Introduction
Intervertebral disc degeneration (IVDD) is a
degenerative disease and is the leading cause of lower back pain
and disability worldwide (1,2).
IVDD is also the most common cause of injury and disability in
older adults, with symptoms such as stiff spine, stiff neck and
back pain (2). The aging
population exacerbates the incidence rate and economic burden of
IVDD (3). It is well-known that
nucleus pulposus (NP) dysfunction has been associated with IVDD. In
NP tissues, NP cells play an essential role in maintaining
extracellular matrix (ECM) homeostasis (4-6).
It has been demonstrated that excessive apoptosis of the NP cells,
caused by oxidative stress, inflammation and mechanical load, which
subsequently leads to exacerbated ECM degradation, could be a
critical pathogenic mechanism of disc degeneration (7). Oxidative stress typically refers to
tissue damage caused by the accumulation of reactive oxygen species
(ROS) in cells and tissues following the production of free
radicals in the body and exceeds its scavenging rate (8). Therefore, the balance is broken. ROS
plays a vital role in the development of many degenerative
diseases, including intervertebral disc degeneration, neuropathy
(9), diabetes (10), senile dementia (11) and degenerative eye disease
(12). It has been shown that high
levels of ROS in the intervertebral disc not only led to NP cell
apoptosis, but also accelerated cell senescence, ultimately leading
to disc degeneration (13).
Therefore, reducing oxidative stress in NP cells could slow ECM
degradation and may be key to improving disc degeneration.
Carthamus tinctorius L, also known as grass
safflower, is a dried flower of Compositae plants. It is a type of
traditional Chinese medicine that promotes blood circulation and
provides pain relief. Hydroxysafflor yellow A (HSYA), a mono
chalcone glycoside derived from safflower, is the main component of
safflower yellow pigment (14).
Lee et al (15) found that
HSYA could protect the kidney by inhibiting oxidative stress injury
and renal cell apoptosis induced by nephropathy in type 2 diabetic
rats. In addition, HSYA could protect neuronal cells from cellular
damage induced by oxidative stress in rats with traumatic brain
injury (14). Furthermore, HSYA
also plays a role in the regulation of ECM balance. HSYA could
reduce the protein expression level of matrix metalloproteinase 13
(MMP-13) and thrombospondin motif 5 (ADAMTS5) to attenuate
IL-1β-induced ECM degradation, thereby delaying the
lipopolysaccharide-induced increase in disc matrix metabolism and
inflammatory response, and playing a protective role (16). Therefore, we hypothesized that HSYA
may alleviate the degenerative process by improving anti-oxidative
stress and the ECM balance regulation ability of the NP cells.
Tert-butyl hydroperoxide (TBHP) is an exogenous
inducer of oxidative stress. Due to its high stability and
sustained release ability, it has been used in various models of
degenerative diseases, including NP cell osteoarthritis oxidative
stress models (17). HSYA is
considered to exert good antioxidant activity and to regulate ECM
balance. In the present study, cell experiments were conducted to
verify whether HSYA could inhibit the oxidative stress-induced
apoptosis of nucleus pulposus cells and collagen degradation by
activating CA XII, thus weakening IVDD. CA XII plays a vital role
in intervertebral disc degeneration (18-20).
Materials and methods
Primary cell culture of the NP
cells
NP tissue was obtained from 4-week-old
Sprague-Dawley rats (sacrificed by cervical dislocation). NP
tissues were digested with 0.2% collagenase type II (Beijing
Solarbio Science and Biotechnology Co., Ltd.) for 25 min at 37˚C,
filtered through a 100-mesh cell filtration screen, resuspended in
DMEM/F12 [Gibco; Thermo Fisher Scientific, Inc.; containing 10% FBS
(Hyclone; Cytiva) and 1% penicillin/streptomycin], and placed in an
incubator at 37˚C with 5% CO2. Subcultures were
performed when the NP cells reached 80-90% confluence. All
protocols conformed to the Guide for the Care and Use of Laboratory
Animals published by the American Physiological Society (21). The current study was approved by
the Affiliated Hospital of Zunyi Medical University Ethics
Committee for the Use of Experimental Animals (approval no.
KLL-2020-317).
NP cell identification
The primary NP cells were cultured in 6-well plates,
at a density of 1x105, and a coverslip was placed in the
bottom layer for cell climbing. After 24 h, the coverslips were
removed and the NP cells were washed with cold PBS, fixed in 4%
paraformaldehyde at 4˚C for 15 min, permeabilized with
0.5% Triton X-100 for 20 min, then blocked with 1% BSA (Proliant
Biologicals, LLC) for 30 min at room temperature. Next, the samples
were incubated with primary antibodies against collagen II (1:100;
cat no. ER1906-48) and aggrecan (1:50; cat no. ET1704-57) (both
from Huabio) overnight at 4˚C. The following day, the samples were
incubated with HRP-labeled goat anti-rabbit IgG antibody (1:100;
cat no. HA1014; Huabio) for 1 h at room temperature, then stained
with DAPI at room temperature for 10 min. Finally, images were
captured under a fluorescence microscope (DM500; Leica
Microsystems, Inc.).
Cytotoxicity assay
According to the manufacturer's instructions, cell
viability was measured using a Cell Counting Kit (CCK)-8 assay (cat
no. C0037; Beyotime Institute of Biotechnology). The NP cell line
was seeded in 96-well plates, at a density of 5x103
cells/well, and incubated with different concentrations (0, 5, 10,
15, 20, 25, 30, 35, 40, 45 and 50 µM) of HSYA (cat no. 78281-02-4)
and TBHP (0, 20, 40, 60, 80, 100, 120, 140, 160, 180 and 200 µM),
cat no. 75-91-2) (both from Shanghai Macklin Biochemical Co., Ltd.)
for 24 h. CCK-8 solution (10 µl) was added to each well and the NP
cells were cultured for another 4 h at 37˚C. The absorbance of each
sample was measured at 450 nm using a microplate reader (Thermo
Fisher Scientific, Inc.).
