Introduction
TNF receptor-associated factors (TRAFs) were
initially identified as adaptor proteins in the TRAF family
signaling pathways (1). TRAF6,
which can activate IL-1 receptor/Toll-like receptor (TLR)
superfamilies, was indicated to play an essential role in cell
survival and apoptosis (2,3). TRAF6 consists of a RING finger domain,
a series of zinc finger motifs, a coiled-coil domain and a highly
conserved TRAF-C domain (4). The
RING domain performs an essential function in ubiquitin ligase
activity (5-7),
while the TRAF-C domain regulates CD40 binding in the immune
response (8). Together with
ubiquitin-conjugating enzyme E2 13/ubiquitin-conjugating enzyme E2
variant 1A (Ubc13/Uev1A), TRAF6 was reported to regulate AKT and
TGF-β-activated kinase 1 (TAK1) activation, and induced cancer cell
apoptosis (9-11).
AKT promotes cell survival against several apoptotic
stimuli through growth factors, and plays a significant role in
tumor development and its potential response to cancer treatment
(12,13). In cancer cells, AKT is activated by
phosphorylation at Thr-308 of the catalytic domain by
phosphoinositide-dependent kinase (PDK)-1 and at Ser-473 of the
C-terminal hydrophobic region by PDK-2(14). Blocking AKT activity with LY294002
induced cell death and cell cycle arrest in HTLV-1-transformed
cells (15).
TAK1 is a serine/threonine kinase playing a critical
role in pro-inflammatory cytokine- and TLR-mediated signaling
pathways (16,17). A previous study indicated that
ubiquitin-activated TAK1 phosphorylates mitogen-activated protein
kinase kinase (MKK), leading to the activation of the JNK and p38
kinase pathways (18). Upon
activation, TAK1 was indicated to phosphorylate the IKK complex,
p38 and JNK, leading to activation of the NF-κB and MAPK signaling
pathways (19).
AKT and TAK1 can also facilitate the activation of
downstream NF-κB via the phosphorylation of NF-κB inhibitor, thus
subsequently affecting the expression levels of apoptosis-related
Bax/Bcl-2(20), and activating
caspase-9 and downstream caspase-3. Therefore, AKT and TAK1 are
involved in the initiation and mediation of cell apoptosis
(21).
Previous studies have suggested that TRAF6 may
directly catalyze AKT ubiquitination, which is essential for AKT
membrane recruitment and its phosphorylation at Thr-308 and Ser-473
(22,23) TRAF6 deficiency was indicated to lead
to constitutive inactivation of the crucial downstream targets of
AKT such as NF-κB and Bax/Bcl-2 (20,21,24).
TAK1 is activated in a polyubiquitin and TRAF6-dependent manner.
The complex formed by TRAF6, Ubc13 and Uev1A induces
Lys-63-dependent ubiquitination on TAK1 binding protein and
MAP3K7-binding protein 2, which results in TAK1 autophosphorylation
(23). Moreover, the polyubiquitin
chains, TRAF6 and Ubc13/Uev1A synthesize, which can promote the
autophosphorylation of TAK1 at Thr-184/187, resulting in its
activation (18).
Considering the important associations between TRAF6
and activations of both AKT and TAK1, the present study further
