cardiovascular
diseases in VSMCs. miR-208a-3p, the main sequence of miR-208a, has
been reported to be associated with cardiovascular disease
(15). Montgomery et al
(16) have reported that
downregulation of miR-208a improved cardiac function. In addition,
miR-208 has been reported to be involved in various diseases,
including colorectal cancer, gastric cancer and coronary artery
disease (15,17,18).
However, the molecular mechanism of miR-208a-3p in CHD remains to
be elucidated.
B-cell translocation gene 1 (BTG1) is member of the
BTG anti-proliferative protein family, which may promote cytochrome
c translocation in mitochondria (19). In addition, it is a vital cofactor
regulating certain biological behaviors, including cell
proliferation and apoptosis (19).
However, the potential role of BTG1 in VSMCs remains unclear.
The aim of the present study was to investigate the
role of miR-208a-3p in CHD by examining its functional role in the
regulation of hVSMC proliferation, and further examine the
molecular mechanism. This study may provide a novel diagnostic and
treatment target in CHD therapy.
Materials and methods
Clinical specimen collection
Peripheral blood samples (2 ml/individual) were
extracted from 30 patients with CHD (age range, 43-75 years; 20
males and 10 females) and 30 healthy donors (age range, 46-77
years; 20 males and 10 females) at Shanxi Dayi Hospital between
February 2017 and August 2018. The samples were immediately frozen
and stored at -80˚C until further use. Written informed consent was
provided from all participants approving the use of their samples
in the present study. All study protocols were approved by the
Ethics Committee of Shanxi Bethune Hospital (Taiyuan, China). The
inclusion criteria were as follows: Coronary angiography suggested
that at least one main coronary artery or main branch had obvious
coronary artery stenosis ≥50%, and the conclusion was judged by at
least two experienced cardiologists. The exclusion criteria were as
follows: i) Patients with myocardial bridge, cardiomyopathy,
valvular heart disease or acute myocardial infarction; ii) patients
with hypothyroidism, hyperthyroidism, hypothalamic or pituitary
disease and other endocrine diseases; iii) patients with cancer,
acute cerebrovascular disease, severe infection, liver or kidney
dysfunction and hereditary hyperlipidemia; and iv) patients with
history of mental illness or family history of mental illness.
Cell culture
hVSMCs were purchased from American Type Culture
Collection. Cells were maintained in Dulbecco's modified Eagle's
medium (HyClone; Cytiva) supplemented with 10% fetal bovine serum
(FBS; Gibco; Thermo Fisher Scientific, Inc.), 100 U/ml penicillin
and 100 mg/ml streptomycin at 37˚C with 5% CO2.
Cell transfection and reagents
A total of 100 nM miR-208a-3p inhibitor
(5'-ACAAGCUUUUUGCUCGUCUUAU-3'; Shanghai GenePharma Co., Ltd.), 100
nM inhibitor control (5'-CAGUACUUUUGUGUAGUACAA-3'; Shanghai
GenePharma Co., Ltd.), 1 µM control-small interfering (si)RNA (cat.
no. sc-36869; Santa Cruz Biotechnology, Inc.), 0.5 µM BTG1-siRNA
(cat. no. sc-43644; Santa Cruz Biotechnology, Inc.), 100 nM
miR-208a-3p inhibitor + 1 µM control-siRNA or 100 nM miR-208a-3p
inhibitor + 0.5 µM BTG1-siRNA was transfected in hVSMCs
(5x104 cells/well) using Lipofectamine® 2000
Reagent (Invitrogen; Thermo Fisher Scientific, Inc.) for 48 h,
according to the manufacturer's protocols. The transfection
efficiency was evaluated via reverse transcription-quantitative PCR
(RT-qPCR).
MTT assay
An MTT assay was performed to assess the cell
viability of hVSMCs. The hVSMCs (6x103 cells/well) were
cultured in 96-well plates (BD Biosciences) in triplicate for 24 h
at 37˚C. DMEM was removed and miR-208a-3p inhibitor, inhibitor
control, miR-208a-3p inhibitor + control-siRNA, or miR-208a-3p
inhibitor + BTG1-siRNA in 100 µl of fresh medium were added, and
cells were subsequently cultured for 48 h at 37˚C. MTT solution (10
µl) was added to each well and incubated for a further 4 h. A total
of 100 µl DMSO was added to solubilize the formazan product. The
optical density (OD) of hVSMCs was determined at a wavelength of
490 nm by a Synergy™ 2 Multi-function microplate reader (Bio-Tek
Instruments, Inc.).
