Introduction
Laryngeal squamous cell carcinoma (LSCC) is one of
the most common types of malignant tumors in the larynx (1). According to Global Cancer Statistics
2018, there are 94,771 deaths from laryngeal carcinoma worldwide,
which presents 1% of all cancer types (2). Due to rapid progression, invasive
growth, frequent lymph node and distant metastasis and poor
prognoses, LSCC severely affects the quality of life (3,4).
However, the molecular mechanism and pathogenesis underlying LSCC
remain poorly understood. Therefore, understanding the molecular
mechanisms that are involved in LSCC development and progression,
in addition to the identification of novel therapeutic targets are
of utmost importance.
Long non-coding RNAs (lncRNAs) belong to a family of
RNAs that is >200 nucleotides in length and can function as
either oncogenes or tumor suppressors (5-7).
Accumulating evidence has suggested that lncRNAs serve a role in
cell proliferation, metastasis, apoptosis and angiogenesis in LSCC
(8,9). Elucidation of the role of lncRNAs in
LSCC may provide novel insights into the occurrence and progression
of LSCC. lncRNA nuclear enriched abundant transcript 1 (NEAT1)
expression was found to be upregulated in LSCC tissues and cell
lines, which negatively correlated with miR-107 expression in LSCC
tissues (10). NEAT1 was
subsequently reported to promote the proliferation and cell cycle
arrest at the G1 phase, and inhibit the apoptosis of
LSCC cells by regulating the microRNA (miR)-107/cyclin-dependent
kinase (CDK) 6 pathway (10). In
addition, lncRNA antisense non-coding RNA in the INK4 locus has
been reported to server a role in the proliferation, apoptosis,
invasion and migration of LSCC cells by sponging miR-181a (11).
Myocardial infarction associated transcript (MIAT)
is a novel lncRNA located on chromosome 22q12 that was originally
identified to be involved in the pathogenesis myocardial infarction
(12). Previous studies indicate
that lncRNA MIAT serves an important role in the progression of
various types of cancer (13-16).
A study in gastric cancer suggested that MIAT knockdown inhibited
gastric cancer cell growth and metastasis both in vitro and
in vivo (13). Another study
previously reported that the expression of MIAT in non-small-cell
lung cancer (NSCLC) tissues was upregulated, whereby knockdown of
MIAT substantially inhibited the invasive ability of NSCLC cells
(14). Additionally, Liu et
al (16) documented that the
level of MIAT expression was upregulated in colorectal cancer (CRC)
tissues and cells, such that MIAT knockdown inhibited
proliferation, migration and invasion whilst enhancing apoptosis in
CRC cells (16). However, to date,
the precise function of lncRNA MIAT in LSCC remains poorly
understood. Therefore, the present study aimed to investigate the
expression and roles of MIAT in LSCC tumorigenesis. The current
study detected the expression of MIAT in LSCC tissues and cells
through reverse transcription-quantitative PCR (RT-qPCR). The
proliferation, migration and invasion of cells was assessed by MTT,
wound healing and Transwell assays, respectively. Furthermore,
experiments were performed to clarify the mechanism of MIAT in LSCC
progression.
Materials and methods
Patient tissue samples and LSCC cell
lines
A total of 32 pairs of human LSCC tissue samples and
corresponding adjacent non-tumor tissues (>2 cm away from the
tumor site) were collected from the Tianjin Union Medical Center
(Tianjin, China) between June 2015 and January 2018. The patients
included 26 males and 6 females, with an average age of 59 years
(59±7.8). The categories of all LSCC tissues was confirmed using
pathological analysis according to the WHO pathology and genetic
classification of tumors of the head and neck (17). The tissue samples were immediately
frozen in liquid nitrogen and then stored at -80˚C for further
research. The patients were diagnosed with LSCC without
radiotherapy or chemotherapy prior to surgery. The exclusion
criteria for patients included hepatic and renal insufficiency,
immune deficiency and other systemic malignancies. All patients
signed informed consent prior to the use of their tissues for the
present study according to the principles of the Declaration of
Helsinki. The present study was approved by the Ethics Committee of
Tianjin Union Medical Center.
LSCC cell lines TU-177 and AMC-HN-8 and the normal
human keratinocyte cell line HaCaT were purchased from the American
Type Culture Collection. HaCaT cells are immortalized human
epidermal cells that has been previously used as the control cell
line for laryngeal squamous cell carcinoma cells (18,19).
The LSCC cell line TU686 was purchased from the Cell Center of Life
Science of Chinese Academy of Science.