examined the role of TRAF6 on cell survival and oncogenic signaling
through the changes in AKT and TAK1 expression. Through
computer-assisted drug screening,
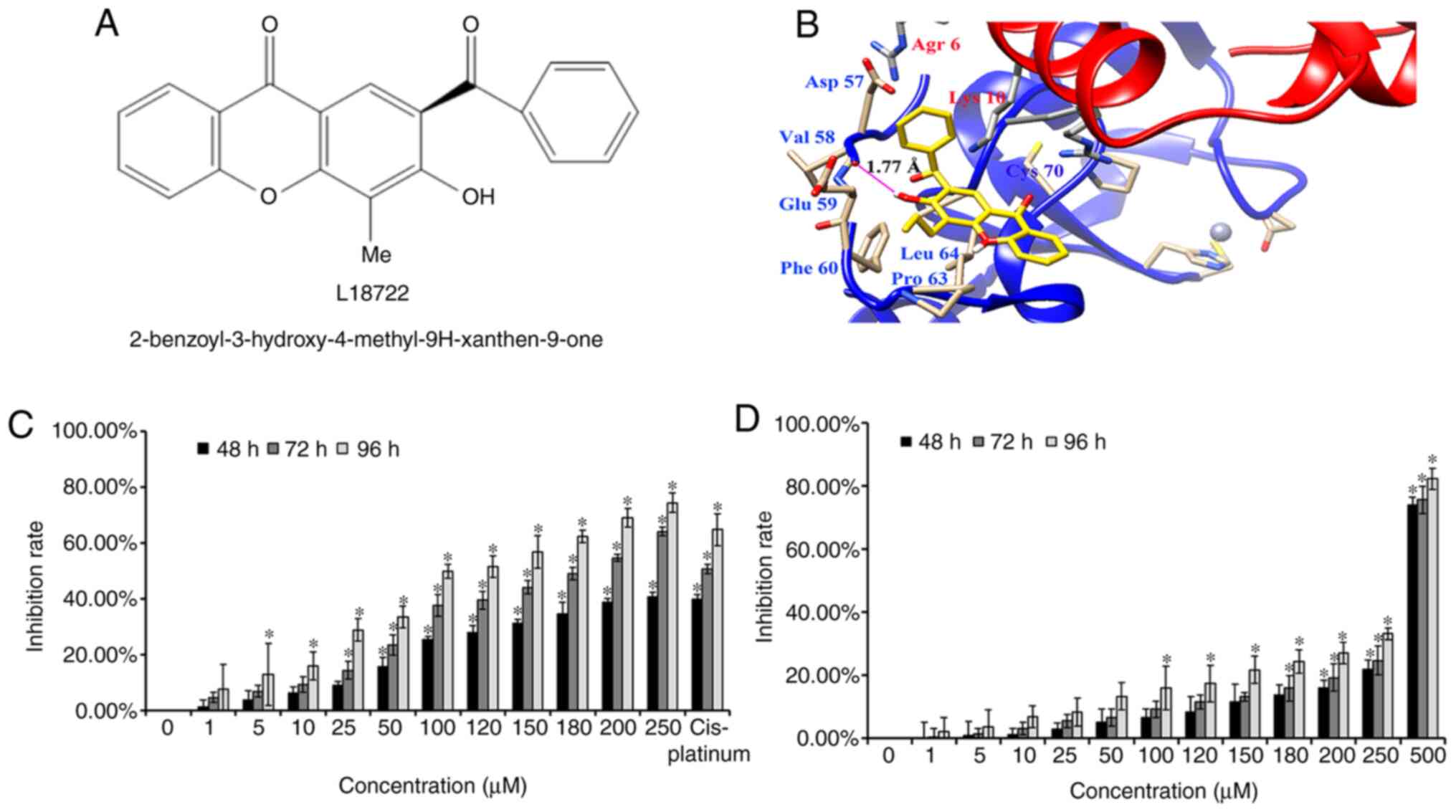
2-benzoyl-3-hydroxy-4-methyl-9H-xanthen-9-one (L18722) was reported
to compete with TRAF6. The suppressive effect of L18722 on the
activation of AKT and TAK1 was further explored.
Materials and methods
Materials
MCF-7 cells were provided by the Tianjin
International Joint Academy of Biomedicine, while normal human
dermal fibroblast (NHDF) cells were gifted by Professor Lijun Zhou
(Tianjin University, Tianjin, China). RPMI-1640 medium, DMEM and
FBS were purchased from Corning Life Sciences. L18722 (Xi'Ensi
Biochemical Technology Co., Ltd.) was dissolved in DMSO (Millipore
Sigma). MTT reagent was obtained from Millipore Sigma.
Cis-platinum was obtained from Jiangsu Hanson Pharmaceutical
Co., Ltd. Protease inhibitors and phosphatase inhibitors were
purchased from Millipore Sigma. PVDF membranes were acquired from
Millipore Sigma. Protein A-Agarose beads were obtained from Pierce
(Thermo Fisher Scientific, Inc.). ECL chemiluminescence detection
kit (SuperSignal HRP) was purchased from Pierce (Thermo Fisher
Scientific, Inc.). The caspase-3 detection assay kit (cat. no.
C1116) and the caspase-9 detection assay kit (cat. no. C1158) were
obtained from Beyotime Institute of Biotechnology.
The following antibodies were used in the present
study: Mouse ubiquitination antibody (cat. no. SC8017) and mouse
anti-TRAF6 polyclonal antibody (cat. no. SC8409) (Santa Cruz
Biotechnology, Inc.); rabbit polyclonal antibody against total AKT
(cat. no. 4685), phosphorylated (p)-AKT (Thr-308) (cat. no. 8205),
p-AKT (Ser-473) (cat. no. 8200), total TAK1 (cat. no. 4505), p-TAK1
(Thr-184/Thr-187) (cat. no. 4508), Bax (cat. no. 5023), Bcl-2 (cat.
no. 3498), p65 (cat. no. 8242), p-p65 (cat. no. 3033), caspase-3
(cat. no. 14220) and caspase-9 (cat. no. 9054) (all Cell Signaling
Technology, Inc.); rabbit anti-β-actin polyclonal antibody (cat.
no. K101527P), corresponding secondary HRP-conjugated antibodies
(cat. no. SE205) (all Beijing Solarbio Science & Technology
Co., Ltd.).
Cell culture
MCF-7 cells were cultured in RPMI-1640 medium, while
NHDF cells were cultured in DMEM; both media were supplemented with
10% FBS. All cells were cultured in a humidified atmosphere
containing 5% CO2 at 37˚C. STR profiling was performed
on NHDF cells to confirm their authenticity.
Docking study
Firstly, the structure of the RING and zinc finger
domains of TRAF6 was obtained from the Protein Data Bank database
(25). A CHARMM-like force field
was used to screen suitable small molecules which could bind to the
RING domain of TRAF6. Subsequently, AutoDock 4.10 software (The
Scripps Research Institute) with the default parameters was used to
dock TRAF6 and the available compounds from the structure library.
The compounds were then filtered according to the predicted binding
free energy.