Cell apoptosis assay
hVSMCs were transfected with miR-208a-3p inhibitor,
inhibitor control, miR-208a-3p inhibitor + control-siRNA or
miR-208a-3p inhibitor + BTG1-siRNA for 48 h, and cells were
subsequently harvested. Cell apoptosis analysis was conducted using
Annexin V-FITC/propidium iodide dual staining at 4˚C for 30 min in
the dark, according to the manufacturer's protocols. Cell apoptosis
(early apoptosis + late apoptosis) was analyzed using a FACS flow
cytometer (BD Biosciences) and the results were analyzed using
FlowJo™ v7.6.1 software (BD Biosciences).
Dual-luciferase reporter assay
Bioinformatics analysis using TargetScan (version
7.2; http://www.targetscan.org/vert_72/) was performed to
predict the target gene of miR-208a-3p. The binding site between
the 3'-UTR of BTG1 and miR-208a-3p was subsequently investigate. To
validate the binding site, a dual-luciferase reporter assay was
performed. DNA segments in the 3'-UTR of BTG1 containing the
miR-208a-3p binding site were cloned into a pmirGLO dual-luciferase
vector (Promega Corporation) to conduct the pmirGLO-BTG1 wild-type
plasmid (BTG1-WT). The mutant (MUT) 3'-UTR of BTG1 was also cloned
into pmirGLO to generate BTG1-MUT plasmids. 1 µg BTG1-WT and 1 µg
BTG1-MUT were co-transfected into hVSMCs (5x104 cells)
with 100 nM miR-208a-3p mimic (sense, 5'-AUAAGACGAGCAAAAAGCUUGU-3';
and antisense, 5'-AGCUUUUUGCUCGUCUUAUUU-3'; Shanghai GenePharma
Co., Ltd.) or 100 nM mimic control (sense,
5'-UUCUCCGAACGUGUCACGUTT-3'; and antisense,
5'-ACGUGACACGUUCGGAGAATT-3'; Shanghai GenePharma Co., Ltd.) using
Lipofectamine 2000 according to the manufacturer's protocols.
Following 48 h of transfection, luciferase activity was detected
using a Dual-Luciferase Reporter Assay System (Promega
Corporation). Firefly luciferase activities were normalized to
Renilla luciferase activity.
RNA isolation and RT-qPCR
Total RNA from cultured cells or blood specimens was
extracted using TRIzol® reagent (Invitrogen; Thermo
Fisher Scientific, Inc.) according to the manufacturer's protocol.
Total RNA (200 ng) was used to produce cDNA using PrimeScript RT
Master Mix (Takara Biotechnology Co., Ltd.) using the RT primer
5'-CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGACAAGCTT-3'. A SYBR
PrimeScript RT-PCR kit (Takara Biotechnology Co., Ltd.) was used to
evaluate the miR-208a-3p and BTG1 expression levels. The relative
expression of mRNA or miR-208a-3p was normalized to GAPDH or U6,
respectively. The thermocycling conditions were as follows: Initial
denaturation at 95˚C for 5 min, followed by 35 cycles of 94˚C for
15 sec, 50˚C for 30 sec, 72˚C for 30 sec; and final extension at
72˚C for 10 min. Primer sequences for PCR were listed as follows:
miR-208a-3p forward, 5'-GGGCCATAAGACGAGCAAA-3' and miR-208a-3p
stemloop reverse, 5'-CTCAACTGGTGTCGTGGAGTC-3'; U6 forward,
5'-TGCGGGTGCTCGCTTCGCAGC-3' and reverse, 5'-CCAGTGCAGGGTCCGAGGT-3';
GAPDH forward, 5'-CGGAGTCAACGGATTTGGTCGTAT-3' and reverse,
5'-AGCCTTCTCCAGGTGGTGAAGAC-3'; BTG1 forward,
5'-CATCTCCAAGTTTCTCCGCACC-3' and reverse,
5'-GCGAATACAACGGTAACCCGATC-3'. The results were calculated using
the 2-ΔΔCq method (20).