Cellular model of oxidative
stress
The primary NP cell line was subjected to oxidative
stress model induction using TBHP (40 µM). The cells treated with
TBHP only were used as the model group. The cells treated with HSYA
(10 µM) and TBHP (40 µM) were the HSYA group. The control group was
untreated.
ROS detection using flow
cytometry
A ROS assay kit (cat no. CA1410; Beijing Solarbio
Science and Technology, Co., Ltd.) was used to detect intracellular
ROS levels. NP cells (treated with 40 µM THBP or 10 µM HSYA) were
incubated with 7-dichloro-dihydrofluorescein diacetate at a final
concentration of 10 µM at 37˚C for 30 min, and were
mixed upside down every 5 min. The NP cells were then washed three
times with cold PBS and analyzed using flow cytometry (Attune NxT;
Applied Biosystems; Thermo Fisher Scientific, Inc.). Flow cytometry
results were analyzed using FlowJo™ v10.8 Software (BD
Biosciences).
ELISA
NP cells (treated with 40 µM THBP or 10 µM HSYA)
were collected, then cell lysis was performed using cell lysis
buffer (Zhejiang Ruyao Biotech Co., Ltd.). Following centrifugation
at 2,000 x g at room temperature for 5 min, the pellet was removed
and the supernatant was collected for further use. The superoxide
dismutase (SOD; cat. no. BC0170), malondialdehyde (MDA; cat. no.
BC0025), catalase (CAT; cat. no. BC0200), and glutathione
peroxidase (GSH-Px; cat. no. BC1175) concentrations were detected
according to the manufacturer's instructions (Beijing Solarbio
Science and Technology, Co., Ltd.).
Analysis of apoptosis
An Annexin V-FITC/PI apoptosis kit (cat no.
70-AP101-100; Multi sciences (Lianke) Biotech Co., Ltd.) was used
in this experiment. The NP cells (5x105) were seeded in
6-well plates for cell culture overnight at 37˚C. The
cells were treated according to the aforementioned groups. After
incubation with 40 µM TBHP and 10 µM HSYA for 24 h, the cell
supernatant was removed to preserve the floating cells. Adherent
cells were digested with trypsin and washed with PBS. The cell
precipitates were obtained after centrifugation at 800 x g at room
temperature for 5 min and then suspended in PBS. They were washed
twice with precooled PBS and resuspended in binding buffer, at a
final concentration of 1x106 cells/ml. A total of 5 µl
Annexin V-FITC and 5 µl PI were added to 100 µl cells containing
1x105 cells. The cells were mixed by gentle rotation and
incubated in the dark at room temperature for 15 min. A total of
400 µl binding buffer was added for analysis using a Life Attune
sonic focusing flow cytometer (Attune NxT; Applied Biosystems;
Thermo Fisher Scientific, Inc.). The flow cytometry results were
analyzed using FlowJo™ v10.8 Software (BD Biosceinces). All the
samples were performed in triplicate and repeated three times.
Nuclear morphological analysis
The NP cells (3.5-5x105) were seeded in
6-well plates for cell culture at 37˚C and a coverslip
was placed at the bottom of the plate for cell climbing, and
attachment overnight. The cells were treated according to the
aforementioned groups. After 24 h, the coverslips were removed,
fixed with methanol precooled at 4˚C for 10 min and permeabilized
for 10 min. After washing three times with PBS containing 0.5%
Tween-20, they were washed once with PBS, followed by staining with
1 µg/ml DAPI (dissolved in PBS) reagent at room temperature for 10
min. Subsequently, the samples were washed three times with PBS,
followed by an anti-fluorescence quencher and mounted with neutral
gum. The results were analyzed using a fluorescence microscope
(DM500; Lecia Microsystems, Inc.). The number of nuclear pyknotic
cells in three different visual fields was counted by fluorescence
microscopic observation (magnification, x400).
Western blot analysis
The treated NP cells were lysed on ice using RIPA
buffer (cat no. P0013B; Beyotime Institute of Biotechnology),
containing protease and phosphatase inhibitors (100:1 by volume).
The supernatant was obtained following centrifugation at 10,000 x g
for 10 min at 4˚C, then the samples were quantified using a BCA
protein assay kit (Beyotime Institute of Biotechnology). Total
protein (20 µg) from each treatment group were separated using 10%
SDS-PAGE and transferred to PVDF membranes. After blocking with 5%
skimmed milk for 2 h at room temperature, the membranes were
incubated with primary antibodies against Bcl-2 (1:2,000; cat. no.
60178-1-Ig; ProteinTech Group, Inc.), Bax (1:2,000; cat. no.
60267-1-Ig; ProteinTech Group, Inc.), caspase-3 (1:1,000; cat. no.
9662S; Cell Signaling Technology, Inc.), cleaved caspase-3
(1:1,000; cat. no. 9664T; Cell Signaling Technology, Inc.),
caspase-9 (1:1,000; cat. no. A2636; ABclonal Biotech Co., Ltd.),
cleaved caspase-9 (1:1,000; cat. no. ER60008; Huabio) MMP-13
(1:1,000; cat. no. 18165-1-AP; ABclonal Biotech Co., Ltd.),
collagen II (1:1,000; cat. no. ER1906-48; Huabio), aggrecan
(1:1,000; cat. no. ET1704-57; Huabio), CA XII (1:1,000; cat. no.