Cell proliferation assay
The effect of L18722 on MCF-7 cell viability and
proliferation was determined using an MTT assay. Briefly, cells
were cultured in 96-well plates overnight at density of
4.1x103 cells/well. overnight. After cellular adhesion,
different concentrations (1, 5, 10, 25, 50, 100, 120, 150, 180,
200, 250 and 500 µM) of L18722 were added into the wells;
cis-platinum (16.7 µM) was used as a positive control. After
incubating cells at 37˚C with L18722 for 48, 72 and 96 h, 20 µl of
MTT (5 mg/ml) was added to each well, and the plates were incubated
at 37˚C for an additional 4 h. The medium was then removed, and
formazan crystals were dissolved in 150 µl DMSO. Optical density
was measured at 490 nm. The inhibition ratio was calculated using
the following formula: Inhibition ratio (%) = (A control-A
treated/A control) x100%. A regression curve was used to calculate
the half-maximal inhibitory concentration.
Determination of early apoptosis via
flow cytometry
The effect of L18722 (100, 150 and 200 µM) on the
early apoptosis of MCF-7 cells (2x104 cells/ml) was
investigated via flow cytometry. At 48 h post-treatment with L18722
or cis-platinum (16.7 µM) at 37˚C, the cells were harvested
and washed twice with cold PBS. Subsequently, each sample was
resuspended in 100 µl 1X binding buffer, in which 5 µl Annexin
V-FITC and PI (BD FITC Annexin V Apoptosis Detection kit) were
added according to the manufacturer's instructions. The mixture was
incubated for 15 min in the dark at room temperature. After the
addition of 400 µl Annexin-V binding buffer per sample, the cells
were analyzed using a FACScalibur flow cytometer and CellQuest Pro
5.1 (BD, Biosciences).
Determination of invasive ability of
MCF-7 cells
The invasive ability of MCF-7 cells was assessed via
Transwell assay based on the number of cells passing through the
polycarbonate membrane. A serum-free RPMI-1640 was used to wash the
upper and lower chambers. Matrigel (1:7) was added to the upper
chamber of the insert at 37˚C and stand for 2 h. Subsequently,
4x105 cells were seeded in the upper chamber with
RPMI-1640 containing 10% FBS, and he same medium was added into the
lower chamber. Then, the chamber was placed into the incubator at
37˚C for 48 h with 150 mM L18722. and cis-platinum-treated
(16.4 µM) group was used as the positive group. Subsequently, the
Transwell chamber was taken out, and the culture medium in the hole
was discarded. The cells were fixed with methanol or formaldehyde
for 30 min. Subsequently, 1% crystal violet was used to stain the
cells present in the lower chamber at room temperature for 30-60
min. A light microscope was used to observe the cells in five
fields (magnification, x400). Crystal violet-stained cells and the
quantified results are presented as the mean ± SD, and the
experiment was repeated three times for each group.
Immunoprecipitation
MCF-7 cells were cultured with L18722 (200 µM) 37˚C
for 24 h, after which the total cellular protein was extracted with
lysis buffer containing 50 mM HEPES (pH 7.4), 150 mM NaCl, 1%
NP-40. The protein was harvested after centrifugation at 12,000 x g
for 20 min at 4˚C. The samples were incubated with antibodies
against AKT (1:1,000) and TAK1 (1:1,000) overnight at 4˚C.
Subsequently, 20 µl of protein A-agarose beads (Pierce; Thermo
Fisher Scientific, Inc.) were added, and samples were incubated for
4-6 h at 4˚C with gentle rotation. The supernatant was discarded
after centrifuging three times at 800 x g for 3 min at 4˚C. Western
blotting was performed to visualize the protein bands, as reported
by Schnetzke et al (26).
Western blotting
MCF-7 cells were treated with L18722 at a
concentration of 200 µM for different durations (1, 2, 3, 6, 12,
24, 48 and 72 h). Subsequently, total proteins of MCF-7 cells were
extracted and homogenized in a lysis buffer (containing 50 mM HEPES
(pH 7.4), 150 mM NaCl, 1% NP-40) and BCA Protein Assay kit was used
to quantify the protein. Total proteins were separated via 10%
SDS-PAGE and transferred to PVDF membranes. The membranes were
blocked in 5% non-fat milk at room temperature for 2 h, which was
supplemented with TBS containing 0.1% Tween-20. Subsequently, the
membranes were incubated with antibodies against TRAF6 (1:1,000),
AKT (1:1,000), p-AKT (1:1,000), TAK1 (1:1,000), p-TAK1 (1:1,000),
ubiquitin (1:1,000), Bax (1:1,000), Bcl-2 (1:1,000), caspase-3
(1:1,000), caspase-9 (1:1,000), and β-actin (1:1,000) at 4˚C
overnight. Next, the membranes were probed with their corresponding
secondary HRP-conjugated antibodies (1:5,000) for 1 h at room
temperature. Finally, an ECL chemiluminescence kit (Thermo Fisher
Scientific, Inc.) was used to detect the expression of the
proteins. The density of the blots was quantified using ImageJ
software v.1.48u (National Institutes of Health), with β-actin as a
loading control.
Caspase-3 and caspase-9 activity
assay
MCF-7 cells were firstly treated with L18722 at a
concentration of 200 µM for 48 h. The cells were collected and
treated with ice-cold lysis buffer after 0, 24, 48 or 72 h of
L18722 treatment. The supernatants were then collected and
centrifuged at 20,000 x g for 15 min at 4˚C. According to the
manufacturer's instructions for the caspase-3 and caspase-9
detection assay kit (Beyotime Institute of Biotechnology), 10 µl of
supernatant and 10 µl of acetyl-DEVD-p-nitroanilide were added to
80 µl of reaction buffer. The mixed samples were incubated at 37˚C
for 2 h, and the enzyme-catalyzed release of p-nitroanilide was
quantified at 405 nm using a Microplate Reader (Tecan Group,
Ltd.).