Western blot analysis
hVSMCs were harvested and lysed using RIPA lysis
buffer (Beyotime Institute of Biotechnology). The concentration of
total protein was calculated using a BCA Protein Assay kit (Thermo
Fisher Scientific, Inc.). The protein fractions were subsequently
mixed with 5X SDS, boiled and centrifuged at 10,000 x g at 4˚C for
5 min. A total of 40 µg proteins per lane were subjected to 10%
SDS-PAGE. The proteins were transferred onto PVDF membranes and
were subsequently incubated with primary antibodies against BTG1
(cat. no. ab151740; Abcam), Bcl-2 (cat. no. ab185002; Abcam), Bax
(cat. no. ab32503; Abcam), phosphorylated (p)-AKT (cat. no. 4060;
Cell Signaling Technology, Inc.), AKT (cat. no. 4691; Cell
Signaling Technology, Inc.) and GAPDH (cat. no. 5174; Cell
Signaling Technology, Inc.) at a dilution of 1:1,000 at 4˚C
overnight. After washing with PBS-0.1% Tween 20, the membranes were
incubated with HRP-conjugated goat anti-rabbit secondary antibody
(1:2,000; cat. no. 7074; Cell Signaling Technology, Inc.) for 1 h
at room temperature. The proteins bands were analyzed using Pierce™
ECL Western Blotting Substrate (Pierce; Thermo Fisher Scientific,
Inc.), according to the manufacturer's protocol. The intensity of
protein bands was quantified using Image Lab™ Software (version
5.2.1; Bio-Rad Laboratories, Inc.).
Statistical analysis
Each experiment was repeated in triplicate. Data
were presented as the mean ± standard deviation. Statistical
analysis was carried out using SPSS software version 16.0 (SPSS,
Inc.). The differences between two groups were assessed by
Student's t-test, and comparisons between multiple groups were
detected using one-way ANOVA followed by Tukey's post hoc test.
P<0.05 was considered to indicate a statistically significant
difference.
Results
miR-208a-3p expression is upregulated
in the peripheral blood of patients with CHD
In order to explore the clinical relevance of
miR-208a-3p in CHD, the expression level of miR-208a-3p was first
determined in the peripheral blood samples of 30 patients with CHD
and 30 healthy volunteers via RT-qPCR analysis. The results showed
that significantly higher miR-208a-3p expression was detected in
the peripheral blood samples of patients with CHD compared with
healthy donors, indicating that miR-208a-3p may participate in CHD
development (Fig. 1).
miR-208a-3p directly targets to the
3'-UTR of BTG1
Bioinformatics analysis was used to identify
potential miR-208a-3p target genes. It was found that miR-208-3p
has hundreds of potential target genes, including BTG1, with a
putative binding site within its 3'-UTR for miR-208a-3p (Fig. 2A). As abnormal growth of VSMCs is
crucial in the progression of CHD (5), and BTG1 serves important roles in
regulating cell proliferation (17). However, the potential mechanism of
BTG1 in VSMCs remains unclear. Thus, BTG1 was selected for further
investigation. A luciferase reporter assay was subsequently
conducted to verify whether miR-208a-3p could directly target to
BTG1. Results indicated that miR-208a-3p mimic significantly
enhanced miR-208a-3p expression in hVSMCs compared with the mimic
control (Fig. 2B). Furthermore,
miR-208a-3p overexpression significantly inhibited the luciferase
activity of BTG1-WT. However, the luciferase activity of BTG1-MUT
was not notably affected by co-transfection with miR-208-3p mimic
compared with mimic control, suggesting that miR-208a-3p directly
targeted the 3'-UTR of BTG1 (Fig.
2C).
To further investigate BTG1 expression in patients
with CHD, RT-qPCR analysis was conducted to evaluate the mRNA
expression levels of BTG1 in the blood samples of patients with
CHD. The findings suggested that the BTG1 mRNA level was
significantly downregulated in patients with CHD compared with
healthy individuals (Fig. 2D).
Therefore, BTG1 was hypothesized to serve as a functional target of
miR-208a-3p in CHD.
miR-208a-3p negatively regulates BTG1
expression in hVSMCs
The effect of miR-208a-3p on BTG1 expression was
subsequently investigated in hVSMCs. Control-siRNA, BTG1-siRNA,
miR-208a-3p inhibitor or inhibitor control were transfected into
hVSMCs for 48 h and transfection efficiency was evaluated via
RT-qPCR analysis. As shown in Fig.
3A, miR-208a-3p inhibitor significantly suppressed miR-208a-3p
levels in hVSMCs compared with the control group. In addition, in
hVSMCs, BTG1-siRNA significantly downregulated the mRNA expression
of BTG1 compared with the control group (Fig. 3B). Further experiments demonstrated
that miR-208a-3p inhibitor was able to upregulate BTG1 at both the
mRNA and the protein level, and this upregulation was reversed by
BTG1-siRNA (Fig. 3C-E). These
findings suggested that miR-208a-3p negatively regulated BTG1
expression in hVSMCs.