ER62856; Huabio), and β-tubulin (1:1,000; cat. no. 2128S; Cell
Signaling Technology, Inc.) overnight at 4˚C. Subsequently, the
membranes were incubated with goat anti-rabbit IgG/HRP antibody
(1:2,000; cat. no. SE134; Beijing Solarbio Science and Technology,
Co., Ltd.) at room temperature for 2 h. An ECL kit (cat. no.
SW2030; Beijing Solarbio Science and Technology, Co., Ltd.) was
used analyze the expression level along with a UVP gel imager.
ImageJ v1.8.0 software (National Institutes of Health) was used for
densitometry.
HSYA target protein prediction
The 3D structure of HSYA was downloaded from the
Chemspider database (http://www.chemspider.com/). The 3D structure of the
protein was obtained from the RCSB Protein Data Bank (PDB;
http://www.rcsb.org/). The target proteins
predicted by HSYA were intersected with genes associated with disc
degeneration to determine the most likely target proteins. The
remaining target proteins were simulated and constructed using
Pymol software (v2.3.0 software (Schrodinger Technologies Inc.).
Pymol software was used to remove water molecules and obtain
ligands from the protein structure, which were stored in PDBQT
format, respectively. Molecular docking analysis was performed
using AutoDock 4 (https://autodock.scripps.edu/). Depending on the
location of the protein ligand, the protein active site coordinates
were determined and default values were used for all other
parameters. Eventually, the conformation with the lowest docking
binding energy was selected for docking binding mode analysis and
plotted using the visualization function of the Pymol molecular
simulation software.
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA, from the treated cells, was extracted
using TRIzol®, then the Reverse transcription kit (cat.
no. E047-01B; Novoprotein Technology Co., Ltd.) was used to
generate cDNA. After adding the components according to the
instructions, samples were incubated at 25˚C for 5 min.
Then it was incubated at 42˚C for 30 min. Subsequently, CA XII
expression level was detected using a NovoStart® SYBR
qPCR SuperMix Plus kit (Novoprotein Technology Co., Ltd.) according
to the manufacturer's instructions. The relative expression level
of CA XII was calculated using the 2-ΔΔCq method
(22) and βIII-tubulin as an
internal reference. Each sample was analyzed in triplicate. The
following primers were used: CA XII forward,
5'-TGGACCTACATTGGTCCTGC-3', and reverse, 5'-GCT
CTGTACTGGTGAGGCTG-3'; and βIII-tubulin forward,
5'-CAATGAGGCCTCCTCTCACA-3, βIII-tubulin and reverse,
5'-TGTATAGTGCCCTTTGGCCC-3'.
CA XII knockdown using small
interfering RNA (siRNA)
The coding sequence of the CA XII mRNA was
downloaded from National Center for Biotechnology Information
(https://www.ncbi.nlm.nih.gov/), and the
CA XII siRNA sequences were designed using siDirect version 2.0
(http://sidirect2.rnai.jp/), and the
three top-scoring sequences were used for validation. The primers
were synthesized by Jiangsu Genewiz Biotechnology Co., Ltd. The
cells were plated in 24-well plates to ensure that transfection was
performed when the confluence of the cells was 30%. The siRNA
duplexes (6 pmol) were first solubilized and diluted with 50 µl
serum-free Opti-MEM, then 1 µl Lipofectamine® RNAimax
mixture (cat no. 13778030; Thermo Fisher Scientific, Inc.) was
added for 10 min at room temperature. The siRNA duplex/RNAimax
complex was added to the cell transfection medium at a final siRNA
concentration of 10 nM and co-incubated at 37˚C for 48
h. After 48 h, the gene and protein expression levels of CA XII
were examined using RT-qPCR and western blot analysis to verify
successful knockdown of expression. The following siRNA sequences
were used: siRNA1 forward, 5'-UUCAAUAGC UUUUCAACAGAC-3' and
reverse, 5'-CUGUUGAAAAGCUAUUGAACC-3'; siRNA2 forward, 5'-AUUGGUUAG
GUUCAAUAGCUU-3' and reverse, 5'-GCUAUUGAACCU AACCAAUGA-3'; siRNA3
forward, 5'-ACUGAAUGGCCA UCAUUGGUU-3' and reverse,
5'-CCAAUGAUGGCCAUUCAGUGA-3'; and negative control siRNA
(non-targeting sequence) forward, 5'-AUCGUACGUACCGUCGUAU-3' and
reverse, 5'-AUACGACGGUACGUACGAU-3'.
Immunocytochemistry
The primary NP cells were cultured in 6-well plates,
at a density of 1x105. A coverslip was placed in the
bottom for cell migration, then it was removed after 24 h. The
cells were washed with cold PBS, fixed with 4% paraformaldehyde at
4˚C for 15 min, permeabilized with 0.5% Triton X-100 for
20 min. The endogenous peroxidase activity was blocked by
incubation with 3% H2O2 at room temperature
for 20 min, and then blocked with 1% BSA (Proliant Biologicals,
LLC) for 30 min at room temperature. Next, the samples were
incubated with collagen II (1:100, cat. no. ER1906-48; HUABIO),
aggrecan (1:50, cat. no. ET1704-57; HUABIO) and MMP-13 (1:50, cat.
no. 18165-1-AP; ABclonal Biotech Co., Ltd.) primary antibodies
overnight at 4˚C. Subsequently, the samples were incubated with
goat anti-rabbit IgG/HRP antibody (1:100, cat. no. SE134; Beijing
Solarbio Science & Technology, Co., Ltd.) for 1 h at room
temperature, stained with diaminobenzidine (DAB, cat. no. P0202;
Beyotime Institute of Biotechnology) chromogenic solution (at room
temperature for 3 min) and hematoxylin (at room temperature for 1
min), dehydrated in a gradient alcohol series, immersed in xylene,
and mounted with neutral gum. The experimental results were
observed and images were captured using a Leica light microscope
(magnification, x400; DM500; Lecia Microsystems, Inc.). IPP.6.0
(Image Pro Plus 6.0; Media Cybernetics, Inc.) software was used to
quantify the optical density values of the results in each group
under three different randomly selected visual fields
(magnification, x400) to judge the intensity of positive
results.