Statistical analysis
Data analyses were performed using SPSS 18.0
software (SPSS Inc.), and the present results represent at least
three independent experiments, presented as the mean ± SD. Data
were log-transformed to detect differences between groups, using
one-way ANOVA and Tukey's post hoc test. P<0.05 was considered
to indicate a statistically significant difference.
Results
Selection of L18722 through docking
study
The present data indicated that L18722 could bind to
the RING domain of TRAF6 (Fig. 1B).
After analyzing all possible areas and comparing the free energies,
the suitable binding mode for L18722 was discovered, which
possessed the lowest free energy of all the possible compounds. As
presented in Fig. 1B, L18722 was
surrounded by amino acid residues Asp-57, Val-58, Glu-59, Phe-60,
Pro-63 and Leu-64, thereby suggesting interactions with residues
(54-66) preceding TRAF6 RING domain (67-124); the free energy (ΔG)
was -5.37 kcal/mol. More notably, the present results reported that
the hydroxy group (-OH) of L18722 formed a key hydrogen bond with
Val-58, which could effectively bind with TRFA6 (Fig. 1B).
L18722 inhibits MCF-7 cell
proliferation
Western blotting results reported high levels of
endogenous TRAF6 in MCF-7 cells compared with NHDF cells (Fig. S1). MTT assays revealed that L18722
could inhibit MCF-7 cell proliferation in a dose- and
time-dependent manner (Fig. 1C).
However, this effect was not observed after treating NHDF cells
with the same concentrations of L18722 (Fig. 1D). The IC50 of L18722 in
MCF-7 cells at 48 h was >250 µM. The IC50 of L18722
at 72 h was 200 µM, and the IC50 of L18722 at 96 h was
180 µM. In order to obtain more satisfying experimental results by
stimulating cells with compounds, we use IC50 at 72 h,
and the concentration was 200 µM. The inhibition rate of NHDF cells
treated with 200 µM of L18722 was 13.81%, 14.74%, 24.34% for 48, 72
and 96 h, respectively (Fig. 1D).
The inhibition ratio in NHDF cells was significantly lower than in
MCF-7 cells.
L18722 induces the early apoptosis and
inhibits the invasion of MCF-7 cells
The apoptosis of MCF-7 cells was measured via flow
cytometry, using the Annexin V-FITC and PI assay 48 h after L18722
treatment. As indicated in Fig. 2,
the early apoptosis rate of MCF-7 cells treated with L18722
increased compared with the control group. The fraction
corresponding to early apoptosis increased in a dose-dependent
manner; 2.78±1.25 to 30.19±3.27% were observed after treating cells
with 100 µM of L18722; 40.74±3.58% after treatment with 150 µM
L18722; 49.42±4.44% after treatment with 200 µM L18722, all
significantly higher compared with cis-platinum (16.7 µM)
treatment (27.93±2.18%).
The Transwell invasion experiment results indicated
that the number of MCF-7 cells that passed through the membrane in
the L18722-treated group and the cis-platinum (16.7 µM)
treated group were significantly lower compared with in the control
group (Fig. 3). These results
suggested that L18722 reduced invasion and induced apoptosis in
MCF-7 cells.
Changes in AKT and TAK1 pathway
activation
It has been reported that activation of the AKT and
TAK1 signaling pathways can regulate cell proliferation, apoptosis
and migration (27-29).
Thus, AKT and TAK1 signaling pathway expression levels were studied
in MCF-7 cells. The cells were exposed to L18722 (200 µM) for 0, 3,
6, 12 and 24 h. Immunoprecipitation was performed to test the level
of ubiquitination of AKT. Results showed that ubiquitination of AKT
was downregulated after being treated with L18722 for 1, 2 and 3 h.
(Fig. 4A). Western blotting results
suggested that, after L18722 treatment, the phosphorylation level
of AKT at Thr-308 and Ser-473 significantly decreased. (Fig. 4B). In addition, TAK1 phosphorylation
and ubiquitination levels were also reduced (Fig. 4C and D).
Effect of L18722 treatment on Bax,
Bcl-2, caspase-3 and caspase-9 expression
To further confirm the inhibitory effect of L18722
on cell proliferation and apoptosis in MCF-7 cells, the protein
expression of Bcl-2, Bax, caspase-3 and caspase-9 was examined via
western blotting. The present results demonstrated that, after
treating cells with L18722, the expression of Bcl-2 was decreased.
Conversely, the expression of Bax, caspase-3 and caspase-9 was
increased (Fig. 5A and B).
L18722 can downregulate the expression
of p-p65 in MCF-7cells
To further detect the mechanism of apoptosis induced
by L17822 in MCF-7 cells, the expression of p-p65 was examined. The
obtained results showed that after treatment with L18722, the
expression of p-p65 was decreased. (Fig. 5C and 5D).