BTG1-siRNA abolishes the effect of
miR-208a-3p inhibitor on cell viability and apoptosis in
hVSMCs
To further assess the role of miR-208a-3p in hVSMCs,
MTT and flow cytometry assays were performed to determine hVSMC
viability and apoptosis, respectively. miR-208a-3p inhibitor,
inhibitor control, miR-208a-3p inhibitor + control-siRNA or
miR-208a-3p inhibitor + BTG1-siRNA were transfected into hVSMCs.
MTT assay results suggested that miR-208a-3p inhibitor
significantly inhibited cell viability compared with the control
group, while miR-208a-3p inhibitor and BTG1-siRNA co-transfection
significantly promoted VSMC viability compared with the miR-208a-3p
inhibitor group (Fig. 4A).
Furthermore, flow cytometry analysis was carried out to evaluate
the effect of miR-208a-3p inhibitor on hVSMC apoptosis. The results
demonstrated that miR-208a-3p inhibitor induced hVSMC cell
apoptosis compared with the inhibitor control, while these effects
were significantly attenuated by inhibiting BTG1 (Fig. 4B and C). Western blot analysis was also
performed to investigate apoptosis-associated proteins, such as
Bcl-2 and Bax. As shown in Fig.
4D-F, miR-208a-3p inhibitor significantly reduced Bcl-2 protein
expression levels compared with the control, while it increased the
expression level of Bax. However, BTG1-siRNA efficiently reversed
the effects of miR-208a-3p inhibitor on Bax and Bcl-2 protein
expression in hVSMCs. These results suggested that miR-208a-3p was
involved in hVSMC proliferation and apoptosis by targeting
BTG1.
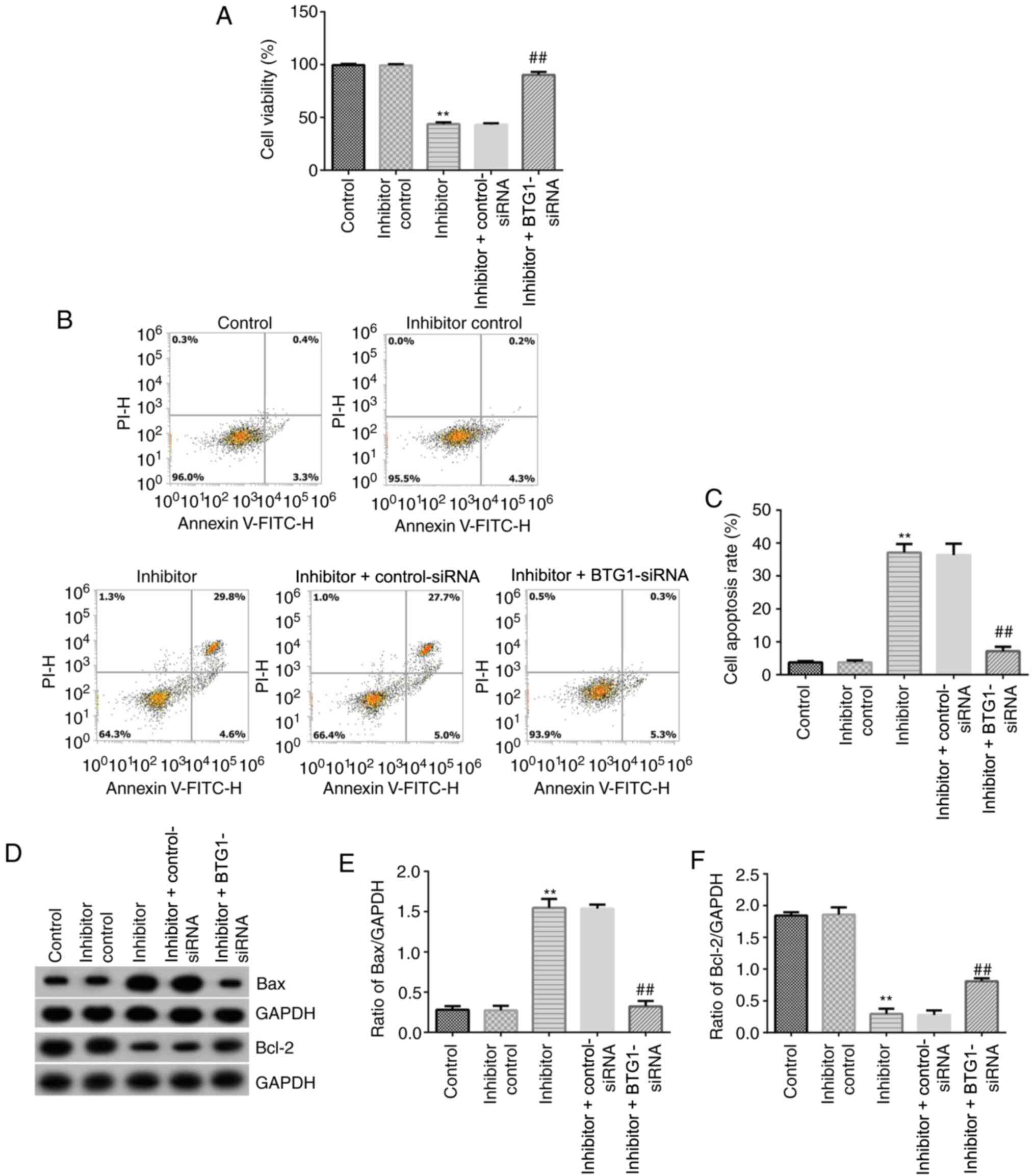 | Figure 4Effects of miR-208a-3p inhibitor or
BTG1-siRNA on the viability and apoptosis of hVSMCs. (A) MTT assay
was conducted to detect the viability of hVSMCs transfected with
inhibitor control, miR-208a-3p inhibitor, miR-208a-3p inhibitor +
control-siRNA or miR-208a-3p inhibitor + BTG1-siRNA for 48 h. (B)
Flow cytometry was performed 48 h after the transfection of hVSMCs
with inhibitor control, miR-208a-3p inhibitor, miR-208a-3p
inhibitor + control-siRNA or miR-208a-3p inhibitor + BTG1-siRNA.
(C) Quantitative analysis of apoptotic cell ratio. (D) Protein
expression levels of Bax and Bcl-2 were detected via western blot
analysis in hVSMCs transfected with inhibitor control, miR-208a-3p
inhibitor, miR-208a-3p inhibitor + control-siRNA or miR-208a-3p