Statistical analysis
All the experiments were repeated independently
three times. The data are expressed as the mean ± standard
deviation. GraphPad Prism 8.0 (GraphPad Software, Inc.) was used to
analyze the data. Differences between groups were analyzed using
one-way ANOVA followed by Tukey's post hoc test. P<0.05 was
considered to indicate a statistically significant difference.
Results
Effect of HSYA on TBHP-induced
oxidative stress injury the NP cells
Disc primary cells were obtained from rat disc NP
tissue and cultured. Immunofluorescence validation of the hallmark
proteins from the primary NP cells, collagen II and aggrecan,
showed that these proteins were highly expressed in the cultured
primary cells (Fig. 1A). The
cytotoxic effect of HSYA on the NP cells was analyzed using a CCK-8
assay and the results showed that HSYA concentration between 0, and
50 µM did not significantly affect the viability of the cells
(Fig. 1B). A CCK-8 assay was also
used to detect the effect of different concentrations of TBHP on NP
cell viability and the results showed that at 40 µM TBHP, cell
viability was significantly decreased compared with that in cells
treated with 0 µM TBHP (P<0.01) (Fig. 1C). Therefore, 40 µM TBHP was
selected for subsequent experiments.
Fig. 2A shows the
detection of cellular ROS levels, which were analyzed using flow
cytometry and further analysis showed that compared with that in
the control group, the ROS level in the model group was
significantly increased (P<0.01) (Fig. 2B). However, after administration of
5, 10, 15 and 20 µM HSYA, the ROS level was significantly decreased
(all P<0.01).
 | Figure 2Effect of HSYA on TBHP-induced
oxidative stress in NP cells. (A) Reactive oxygen species levels
were measured using flow cytometry and the results were (B)
statistically analyzed. (C) SOD, (D) MDA, (E) CAT and (F) GSH-Px
concentrations were analyzed using ELISA. *P<0.05,
**P<0.01 vs. control; #P<0.05,
##P<0.01 vs. model. HSYA, hydroxysafflor yellow A;
NP, nucleus pulposus; TBHP, tert-butyl hydroperoxide; SOD,
superoxide dismutase; MDA, malondialdehyde; CAT, catalase; GSH-Px,
glutathione peroxidase. |
The results of oxidative stress index showed that
treatment with TBHP could significantly inhibit the concentrations
of SOD (Fig. 2C), CAT (Fig. 2E), and GSH-Px (Fig. 2F) in the model group, while the
concentrations of MDA (Fig. 2D)
were significantly increased (compared with that in the control
group, P<0.01). Compared with that in the model group, the
concentrations of SOD, CAT and GSH-Px were significantly increased,
and the MDA concentration was significantly decreased following
HSYA treatment (all P<0.01). With respect to oxidative stress,
10 µM HSYA had a greater effect than 5 µM HSYA. However, there was
no significant difference between 10, 15 and 20 µM concentrations
of HSYA.
Effect of HSYA on TBHP-induced NP cell
apoptosis
The apoptosis level was detected using flow
cytometry (Fig. 3A). The
statistical analysis showed that TBHP induced apoptosis in the NP
cells (Fig. 3B). The apoptosis
level in the model group was significantly increased to 15.712%
compared with 4.895% in the control group (P<0.01). After
treatment with 10 µM HSYA, the level of apoptosis decreased to
7.96%, which was significantly lower compared with that in the
model group (P<0.01). Morphology analysis was subsequently
performed using DAPI (Fig. 3C).
The results showed that the nucleus of the model cell appeared
pyknotic, accompanied by the presence of more typical apoptotic
bodies (indicated using red circles). The cells showed pyknotic
nuclei and there was a decrease in the number of apoptotic bodies
following treatment with 10 µM HSYA. Apoptosis-related protein
expression was detected using western blot analysis (Fig. 3D) and the statistical results
showed that compared with that in the control group, the expression
of the pro-apoptotic proteins Bax, cleaved-caspase-3, and
cleaved-caspase-9 were significantly increased in the model group
(all P<0.05) (Fig. 3E and
F). In addition, the expression
level of the anti-apoptotic protein, Bcl-2 was significantly
decreased (P<0.05). Compared with that in the model group, HSYA
significantly inhibited the protein expression levels of Bax,
cleaved-caspase-3 and cleaved-caspase-9 in the TBHP-induced NP
cells, and increased the protein expression levels of Bcl-2 (all
P<0.05).
Effect of HSYA on ECM-associated
proteins
The expression levels of collagen-related proteins
were detected using western blot analysis (Fig. 4A). Statistical quantification
(Fig. 4B-D) revealed that the
expression levels of collagen II and aggrecan protein, which were
the markers of NP cells in the model group, were significantly
lower than those in control group (both P<0.05). In addition,
the protein expression levels of MMP-13, a protein related to
collagen breakdown (23), were
significantly upregulated (P<0.05). Compared with that in the
model group, treatment with HSYA increased the protein expression
levels of collagen II and aggrecan in the NP cells, while
decreasing the protein expression level of MMP-13, and the
difference was statistically significant (all P<0.05).