L18722 can increase caspase-3 and 9
activities in MCF-7 cells
To further detect the mechanism of apoptosis induced
by L17822 in MCF-7 cells, the activities of caspase-9 and caspase-3
were examined. The obtained results showed that after treatment
with L18722, the actions of caspase-3 and caspase-9 were
significantly increased (Fig.
6).
Discussion
TRAF6 has been indicated to be involved in
carcinogenesis in numerous cancers; overexpression of TRAF6
resulted in the malignant transformation of fibroblasts and tumor
formation (30), whereas its
knockdown reduced cell proliferation and tumor formation (31). The RING domain of TRAF6 is
well-known to possess ubiquitin E3 ligase activity, while the TRAF
domain serves as a protein-protein interaction domain (32). Previous studies have demonstrated
that the E3 ubiquitin ligase activity of TRAF6 exerted a pivotal
function in tumorigenesis (7,32).
Furthermore, Liu et al (33)
have indicated that suppression of TRAF6 could rescue cell
proliferation and induce apoptosis in myeloma cells. Moreover,
previous studies have suggested that TRAF6 played a critical role
in regulating a number of genes involved in cell proliferation and
apoptosis, as well as immune responses to invasion through NF-κB
(16,34).
Based on the vital function of TRAF6 in
tumorigenesis, growth and apoptosis, the present study used a
computational docking program to screen small molecules that could
competitively and effectively bind with the RING domain of TRAF6.
Yin et al (35) identified a
salt bridge between Glu-69 of the RING domain and Arg-14 of Ubc13,
and demonstrated that the salt bridges between Asp-57 and Lys-10 of
Ubc13 were essential for the interaction between TRAF6 and Ubc13.
Through computational analysis, L18722 was identified in the
present study to bind with Val-58 of TRAF6, and could block the
interaction between TRAF6 and Ubc13 by preventing the formation of
a salt bridge. Without the salt bridge between TRAF6 and Ubc13,
Ubc13 could not bind with TRAF6 and activate its downstream
signaling pathway.
Next, the effects of L18722 on cell proliferation,
apoptosis and invasion ability were examined in vitro.
L18722 inhibited proliferation of MCF-7 cells in a concentration-
and time-dependent manner; however, this effect was not observed in
non-tumoral NHDF cells with concentration of <250 µM. The
present results suggested that L18722 had low cytotoxicity when
used at lower concentrations. Furthermore, flow cytometry indicated
that treatment with L18722 for 48 h could induce early apoptosis
without causing cell death.
Chaudhry et al (36) demonstrated that TRAF6 was essential
in promoting squamous cell carcinoma invasion. In the present
study, Transwell invasion assay results revealed reduced invasion
by cells treated with L18722 compared with the control group,
suggesting that L18722 may affect TRAF6 activity and subsequently
inhibit the invasive ability of the cells.
As a ubiquitin E3 ligase, TRAF6 mediates numerous
apoptosis-related signaling pathways (37-39).
Additional activation of AKT regulates a wide range of target
proteins that control cell proliferation, survival and growth
(12). Downregulation of TRAF6
using a short hairpin RNA resulted in a significant decrease of AKT
ubiquitination (22) and
phosphorylation at both Thr-308 and Ser-473(26). In addition, a previous report has
indicated that TRAF6, together with Ubc13-Uev1A, could rapidly
active TAK1 and subsequently cause a tumorigenic response (16). The activated TAK1 complex can also
phosphorylate members of the MKK family, leading to JNK and p38
kinase activation (40).
Furthermore, in TRAF6-deficient mouse embryonic fibroblasts,
phosphorylation levels at Thr-184 and Thr-187 of TAK1 were found to
be reduced, which affected the activation of downstream signals
(18,41). Therefore, the present results
demonstrated that L18722 could competitively bind with the RING
domain of TRAF6 and affect the activation of AKT and TAK1 by
targeting TRAF6.
The Bcl-2 and caspase protein families are
well-known regulators of cell apoptosis. The Bcl-2 protein family
comprises anti-apoptotic Bcl-2 and pro-apoptotic Bax (42) . Wu et al (21) reported that caspase-3 and caspase-9
activity measurements were important in determining apoptosis
factors. Bcl-2 could regulate apoptosis through caspase-9 and
caspase-3-dependent pathways (43-45).
In addition, inhibition of Bax and Bcl-2 could activate
caspase-9(46), as well as promote
the activation of caspase-3, leading to apoptosis (47).
To further confirm the inhibitory effect of L18722
on cell proliferation and apoptosis, the expression of Bcl-2, Bax,
caspase-3 and caspase-9 was examined in MCF-7 cells via western
blot analysis. After L18722 treatment, the expression of Bcl-2
decreased, while Bax, caspase-3 and caspase-9 expression levels
increased.
Compounds that could bind to the ubiquitin ligase
active region of TRAF6 were screened from a compound database (a
small library of chemical compounds established by Jkchemical Sigma
and Alfa Aesar that includes 1,792 commercially available
compounds) (48), and L18722 was
identified within this database through computer-aided drug design
and molecular docking studies. Simultaneously, MTT assays in MCF-7
and NHDF cells indicated that L18722 had a strong inhibitory effect
on the proliferation of MCF-7 cells, but not on NHDF cell (it is a
limitation to the present study that these cells were not both
sourced from breast tissues). Therefore, subsequent experiments
were conducted to verify if L18722 could induce apoptosis by
inhibiting the E3 ubiquitin ligase activity of TRAF6. Future
research will be conducted to uncover precise mechanisms associated
with L18722 in MCF-7 cells.