inhibitor + BTG1-siRNA for 48 h. (E) Ratio of Bax/GAPDH was
calculated and presented. (F) Ratio of Bcl-2/GAPDH was calculated
and presented. **P<0.01 vs. control;
##P<0.01 vs. inhibitor. BTG1, B-cell translocation
gene 1; CHD, coronary heart disease; miR-208a-3p, microRNA-208a-3p;
hVSMCs, human vascular smooth muscle cells; siRNA, small
interfering RNA; PI, propidium iodide. |
miR-208a-3p inhibitor affects the
apoptosis of hVSMCs by regulating the PI3K/AKT pathway
The potential molecular mechanism of miR-208a-3p
inhibitor was further investigated in terms of inducing cell
apoptosis. Western blot analysis was conducted to detect relative
protein expression levels, including p-AKT and AKT in hVSMCs
(Fig. 5A). The ratio of p-AKT/AKT
levels (Fig. 5B) was significantly
suppressed in the miR-208a-3p inhibitor group compared with the
control group; BTG1-siRNA significantly reversed these effects.
Effects of BTG1-siRNA on cell
viability, apoptosis and the PI3K/AKT pathway in hVSMCs
To further explore the role of BTG1 in hVSMCs, the
effect of BTG1-siRNA transfection alone on cell viability,
apoptosis and PI3K/AKT activity were investigated in hVSMCs. It was
observed that compared with the control group, BTG1-siRNA
significantly promoted cell viability (Fig. 6A), inhibited cell apoptosis
(Fig. 6B and C), and enhanced AKT phosphorylation
(Fig. 6D and E) in hVSMCs. These findings further
suggested that BTG1 served a role in CHD development through
effects on hVSMC function.
Discussion
It has been previously reported that abnormal
regulation of hVSMCs may lead to the development of cardiovascular
disease (21). Studies have
demonstrated that miRNAs participate in the function of VSMCs, such
as miRNA-21(22) and
miRNA-214(23). Therefore, it is
necessary to identify the specific miRNAs associated with the
disease and their respective targets to further understand their
role and examine novel therapeutic strategies for disease
treatment. Previous reports have indicated that miR-208a-3p is
involved in a number of diseases, including atrial fibrillation
(24) and pathological cardiac
hypertrophy (25). In addition, a
previous study has suggested that miR-208a-3p is involved in acute
cardiac injury (26). However, to
the best of our knowledge, there are a limited number of reports
investigating the expression levels and roles of miR-208a-3p in
CHD.