HSYA regulates the expression level of
collagen-associated proteins in NP cells via CA XII
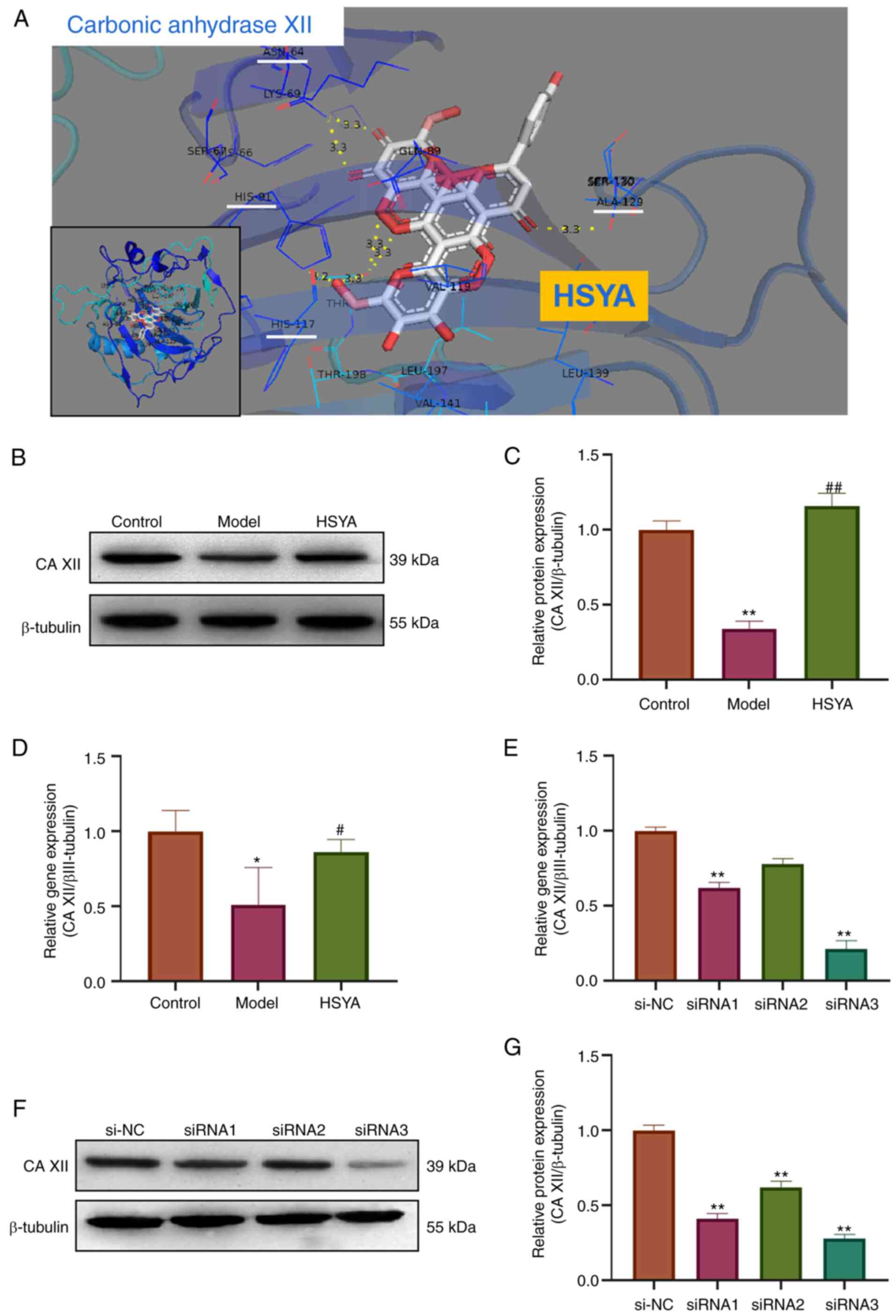
Using bioinformatics analysis and Pymol molecular
simulation docking software, the interaction between HSYA and CA
XII was analyzed (Fig. 5A). The
docking results showed that HSYA had a binding site for CA XII.
There were six binding bond energies between HSYA and CA XII
protein amino acid residues (HIS-117, HIS-91, ALA129 and ASN-64),
with a binding energy of -10.7 kcal/mol, providing a stable spatial
structure domain. The protein expression level of CA XII in the
treated NP cells was subsequently detected using western blot
analysis (Fig. 5B). The
densitometry results showed that compared with that in the control
group, the protein expression level of CA XII in the model group
was significantly inhibited, while the protein expression level of
CA XII in the HSYA group was significantly higher compared with
that in the model group (all P<0.05) (Fig. 5C). The mRNA expression level of the
CA XII was also detected using RT-qPCR and the results showed that
the mRNA expression level of CA XII in the model group was
decreased (compared with that in the control group), while the gene
expression level of CA XII in the HSYA group was significantly
higher compared with that in the model group (all P<0.05)
(Fig. 5D). RT-qPCR was also used
to investigate the transfection efficiency of siRNA in inhibiting
CA XII expression and the results showed that siRNA3 inhibited the
expression level more significantly, with a inhibition rate of 77%
(Fig. 5E). These results were
confirmed using western blot analysis (Fig. 5F and G). Therefore, siRNA3 was used in
subsequent experiments.
The results from western blot analysis, in Fig. 6A and B, showed that si-CA XII could reverse the
increase in CA XII protein expression by HSYA. Immunocytochemistry
results demonstrated that Aggrecan (Fig. 6C) and collagen II (Fig. 6D) proteins in the HSYA group were
higher than those in the Model group, while MMP-13 (Fig. 6E) proteins were lower than those in
the TBHP group. However, si-CA12 was able to reverse the effects of
HSYA on aggrecan, collagen II and MMP-13 protein expression.
Further validation was performed using western blot analysis
(Fig. 6F, and the results were
consistent with immunocytochemistry. si-CA XII could reverse the
increase of aggrecan and collagen II protein expression by HSYA, as
well as the inhibitory effect on MMP-13 protein expression (all
P<0.05) (Fig. 6G-I).
Discussion
Currently, pharmacological treatment strategies for
degenerative disc disease aim to relieve the patients' symptoms.