Furthermore, the present research aimed to find
small molecule compounds that could target TRAF6 and inhibit tumor
cell proliferation. Western blot assay results indicated that MCF-7
cells presented a high expression level of TRAF6 compared with NHDF
cells. It was previously demonstrated that small molecule compounds
could significantly inhibit tumor cells with increased expression
of TRAF6, and with little effect on healthy tissue cells (4). In the present study, the use of MCF-7
and NHDF cells demonstrated the antitumor activity of the small
molecule compound L18722. However, analysis of the overexpression
and knockdown of TRAF6 would also be essential for the study of
compounds and their effect on downstream signal pathways. In future
research, small interfering RNAs will be used to knock down TRAF6
in MCF-7 cells. L18722 will be used to treat the cells to certify
that the compound has little effect on knockout of TRAF6 cells. In
addition, TRAF6 overexpression using a recombinant plasmid into
cells may be considered, in order to verify if high-expressing
TRAF6 cells may have a heightened sensitivity for L18722.
In conclusion, the present data suggested that the
compound L18722 could competitively bind with TRAF6 and inhibit its
ubiquitination activity. Immunoprecipitation and western blot
assays demonstrated that L18722 could decrease AKT and TAK1
phosphorylation levels, thus inactivating them. The decrease in AKT
and TAK1 activity could lead to subsequent suppression of
anti-apoptotic protein Bcl-2, while elevating pro-apoptotic protein
Bax. In addition, caspase-3 and caspase-9 expression levels
increased, suggesting that L18722 could play key roles in cell
apoptosis. The RING domain of TRAF6 could be considered a
potentially viable antitumoral target, and future research will
investigate its potential for a practical approach for treating
tumors.
Supplementary Material
Expression level of TRAF6 in MCF-7 and
NHDF cells. Cells were cultured in a 60-mm plate. When the
confluence of the cells reached 60-70%, the cells were collected.
Western blot analysis was performed to detect the expression level
of TRAF6 in MCF-7 and NHDF cells. The results shown are the mean ±
SD of three independent experiments. *P<0.05 vs.
control group. TRAF6, TNF receptor-associated factor 6.
Acknowledgements
Not applicable.
Funding
Funding: This research was funded by Shijiazhuang University
Doctoral Research Startup Fund Project (grant no. 20BS004).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
YQ conceived and designed the experiments. YQ, XZ
and QY participated in the design of the study and drafting of the
manuscript. XZ carried out most of the experiments. XZ, XW and GH
participated in the cell culture and MTT assays. LR and SW
participated in the flow cytometry and detection of signaling
pathways. YQ and XZ confirm the authenticity of all the raw data.
All authors have read and approved the final manuscript.
Ethics approval and consent to
participate
The requirement for ethics approved for the use of
NHDFs initially derived from the National Biomedical Experimental
Cell Resource Bank was waived by the Ethics Committee of
Shijiazhuang University.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Inoue JI, Ishida T, Tsukamoto N, Kobayashi
N, Natio A, Azuma S and Yamamoto T: Tumor necrosis factor
receptor-associated factor (TRAF) family: Adapter proteins that
mediate cytokine signaling. Exp Cell Res. 254:14–24.
2000.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Han F, Zhang L, Qiu W and Yi X: TRAF6
promotes the invasion and metastasis and predicts a poor prognosis
in gastric cancer. Pathol Res Pract. 212:31–37. 2016.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Tian X, Zhao H, Zhang Z, Guo Z and Li W:
Intestinal mucosal injury induced by obstructive jaundice is
associated with activation of TLR4/TRAF6/NF-κB pathways. PLoS One.
14(e0223651)2019.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Qi Y, Zhao X, Chen J, Pradipta AR, Wei J,
Ruan H, Zhou L, Hsung RP and Tanaka K: In vitro and in vivo cancer
cell apoptosis triggered by competitive binding of Cinchona
alkaloids to the RING domain of TRAF6. Biosci Biotechnol Biochem.
83:1011–1026. 2019.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Frede S, Berchner-Pfannschmidt U and
Fandrey J: Regulation of hypoxia-inducible factors during
inflammation. Methods Enzymol. 435:405–419. 2007.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Grivennikov SI, Greten FR and Karin M:
Immunity, inflammation, and cancer. Cell. 140:883–899.
2010.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Zucchelli S, Codrich M, Marcuzzi F, Pinto
M, Vilotti S, Biagioli M, Ferrer I and Gustincich S: TRAF6 promotes
atypical ubiquitination of mutant DJ-1 and alpha-synuclein and is
localized to Lewy bodies in sporadic Parkinson's disease brains.
Hum Mol Genet. 19:3759–3770. 2010.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Lamothe B, Besse A, Campos AD, Webster WK,
Wu H and Darnay BG: Site-specific Lys-63-linked tumor necrosis
factor receptor-associated factor 6 auto-ubiquitination is a
critical determinant of I kappa B kinase activation. J Biol Chem.