To understand the function of miR-208a-3p in CHD,
the expression levels of miR-208a-3p were initially determined in
the peripheral blood samples of 30 patients with CHD and healthy
volunteers. The results showed that miR-208a-3p expression levels
were significantly upregulated in patients with CHD compared with
healthy individuals, suggesting that miR-208a-3p may serve
important roles in CHD progression. This study also revealed that
miR-208a-3p directly targeted the BTG1 3'-UTR by performing a
dual-luciferase reporter assay. BTG1, which is a member of the
pro-apoptotic Bcl-2 family, has been reported to be involved in
multiple biological behaviors, including apoptosis and factor
translocation regulation (27).
Studies have shown that BTG1 plays an important role in regulating
cell proliferation and apoptosis (19,28),
and it has been found to be a target gene of multiple miRNAs
(28,29). Furthermore, the present study found
that BTG1 was downregulated in patients with CHD. Thus, it was
hypothesized that miR-208a-3p may participate in CHD by regulating
hVSMC function via targeting of BTG1.
Then, the present study examined the functional
mechanism through which miR-208a-3p regulated hVSMCs. The results
demonstrated that miR-208a-3p inhibitor significantly inhibited
cell viability and promoted apoptosis, whereas downregulation of
BTG1 reversed these effects. Furthermore, miR-208a-3p participated
in the regulation of apoptosis-related proteins, including Bcl-2
and Bax (30). These results
suggested that miR-208a-3p may regulate VSMC proliferation and
apoptosis in CHD. Therefore, the present study examined potential
signaling pathways involved in miR-208a-3p-regulated apoptosis in
hVSMCs, and the PI3K/AKT pathway (31) was analyzed in this study. PI3K/AKT
signaling has also been reported to be involved in CHD development
(32-34).
For example, miR-26a-5p inhibition induced endothelial cell
apoptosis by inhibiting the PI3K/AKT pathway (33). Decoy receptor-3 could regulate
inflammation and apoptosis by regulating the PI3K/AKT signaling
pathway in a mouse model of coronary artery disease (33). The results of the present study
indicated that miR-208a-3p inhibitor could inhibit the PI3K/AKT
signaling pathway in hVSMCs, and in turn, this inhibition was
abolished by BTG1-siRNA.
Finally, to further investigate the reversal effect
of BTG1-siRNA on miR-208a-3p inhibitor-transfected hVSMCs, the
effect of BTG1-siRNA on hVSMCs was determined. The data suggested
that BTG1-siRNA could promote cell viability, inhibit cell
apoptosis and promote the PI3K/AKT signaling pathway in hVSMCs. The
data further suggested that miR-208a-3p may serve a role in CHD
development by targeting BTG1.
In summary, these findings provided novel insight
into the involvement of miR-208a-3p in the progression of CHD by
regulating the proliferation of hVSMCs via downregulation of BTG1.
This study provided a basis for understanding the functional
mechanism of miR-208a-3p in CHD. However, this study is only a
preliminary study of miR-208-3p in CHD. In order to verify the role
of miR-208-3p in CHD, further in-depth studies are required. For
example, the effect of BTG1-siRNA on hVSMCs requires further
investigation. The role of miR-208-3p/BTG1 in CHD should also be
investigated in vivo. The association between the expression
of miR-208-3p/BTG1 and the clinicopathological features of patients
with CHD also requires further exploration. In addition, examining
other targets of miR-208a-3p in CHD therapy is critical.
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
DW contributed to data collection, statistical
analysis, data interpretation and manuscript preparation. CY
contributed to data collection and data interpretation. DW and CY
confirm the authenticity of all the raw data. All authors have read
and approved the final manuscript.
Ethics approval and consent to
participate
Written informed consent was provided from all
participants approving the use of their samples in the present
study. All study protocols were approved by the Ethics Committee of
Shanxi Bethune Hospital (Taiyuan, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Hajek C, Guo X, Yao J, Hai Y, Johnson WC,
Frazier-Wood AC, Post WS, Psaty BM, Taylor KD and Rotter JI:
Coronary heart disease genetic risk score predicts cardiovascular
disease risk in men, not women. Circ Genom Precis Med.