However, it cannot control or reverse the progression of disc
degeneration and it produces some side effects (3,24).
In the late stages of IVDD, conservative management requires
surgical treatment and is accompanied by various surgical risks
(25). Excessive apoptosis of disc
cells, particularly NP cells, leads to changes in disc structure
and function, and is one of the most important pathophysiological
mechanisms in the progression of IVDD (7). As a normal byproduct of oxygen
metabolism formation, ROS plays a crucial role in homeostasis and
cellular signal transduction (26). It has been reported that excess
ROS, in the pathogenesis of IVDD, leads to NP cell apoptosis and
ECM degradation. Therefore, in the present study, the effect of
HSYA on IVDD disease progression was investigated using TBHP
oxidative stress injury induction and ROS accumulation. It was
found that HSYA could reduce TBHP-induced ROS levels in the NP
cells, while increasing the concentrations of the antioxidant
enzymes, SOD, CAT and GSH-Px, decrease the expression levels of
pro-apoptotic proteins, Bax, cleaved caspase-3 and cleaved
caspase-9, and decrease apoptosis in the NP cells. The
investigation of HSYA in oxidative stress and anti-apoptosis has
been reported in other types of diseases. For example, HSYA reduced
oxidative stress and reversed the increase in Bax and caspase-3
from nephropathy in type 2 diabetic rats, and significantly
increased the protein expression of Bcl-2 in renal tissue (15). In addition, in traumatic brain
injury in rats, HSYA protected neural cells against oxidative
stress and anti-apoptosis by inhibiting the protein expression
level of Bax, caspase-3, and caspase-9, while increasing
antioxidant enzyme concentrations (14). All of the aforementioned studies
confirmed that HSYA has an essential role in anti-oxidative stress
and anti-apoptosis.
Furthermore, the degradation of the ECM is also an
important factor driving the progression of IVDD (27). The ECM is a non-cellular 3D
macromolecular network composed of collagen,
proteoglycans/glycosaminoglycans, elastin, fibronectin, laminin and
several other glycoproteins (28).
While collagen II and aggrecan are important in disc NP cells,
their presence provides critical properties for the disc (29). Proteoglycan and type II collagen
content in the ECM is reduced when the disc degenerates (30). In addition, when the surrounding
biochemical conditions change, NP cells lose their normal phenotype
and turn into mast cells, and replace type II collagen with type X
collagen. A previous study confirmed that TBHP could induce ECM
metabolic imbalance in the NP cells via the production of large
amounts of ROS (31). The in
vitro study showed that HSYA significantly increased collagen
II and aggrecan protein expression levels in the disc matrix. In
addition, ECM metabolism in the NP cells is a dynamic process,
including ECM anabolism and ECM catabolism. MMP-13 was shown to be
one of the major catabolic factors of ECM in disc degeneration
(32,33). The synthesis and degradation of the
ECM maintains a dynamic balance in healthy discs, particularly in
NP cells (1). However, oxidative
stress disrupts this homeostasis by increasing the secretion and
expression of proteases, such as MMPs and ADAMTs, and
downregulating the expression of ECM proteins, such as collagen II
and aggrecan (34). Therefore, the
present results demonstrated that HSYA promoted the ability of NP
cells to synthesize ECM components, inhibit the production of
MMP-13 and reduce the catabolic level of ECM. To the best of our
knowledge, we hypothesize for the first time that HSYA plays an
important role in the balance between synthesis and metabolism of
ECM in NP cells.
HSYA has antioxidant and anti-apoptotic effects on
NP cells, and can regulate the balance of ECM synthesis and
metabolism. SEA, Pharmmappe and Moltarpred databases were used to
predict HSYA target proteins. Drugbank, TTD and Genecards screened
the critical proteins of disc degeneration, and those that were
predicted were selected as target proteins. CA IX and CA XII were
filtered from the predicted data, after which Pymol software was
used to predict the binding energy of the two proteins and models,
respectively. The results revealed that the highest binding energy
was between HSYA and CA12. The predicted binding energy value
between small molecule and protein reached-10.7 kcal/mol, which
provided better conformational stability. CA XII is a transmembrane
zinc metalloenzyme, from the carbonic anhydrase family that
catalyze the reversible hydration of carbon dioxide into
bicarbonate and hydrogen ions (35,36).
CA XII is considered as a marker gene of human NP cells (20,35)
and is enriched in human NP tissue compared with that in other
tissues from the disc, including fibrosis and cartilage (20,37).
The study by Chen et al (18) revealed that CA XII gene and protein
expression were decreased in patients with disc degeneration, while
increase of CA XII expression could reduce apoptosis of NP cells
and prevent degenerative disc disease. In addition, they also found
that overexpression of CA XII could rescue IL-1β-stimulated
endplate chondrocyte apoptosis, both in vitro and in
vitro (18). Furthermore, CA
XII could also protect inner plate cartilage from the degradation
of ECM and collagen regulated by the IGF-1/PI3K/CREB signaling
pathway (38). CA XII inhibition
may not only disrupt the pH, which leads to lactate accumulation in
NP cells, but also reduce the protein expression level of collagen
II and aggrecan (20).
Therefore, RT-qPCR and western blot analysis was
used to detect the gene and protein expression levels of CA XII in
the NP cells. The results showed that CA XII gene and protein
expression levels were significantly inhibited in the model group,
while administration of HSYA could increase the gene and protein
expression levels of CA XII. Therefore, the NP cells were
transfected with CA XII siRNA. The results confirmed that
transfection of CA XII siRNA could significantly reverse the
increase of collagen II and aggrecan protein expression caused by
HSYA in the NP cells, and weaken the inhibitory effect of HSYA on
MMP-13 protein expression. The results were consistent with those
from Chen et al (18) and
Zhao et al (38).