282:4102–4112. 2007.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Shi J, Liu Z and Xu Q: Tumor necrosis
factor receptor-associated factor 6 contributes to malignant
behavior of human cancers through promoting AKT ubiquitination and
phosphorylation. Cancer Sci. 110:1909–1920. 2019.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Zhi X, Fang C, Gu Y, Chen H, Chen X, Cui
J, Hu Y, Weng W, Zhou Q, Wang Y, et al: Guaiacol suppresses
osteoclastogenesis by blocking interactions of RANK with TRAF6 and
C-Src and inhibiting NF-κB, MAPK and AKT pathways. J Cell Mol Med.
24:5122–5134. 2020.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Jang JH, Kim H and Cho JH: Molecular
cloning and functional characterization of TRAF6 and TAK1 in
rainbow trout, Oncorhynchus mykiss. Fish Shellfish Immunol.
84:927–936. 2019.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Luo J, Manning BD and Cantley LC:
Targeting the PI3K-Akt pathway in human cancer: Rationale and
promise. Cancer Cell. 4:257–262. 2003.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Parsons R: Human cancer, PTEN and the PI-3
kinase pathway. Semin Cell Dev Biol. 15:171–176. 2004.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Scheid MP and Woodgett JR: Unravelling the
activation mechanisms of protein kinase B/Akt. FEBS Lett.
546:108–112. 2003.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Jeong SJ, Pise-Masison CA, Radonovich MF,
Park HU and Brady JN: Activated AKT regulates NF-kappaB activation,
p53 inhibition and cell survival in HTLV-1-transformed cells.
Oncogene. 24:6719–6728. 2005.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Landstrom M: The TAK1-TRAF6 signalling
pathway. Int J Biochem Cell Biol. 42:585–589. 2010.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Takeuchi O and Akira S: Pattern
recognition receptors and inflammation. Cell. 140:805–820.
2010.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Wang C, Deng L, Hong M, Akkaraju GR, Inoue
J and Chen ZJ: TAK1 is a ubiquitin-dependent kinase of MKK and IKK.
Nature. 412:346–351. 2001.PubMed/NCBI View
Article : Google Scholar
|
|
19
|
Ouyang C, Nie L, Gu M, Wu A, Han X, Wang
X, Shao J and Xia Z: Transforming growth factor (TGF)-β-activated
kinase 1 (TAK1) activation requires phosphorylation of serine 412
by protein kinase A catalytic subunit α (PKACα) and X-linked
protein kinase (PRKX). J Biol Chem. 289:24226–24237.
2014.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Pugazhenthi S, Nesterova A, Sable C,
Heidenreich KA, Boxer LM, Heasley LE and Reusch JE: Akt/protein
kinase B up-regulates Bcl-2 expression through cAMP-response
element-binding protein. J Biol Chem. 275:10761–10766.
2000.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Wu R, Tang S, Wang M, Xu X, Yao C and Wang
S: MicroRNA-497 induces apoptosis and suppresses proliferation via
the Bcl-2/Bax-caspase9-caspase3 pathway and cyclin D2 protein in
HUVECs. PLoS One. 11(e0167052)2016.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Yang WL, Wang J, Chan CH, Lee SW, Campos
AD, Lamothe B, Hur L, Grabiner BC, Lin X, Darnay BG and Lin HK: The
E3 ligase TRAF6 regulates Akt ubiquitination and activation.
Science. 325:1134–1138. 2009.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Avila M, Martinez-Juarez A, Ibarra-Sanchez
A and Gonzalez-Espinosa C: Lyn kinase controls TLR4-dependent IKK
and MAPK activation modulating the activity of TRAF-6/TAK-1 protein
complex in mast cells. Innate Immun. 18:648–660. 2012.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Kim SY, Bae S, Choi KH and An S: Hydrogen
peroxide controls Akt activity via ubiquitination/degradation
pathways. Oncol Rep. 26:1561–1566. 2011.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Berman HM, Westbrook J, Feng Z, Gilliland
G, Bhat TN, Weissig H, Shindyalov NI and Bourne PE: The protein
data bank. Nucleic Acids Res. 28:235–242. 2000.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Schnetzke U, Fischer M, Kuhn AK,
Spies-Weisshart B, Zirm E, Hochhaus A, Muller JP and Scholl S: The
E3 ubiquitin ligase TRAF6 inhibits LPS-induced AKT activation in
FLT3-ITD-positive MV4-11 AML cells. J Cancer Res Clin Oncol.
139:605–615. 2013.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Yoon K, Jung EJ, Lee SR, Kim J, Choi Y and
Lee SY: TRAF6 deficiency promotes TNF-induced cell death through
inactivation of GSK3beta. Cell Death Differ. 15:730–738.
2008.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Ling MT, Wang XH, Ouyang XS, Xu K, Tsao SW
and Wong YC: Id-1 expression promotes cell survival through
activation of NF-kappaB signaling pathway in prostate cancer cells.
Oncogene. 22:4498–4508. 2013.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Wen J, Liu X, Qi Y, Niu F, Niu Z, Geng W,
Zou Z, Huang R, Wang J and Zou H: BMP3 suppresses colon
tumorigenesis via ActRIIB/SMAD2-dependent and TAK1/JNK signaling
pathways. J Exp Clin Cancer Res. 38(428)2019.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Starczynowski DT, Lockwood WW, Delehouzee
S, Chari R, Wegrzyn J, Fuller M, Tsao MS, Lam S, Gazdar AF, Lam WL
and Karsan A: TRAF6 is an amplified oncogene bridging the RAS and
NF-κB pathways in human lung cancer. J Clin Invest. 121:4095–4105.