11(e002324)2018.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Olubowale OT, Safford MM, Brown TM, Durant
RW, Howard VJ, Gamboa C, Glasser SP, Rhodes JD and Levitan EB:
Comparison of expert adjudicated coronary heart disease and
cardiovascular disease mortality with the national death index:
Results from the REasons for geographic and racial differences in
stroke (REGARDS) study. J Am Heart Assoc. 6(e004966)2017.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Yoshioka S, Tsukamoto T and Chihara K:
Vascular smooth muscle cells in coronary heart disease. Nihon
Rinsho. 61 (Suppl 4):S80–S85. 2003.PubMed/NCBI(In Japanese).
|
|
4
|
Wang S, Cheng Z and Chen X: Promotion of
PTEN on apoptosis through PI3K/Akt signal in vascular smooth muscle
cells of mice model of coronary heart disease. J Cell Biochem.
120:14636–14644. 2019.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Lacolley P, Regnault V, Segers P and
Laurent S: Vascular smooth muscle cells and arterial stiffening:
Relevance in development, aging, and disease. Physiol Rev.
97:1555–1617. 2017.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Maier KG, Ruhle B, Stein JJ, Gentile KL,
Middleton FA and Gahtan V: Thrombospondin-1 differentially
regulates microRNAs in vascular smooth muscle cells. Mol Cell
Biochem. 412:111–117. 2016.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Cheuk BL and Cheng SW: Identification and
characterization of microRNAs in vascular smooth muscle cells from
patients with abdominal aortic aneurysms. J Vasc Surg. 59:202–209.
2014.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Matuszcak C, Lindner K, Eichelmann AK,
Hussey DJ, Haier J and Hummel R: MicroRNAs: Key regulators of
chemotherapy response and metastatic potential via complex control
of target pathways in esophageal adenocarcinoma. Surg Oncol.
27:392–401. 2018.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Pio G, Ceci M, D'Elia D, Loglisci C and
Malerba D: A novel biclustering algorithm for the discovery of
meaningful biological correlations between microRNAs and their
target genes. BMC Bioinformatics. 14 (Suppl 7)(S8)2013.PubMed/NCBI View Article : Google Scholar
|
|
10
|
De Rosa S and Indolfi C: Circulating
microRNAs as biomarkers in cardiovascular diseases. Exp Suppl.
106:139–149. 2015.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Fernandes T, Soci UP and Oliveira EM:
Eccentric and concentric cardiac hypertrophy induced by exercise
training: MicroRNAs and molecular determinants. Braz J Med Biol
Res. 44:836–847. 2011.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Lin RC, Weeks KL, Gao XM, Williams RB,
Bernardo BC, Kiriazis H, Matthews VB, Woodcock EA, Bouwman RD,
Mollica JP, et al: PI3K(p110 alpha) protects against myocardial
infarction-induced heart failure: Identification of PI3K-regulated
miRNA and mRNA. Arterioscler Thromb Vasc Biol. 30:724–732.
2010.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Wang H, Lu J, Wu S, Yang S, Wang L, Zhou
H, Fu Y and Liu J: Effects of electroacupuncture at different
acupoints on apoptosis and the expression of miRNAs in myocardial
cells in rats model of myocardial ischemia. Zhongguo Zhen Jiu.
36:281–286. 2016.PubMed/NCBI(In Chinese).
|
|
14
|
Seo HH, Lee SY, Lee CY, Kim R, Kim P, Oh
S, Lee H, Lee MY, Kim J, Kim LK, et al: Exogenous miRNA-146a
enhances the therapeutic efficacy of human mesenchymal stem cells
by increasing vascular endothelial growth factor secretion in the
ischemia/reperfusion-injured heart. J Vasc Res. 54:100–108.
2017.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Wu H, Xu L, Chen Y and Xu C: MiR-208a-3p
functions as an oncogene in colorectal cancer by targeting PDCD4.
Biosci Rep. 39(BSR20181598)2019.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Montgomery RL, Hullinger TG, Semus HM,
Dickinson BA, Seto AG, Lynch JM, Stack C, Latimer PA, Olson EN and
van Rooij E: Therapeutic inhibition of miR-208a improves cardiac
function and survival during heart failure. Circulation.