Therefore, we hypothesized that CA XII may be an important target
for HSYA to exert the regulation of ECM homeostasis. However, how
CA XII plays a specific role in disc degeneration requires further
investigation, and there are few reports in the relevant
literature. This will be the focus of subsequent research.
In summary, the present study showed that HSYA can
reverse TBHP-induced NP cell apoptosis and ECM degradation. In
addition, HSYA targets were predicted using bioinformatics analysis
and validated using siRNA targeting CA XII. Therefore, HSYA could
regulate the synthesis and metabolic balance of ECM by increasing
the protein expression level of CA XII.
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
All datasets used and/or analyzed during the current
study are available upon reasonable request from the corresponding
author.
Authors' contributions
SY preformed the experiments, analyzed the data and
drafted the manuscript. SY and WL conceived and designed the study.
WL reviewed and edited the manuscript. All authors have read and
approved the final manuscript. SY and WL confirm the authenticity
of all the raw data.
Ethics approval and consent to
participate
The animal experiments of the current study were
approved by the Ethics Committee of Affiliated Hospital of Zunyi
Medical University (approval no. KLL-2020-317).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Zhang S, Liang W, Abulizi Y, Xu T, Cao R,
Xun C, Zhang J and Sheng W: Quercetin Alleviates Intervertebral
disc degeneration by modulating p38 MAPK-Mediated autophagy. Biomed
Res Int. 2021(6631562)2021.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Trefilova VV, Shnayder NA, Petrova MM,
Kaskaeva DS, Tutynina OV, Petrov KV, Popova TE, Balberova OV,
Medvedev GV and Nasyrova RF: The role of polymorphisms in
Collagen-encoding genes in intervertebral disc degeneration.
Biomolecules. 11(1279)2021.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Patil P, Niedernhofer LJ, Robbins PD, Lee
J, Sowa G and Vo N: Cellular senescence in intervertebral disc
aging and degeneration. Curr Mol Biol Rep. 4:180–190.
2018.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Wu X, Song Y, Liu W, Wang K, Gao Y, Li S,
Duan Z, Shao Z, Yang S and Yang C: IAPP modulates cellular
autophagy, apoptosis, and extracellular matrix metabolism in human
intervertebral disc cells. Cell Death Discov.
3(16107)2017.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Chen S, Qin L, Wu X, Fu X, Lin S, Chen D,
Xiao G, Shao Z and Cao H: Moderate fluid shear stress regulates
Heme Oxygenase-1 expression to promote autophagy and ECM
homeostasis in the nucleus pulposus cells. Front Cell Dev Biol.
8(127)2020.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Gonzales S, Wang C, Levene H, Cheung HS
and Huang CC: ATP promotes extracellular matrix biosynthesis of
intervertebral disc cells. Cell Tissue Res. 359:635–642.
2015.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Clouet J, Fusellier M, Camus A, Le Visage
C and Guicheux J: Intervertebral disc regeneration: From cell
therapy to the development of novel bioinspired endogenous repair
strategies. Adv Drug Deliv Rev. 146:306–324. 2019.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Pizzino G, Irrera N, Cucinotta M, Pallio
G, Mannino F, Arcoraci V, Squadrito F, Altavilla D and Bitto A:
Oxidative stress: Harms and benefits for human health. Oxid Med
Cell Longev. 2017(8416763)2017.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Singh A, Kukreti R, Saso L and Kukreti S:
Oxidative Stress: A key modulator in neurodegenerative diseases.
Molecules. 24(1583)2019.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Johnson J, Jaggers RM, Sreejit G, Dahdah
A, Murphy AJ, Hanssen NMJ and Nagareddy P: Oxidative stress in
neutrophils: Implications for diabetic cardiovascular
complications. Antioxid Redox Signal: Jun 19, 2021 (Epub ahead of
print).
|
|
11
|
Butterfield DA and Boyd-Kimball D:
Oxidative stress, Amyloid-β peptide, and altered key molecular
pathways in the pathogenesis and progression of Alzheimer's
disease. J Alzheimers Dis. 62:1345–1367. 2018.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Masuda T, Shimazawa M and Hara H: Retinal
diseases associated with oxidative stress and the effects of a free
radical scavenger (Edaravone). Oxid Med Cell Longev.
2017(9208489)2017.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Krupkova O, Handa J, Hlavna M, Klasen J,
Ospelt C, Ferguson SJ and Wuertz-Kozak K: The natural polyphenol
epigallocatechin gallate protects intervertebral disc cells from
oxidative stress. Oxid Med Cell Longev.
2016(7031397)2016.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Xu J, Zhan T, Zheng W, Huang YK, Chen K,
Zhang XH, Ren P and Huang X: Hydroxysafflor yellow A acutely
attenuates blood-brain barrier permeability, oxidative stress,
inflammation and apoptosis in traumatic brain injury in rats1. Acta
Cir Bras. 35(e351202)2021.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Lee M, Zhao H, Liu X, Liu D, Chen J, Li Z,
Chu S, Kou X, Liao S, Deng Y, et al: Protective effect of
hydroxysafflor yellow A on nephropathy by attenuating oxidative
stress and inhibiting apoptosis in induced type 2 diabetes in rat.