2011.PubMed/NCBI View
Article : Google Scholar
|
|
31
|
Zhang J, Lei Z, Huang Z, Zhang X, Zhou Y,
Luo Z, Zeng W, Su J, Peng C and Chen X: Epigallocatechin-3-gallate
(EGCG) suppresses melanoma cell growth and metastasis by targeting
TRAF6 activity. Oncotarget. 7:79557–79571. 2016.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Mercier P, Lewis MJ, Hau DD, Saltibus LF,
Xiao W and Spyracopoulos L: Structure, interactions, and dynamics
of the RING domain from human TRAF6. Protein Sci. 16:602–614.
2007.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Liu H, Tamashiro S, Baritaki S, Penichet
M, Yu Y, Chen H, Berenson J and Bonavida B: TRAF6 activation in
multiple myeloma: A potential therapeutic target. Clin Lymphoma
Myeloma Leuk. 12:155–163. 2012.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Naito A, Azuma S, Tanaka S, Miyazaki T,
Takaki S, Takatsu K, Nakao K, Nakamura K, Katsuki M, Yamamoto T and
Inoue J: Severe osteopetrosis, defective interleukin-1 signalling
and lymph node organogenesis in TRAF6-deficient mice. Genes Cells.
4:353–362. 1999.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Yin Q, Lin SC, Lamothe B, Lu M, Lo YC,
Hura G, Zheng L, Rich RL, Campos AD, Myszka DG, et al: E2
interaction and dimerization in the crystal structure of TRAF6. Nat
Struct Mol Biol. 16:658–666. 2009.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Chaudhry SI, Hooper S, Nye E, Williamson
P, Harrington K and Sahai E: Autocrine IL-1β-TRAF6 signaling
promotes squamous cell carcinoma invasion through paracrine TNFα
signaling to carcinoma-associated fibroblasts. Oncogene.
32:747–758. 2013.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Ji YX, Zhang P, Zhang XJ, Zhao YC, Deng
KQ, Jing X, Wang PX, Huang Z and Li H: The ubiquitin E3 ligase
TRAF6 exacerbates pathological cardiac hypertrophy via
TAK1-dependent signaling. Nat Commun. 7(11267)2016.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Li T, Qin JJ, Xia Y, Ji YX, Guo F, Cheng
WL, Wu X, Gong FH, Hong Y, Zhu XY, et al: The ubiquitin E3 ligase
TRAF6 exacerbates ischemic stroke by ubiquitinating and activating
Rac1. J Neurosci. 37:12123–12140. 2017.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Paul PK, Bhatnagar S, Mishra V, Srivastava
S, Darnay BG, Choi Y and Kumar A: The E3 ubiquitin ligase TRAF6
intercedes in starvation-induced skeletal muscle atrophy through
multiple mechanisms. Mol Cell Biol. 32:1248–1259. 2012.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Dey N, Liu T, Garofalo RP and Casola A:
TAK1 regulates NF-κB and AP-1 activation in airway epithelial cells
following RSV infection. Virology. 418:93–101. 2011.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Yamashita M, Fatyol K, Jin C, Wang X, Liu
Z and Zhang YE: TRAF6 mediates Smad-independent activation of JNK
and p38 by TGF-beta. Mol Cell. 31:918–924. 2008.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Kim H, Chung H, Kim HJ, Lee JY, Oh MY, Kim
Y and Kong G: Id-1 regulates Bcl-2 and Bax expression through p53
and NF-kappaB in MCF-7 breast cancer cells. Breast Cancer Res
Treat. 112:287–296. 2008.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Marsden VS, O'Connor L, O'Reilly LA, Silke
J, Metcalf D, Ekert PG, Huang DC, Cecconi F, Kuida K, Tomaselli KJ,
et al: Apoptosis initiated by Bcl-2-regulated caspase activation
independently of the cytochrome c/Apaf-1/caspase-9
apoptosome. Nature. 419:634–637. 2002.PubMed/NCBI View Article : Google Scholar
|
|
44
|
McNutt MC, Lagace TA and Horton JD:
Catalytic activity is not required for secreted PCSK9 to reduce low
density lipoprotein receptors in HepG2 cells. J Biol Chem.
282:20799–20803. 2007.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Rahman M, Chan AP, Tang M and Tai IT: A
peptide of SPARC interferes with the interaction between caspase8
and Bcl2 to resensitize chemoresistant tumors and enhance their
regression in vivo. PLoS One. 6(e26390)2011.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Vegran F, Boidot R, Solary E and
Lizard-Nacol S: A short caspase-3 isoform inhibits
chemotherapy-induced apoptosis by blocking apoptosome assembly.
PLoS One. 6(e29058)2011.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Cain K, Bratton SB and Cohen GM: The
Apaf-1 apoptosome: A large caspase-activating complex. Biochimie.
84:203–214. 2002.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Qi Y, Pradipta AR, Li M, Zhao X, Lu L, Fu
X, Wei J, Hsung RP, Tanaka K and Zhou L: Cinchonine induces
apoptosis of HeLa and A549 cells through targeting TRAF6. J Exp
Clin Cancer Res. 36(35)2017.PubMed/NCBI View Article : Google Scholar
|