124:1537–1547. 2011.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Cui HB, Ge HE, Wang YS and Bai XY:
MiR-208a enhances cell proliferation and invasion of gastric cancer
by targeting SFRP1 and negatively regulating MEG3. Int J Biochem
Cell Biol. 102:31–39. 2018.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Hortmann M, Walter JE, Benning L, Follo M,
Mayr RM, Honegger U, Robinson S, Stallmann D, Duerschmied D,
Twerenbold R, et al: Droplet digital PCR of serum miR-499, miR-21
and miR-208a for the detection of functionally relevant coronary
artery disease. Int J Cardiol. 275:129–135. 2019.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Zhao S, Chen SR, Yang XF, Shen DF, Takano
Y, Su RJ and Zheng HC: BTG1 might be employed as a biomarker for
carcinogenesis and a target for gene therapy in colorectal cancers.
Oncotarget. 8:7502–7520. 2017.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Wu ZW, Liu YF, Wang S and Li B: MiRNA-146a
induces vascular smooth muscle cell apoptosis in a rat model of
coronary heart disease via NF-kappaB pathway. Genet Mol Res.
14:18703–18712. 2015.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Li FP, Lin DQ and Gao LY: LncRNA TUG1
promotes proliferation of vascular smooth muscle cell and
atherosclerosis through regulating miRNA-21/PTEN axis. Eur Rev Med
Pharmacol Sci. 22:7439–7447. 2018.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Afzal TA, Luong LA, Chen D, Zhang C, Yang
F, Chen Q, An W, Wilkes E, Yashiro K, Cutillas PR, et al: NCK
associated protein 1 modulated by miRNA-214 determines vascular
smooth muscle cell migration, proliferation, and neointima
hyperplasia. J Am Heart Assoc. 5(e004629)2016.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Li S, Jiang Z, Wen L, Feng G and Zhong G:
MicroRNA-208a-3p contributes to connexin40 remolding in human
chronic atrial fibrillation. Exp Ther Med. 14:5355–5362.
2017.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Wang L, Ye N, Lian X, Peng F, Zhang H and
Gong H: MiR-208a-3p aggravates autophagy through the PDCD4-ATG5
pathway in Ang II-induced H9c2 cardiomyoblasts. Biomed
Pharmacother. 98:1–8. 2018.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Glineur SF, De Ron P, Hanon E, Valentin
JP, Dremier S and Nogueira da Costa A: Paving the route to plasma
miR-208a-3p as an acute cardiac injury biomarker: Preclinical rat
data supports its use in drug safety assessment. Toxicol Sci.
149:89–97. 2016.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Huang Y, Zheng J, Tan T, Song L, Huang S,
Zhang Y, Lin L, Liu J, Zheng P, Chen X, et al: BTG1 low expression
in pancreatic ductal adenocarcinoma is associated with a poorer
prognosis. Int J Biol Markers. 33:189–194. 2018.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Su C, Huang DP, Liu JW, Liu WY and Cao YO:
MiR-27a-3p regulates proliferation and apoptosis of colon cancer
cells by potentially targeting BTG1. Oncol Lett. 18:2825–2834.
2019.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Zhao X, Chen GQ and Cao GM: Abnormal
expression and mechanism of miR-330-3p/BTG1 axis in hepatocellular
carcinoma. Eur Rev Med Pharmacol Sci. 23:6888–6898. 2019.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Hassan M, Watari H, AbuAlmaaty A, Ohba Y
and Sakuragi N: Apoptosis and molecular targeting therapy in
cancer. Biomed Res Int. 2014(150845)2014.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Reddy D, Kumavath R, Tan TZ, Ampasala DR
and Kumar AP: Peruvoside targets apoptosis and autophagy through
MAPK Wnt/β-catenin and PI3K/AKT/mTOR signaling pathways in human
cancers. Life Sci. 241(117147)2020.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Feng H, Wang Z, Wang C, Zhu X, Liu Z, Liu
H, Guo M, Hou Q and Chu Z: Effect of furostanol saponins from
allium macrostemon bunge bulbs on platelet aggregation rate and
pi3k/akt pathway in the rat model of coronary heart disease. Evid
Based Complement Alternat Med. 2019(9107847)2019.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Jing R, Zhong QQ, Long TY, Pan W and Qian
ZX: Downregulated miRNA-26a-5p induces the apoptosis of endothelial
cells in coronary heart disease by inhibiting PI3K/AKT pathway. Eur
Rev Med Pharmacol Sci. 23:4940–4947. 2019.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Chen X, Wang R, Chen W, Lai L and Li Z:
Decoy receptor-3 regulates inflammation and apoptosis via PI3K/AKT
signaling pathway in coronary heart disease. Exp Ther Med.
17:2614–2622. 2019.PubMed/NCBI View Article : Google Scholar
|




