Oxid Med Cell Longev. 2020(7805393)2020.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Hu ZC, Xie ZJ, Tang Q, Li XB, Fu X, Feng
ZH, Xuan JW, Ni WF and Wu AM: Hydroxysafflor yellow A (HSYA)
targets the NF-κB and MAPK pathways and ameliorates the development
of osteoarthritis. Food Funct. 9:4443–4456. 2018.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Zhang Z, Wang C, Lin J, Jin H, Wang K, Yan
Y, Wang J, Wu C, Nisar M, Tian N, et al: Therapeutic potential of
naringin for intervertebral disc degeneration: Involvement of
autophagy against oxidative stress-induced apoptosis in nucleus
pulposus cells. Am J Chin Med: Oct 4, 2018 (Epub ahead of
print).
|
|
18
|
Chen S, Fang XQ, Wang Q, Wang SW, Hu ZJ,
Zhou ZJ, Xu WB, Wang JY, Qin A and Fan SW: PHD/HIF-1 upregulates
CA12 to protect against degenerative disc disease: A human sample,
in vitro and ex vivo study. Lab Invest. 96:561–569. 2016.PubMed/NCBI View Article : Google Scholar
|
|
19
|
van den Akker GG, Surtel DA, Cremers A,
Rodrigues-Pinto R, Richardson SM, Hoyland JA, van Rhijn LW, Welting
TJ and Voncken JW: Novel immortal human cell lines reveal
subpopulations in the nucleus pulposus. Arthritis Res Ther.
16(R135)2014.PubMed/NCBI View
Article : Google Scholar
|
|
20
|
Power KA, Grad S, Rutges JP, Creemers LB,
van Rijen MH, O'Gaora P, Wall JG, Alini M, Pandit A and Gallagher
WM: Identification of cell surface-specific markers to target human
nucleus pulposus cells: Expression of carbonic anhydrase XII varies
with age and degeneration. Arthritis Rheum. 63:3876–3886.
2011.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Bayne K: Revised guide for the care and
use of laboratory animals available. American Physiological
Society. Physiologist. 39:199. 208–211. 1996.PubMed/NCBI
|
|
22
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Hu W, Zhang W, Li F, Guo F and Chen A:
Bortezomib prevents the expression of MMP-13 and the degradation of
collagen type 2 in human chondrocytes. Biochem Biophys Res Commun.
452:526–530. 2014.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Lyu FJ, Cheung KM, Zheng Z, Wang H, Sakai
D and Leung VY: IVD progenitor cells: A new horizon for
understanding disc homeostasis and repair. Nat Rev Rheumatol.
15:102–112. 2019.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Swann MC, Hoes KS, Aoun SG and McDonagh
DL: Postoperative complications of spine surgery. Best Pract Res
Clin Anaesthesiol. 30:103–120. 2016.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Forrester SJ, Kikuchi DS, Hernandes MS, Xu
Q and Griendling KK: Reactive oxygen species in metabolic and
inflammatory signaling. Circ Res. 122:877–902. 2018.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Ge J, Wang Y, Yan Q, Wu C, Yu H, Yang H
and Zou J: FK506 Induces the TGF-β1/Smad 3 pathway independently of
calcineurin inhibition to prevent intervertebral disk degeneration.
Front Cell Dev Biol. 8(608308)2020.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Theocharis AD, Skandalis SS, Gialeli C and
Karamanos NK: Extracellular matrix structure. Adv Drug Deliv Rev.
97:4–27. 2016.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Guo S, Cui L, Xiao C, Wang C, Zhu B, Liu
X, Li Y, Liu X, Wang D and Li S: The Mechanisms and functions of
GDF-5 in intervertebral disc degeneration. Orthop Surg. 13:734–741.
2021.PubMed/NCBI View
Article : Google Scholar
|
|
30
|
Xu J, E XQ, Wang NX, Wang MN, Xie HX, Cao
YH, Sun LH, Tian J, Chen HJ and Yan JL: BMP7 enhances the effect of
BMSCs on extracellular matrix remodeling in a rabbit model of
intervertebral disc degeneration. FEBS J. 283:1689–1700.
2016.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Gan JC, Ducheyne P, Vresilovic EJ, Swaim W
and Shapiro IM: Intervertebral disc tissue engineering I:
Characterization of the nucleus pulposus. Clin Orthop Relat Res.
411:305–314. 2003.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Le Maitre CL, Freemont AJ and Hoyland JA:
Localization of degradative enzymes and their inhibitors in the
degenerate human intervertebral disc. J Pathol. 204:47–54.
2004.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Wang J, Hu J, Chen X, Huang C, Lin J, Shao
Z, Gu M, Wu Y, Tian N, Gao W, et al: BRD4 inhibition regulates
MAPK, NF-κB signals, and autophagy to suppress MMP-13 expression in
diabetic intervertebral disc degeneration. FASEB J. 33:11555–11566.
2019.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Chen Y, Zheng Z, Wang J, Tang C, Khor S,
Chen J, Chen X, Zhang Z, Tang Q, Wang C, et al: Berberine
suppresses apoptosis and extracellular matrix (ECM) degradation in
nucleus pulposus cells and ameliorates disc degeneration in a
rodent model. Int J Biol Sci. 14:682–692. 2018.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Waheed A and Sly WS: Carbonic anhydrase
XII functions in health and disease. Gene. 623:33–40.
2017.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Hynninen P, Hämäläinen JM, Pastorekova S,
Pastorek J, Waheed A, Sly WS, Tomas E, Kirkinen P and Parkkila S:
Transmembrane carbonic anhydrase isozymes IX and XII in the female
mouse reproductive organs. Reprod Biol Endocrinol.
2(73)2004.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Minogue BM, Richardson SM, Zeef LA,
Freemont AJ and Hoyland JA: Characterization of the human nucleus
pulposus cell phenotype and evaluation of novel marker gene
expression to define adult stem cell differentiation. Arthritis
Rheum. 62:3695–3705. 2010.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Zhao X, Shen P, Li H, Yang Y, Guo J, Chen
S, Ma Y, Sheng J, Shen S, Liu G and Fang X: Carbonic Anhydrase 12
protects endplate cartilage from degeneration regulated by
IGF-1/PI3K/CREB signaling pathway. Front Cell Dev Biol.
8(595969)2020.PubMed/NCBI View Article : Google Scholar
|