Cell culture and transfection
All cells were cultured in DMEM (Invitrogen; Thermo
Fisher Scientific, Inc.) supplemented with 10% FBS (Gibco; Thermo
Fisher Scientific, Inc.), 100 IU/ml of penicillin and 100 µg/ml of
streptomycin at 37˚C in a humidified atmosphere with 5%
CO2.
Small interfering RNA (siRNA) containing the
specific MIAT interference sequence (si-MIAT,
5'-CCAGGCUCCUUUAAACCAATT-3') and negative control (si-NC,
5'-UUCUCCGAACGUGUCA-3') were purchased from Shanghai GeneChem Co.,
Ltd. miR-613 mimics (5'-AGGAAUGUUCCUUCUUUGCC-3'), miR-NC
(5'-UUCUCCGAACGUGUCACGUTT-3'), anti-miR-613
(5'-GGCAAAGAAGGAACAUUCCT-3') and anti-miR-NC
(5'-CAGUACUUUUGUGUAGUACAA-3') were purchased from Guangzhou Ribobio
Co., Ltd. A total of 100 nM si-MIAT, 100 nM si-NC, 50 nM miR-613
mimics, 50 nM anti-miR-613 and 50 nM of their corresponding
negative controls were transfected into LSCC cells using the
Lipofectamine® 2000 reagent (Invitrogen; Thermo Fisher
Scientific, Inc.) following the manufacturer's protocol. After 48 h
of transfection, the cells were collected for subsequent
experimentation.
RT-qPCR
Total RNA was extracted from the cultured cells or
fresh surgical tissue samples using TRIzol® reagent
(Invitrogen; Thermo Fisher Scientific, Inc.) according to the
manufacturer's protocols. Subsequently, cDNA was synthesized using
the PrimeScript™ RT Reagent kit (Takara Bio, Inc.). The temperature
protcol was as follows: 15 min at 42˚C and 5 sec at 85˚C. qPCR was
performed using the All-in-One™ miRNA qRT-PCR detection kit
(GeneCopoeia, Inc.) for miR-613, where U6 was used as an endogenous
control. The expression level of MIAT mRNA was detected using the
SYBR Premix Ex Taq™ ІІ kit (cat. no. DRR081A; Takara Bio, Inc.) in
accordance with manufacturer's protocol, where GAPDH was used as an
endogenous control. The thermocycling conditions were as follows:
30 sec at 95˚C, followed by 40 cycles for 5 sec at 95˚C and 35 sec
at 60˚C. The sequences of the primers were as follows: MIAT
forward, 5'-TCTTCATGTCAGAACACGCTTTA-3' and reverse,
5'-AAGGTCACCCGAGGTCCAA-3'; GAPDH forward,
5'-AGGTGAAGGTCGGAGTCAACG-3' and reverse,
5'-AGGGGTCATTGATGGCAACA-3'; miR-613 forward,
5'-CGCAGCACACCTGCTTTTTG-3' and reverse, 5'-AGAGATTCGGGTCGATGCTC-3'
and U6 forward, 5'-CTCGCTTCGGCAGCACA-3' and reverse,
5'-AACGCTTCACGAATTTGCGT-3'. The relative expression levels of
detective genes were calculated using the 2-ΔΔCt method
(20).
MTT assay
MTT assay was used to examine cell viability. TU-177
and AMC-HN-8 cells (5x103/well) were seeded into 96-well
plates and allowed to grow for 12, 24, 48 and 72 h at 37˚C.
Subsequently, 20 µl MTT (5 mg/ml; Sigma-Aldrich; Merck KGaA) were
added into each well and the plates were incubated at 37˚C for
additional 4 h. The optical density (OD) value of each well was
detected using an iMark microplate absorbance reader (Bio-Rad
Laboratories, Inc.) at 570 nm. All the experiments were performed
three times.
Colony-formation assay
The LSCC cells were seeded into six-well plates at a
density of 500 cells per well following transfection for 48 h. The
DMEM medium containing 10% FBS was replaced every 3 days. The cells
were then cultured in complete medium for 2 weeks. After cell
colonies formed, the medium was removed. The cells were fixed with
4% paraformaldehyde at 4˚C for 1 h and stained with 0.1% crystal
violet staining solution for 20 min at room temperature. Finally,
the colony number in each well (colony comprising >50 cells) was
counted under a light microscope (Nikon Corporation; magnification,
x10) and the colony-formation rate was calculated.
Wound healing assay
TU-177 and AMC-HN-8 cells were first seeded into
12-well plates and grown until 90% confluence. The cellular
monolayer was then wounded using a sterilized 200-µl pipette tip
and the cells were incubated at 37˚C in serum-free medium.
Microscopic images of the cultures were acquired at 0 and 48 h
under a light microscope (magnification, x100), following which
wound closure was quantified and then assessed from the images
using Scion Image software 4.0.2 (Scion Corporation) and Adobe
Photoshop Program 7.0 (Adobe Corporation) (21).
Transwell invasion assay
For cell invasion assay, the insert (8.0 µm pore
size; BD Biosciences) was coated with 0.2 mg/ml Matrigel
(Sigma-Aldrich; Merck KGaA) at 4˚C for 3-4 h. The TU-177 and
AMC-HN-8 cells (5x104 cells/well) were first plated into
the upper chamber with serum-free medium, whilst the lower chamber
was filled with 600 µl medium containing 10% FBS. Following
incubation at 37˚C for 48 h, the cells that invaded to the lower
filter and then migrated through the membrane were fixed with 75%
ethanol for 30 min and stained with 0.1% crystal violet at room
temperature. Finally, the number of invaded cells was counted
randomly in five fields of each membrane and imaged under a light
microscope (Nikon Corporation; magnification, x100).
Luciferase reporter assay
Firstly, Starbase v2.0 (http://starbase.sysu.edu.cn) was used to predict the
target miRNAs of lncRNA MIAT. Subsequently, the sequences of MIAT
containing the binding sites of miR-613 were amplified and cloned
into the psiCHECK-2 vector (Promega Corporation) and named
MIAT-wild type (Wt). The putative common fragments were replaced
and named MIAT-mutant (Mut). The TU-177 and AMC-HN-8 cells were
seeded at a density of 5x104 into 24-well plates and
then co-transfected with luciferase reporter vectors comprising
MIAT-Wt or MIAT-Mut (0.4 µg), miR-613 mimics (50 nmol/l) or miR-NC
(50 nmol/l) and pRL-TK plasmid (50 ng; Promega Corporation) using
Lipofectamine® 2000 reagent (Invitrogen; Thermo Fisher
Scientific, Inc.). The pRL-TK Renilla luciferase plasmid was used
as an internal control for normalization. Following transfection
for 48 h, the protein-extraction reagent RIPA Lysis buffer (Gibco;
Thermo Fisher Scientific, Inc.) was used to lyse the cells. The
luciferase activities were then detected using the Dual-Luciferase
Reporter Assay System (Promega Corporation).
RNA immunoprecipitation (RIP)
assay
RNA immunoprecipitation (RIP) assay was performed to
further confirm the direct association between lncRNA MIAT and
miR-613. Magna RIP™ RNA Immunoprecipitation kit (EMD Millipore) was
used by following the manufacturer's protocols. Briefly, the LSCC
cells were transfected with the miR-613 mimics or miR-NC (50 nM)
for 48 h before being lysed using the RIP lysis buffer (EMD
Millipore). A total of 50 µl magnetic beads were resuspended in 100
µl RIP washing buffer and swirled. After the supernatant had been
discarded, magnetic beads were resuspended with 100 µl RIP washing
buffer, and 5 µg antibodies were added. The magnetic bead-antibody
complex was washed and resuspended in 900 µl RIP washing buffer
before incubation with cell lysates. The cell lysates were then
incubated with the RIP buffer containing magnetic bead-antibody
complex at 4˚C overnight. Finally, co-precipitated magnetic
bead-protein complex and input were separately detached with
proteinase K to extract RNA for subsequent RT-qPCR detection of
MIAT. The antibodies used for RIP were rabbit anti-human Ago2
(1:500; Abcam; cat. no. ab186733) and rabbit anti-human IgG (1:500;
Abcam; cat. no. ab109489).
Statistical analysis
Statistical analysis was performed using GraphPad
Prism 6.0 (GraphPad Software, Inc.) and SPSS version 20.0 software
(IBM, Corp.). All experiments were independently repeated in
triplicate. Data are presented as the means ± standard deviation.
Differences among multiple groups were analyzed using one-way ANOVA
followed by Tukey's test, whilst differences between two groups
were analyzed using Student's t-test. Correlation between MIAT and
miR-613 expression was estimated using Pearson's correlation
coefficient. P<0.05 was considered to indicate a statistically
significant difference.
Results
lncRNA MIAT expression is upregulated
in LSCC tissues and cell lines
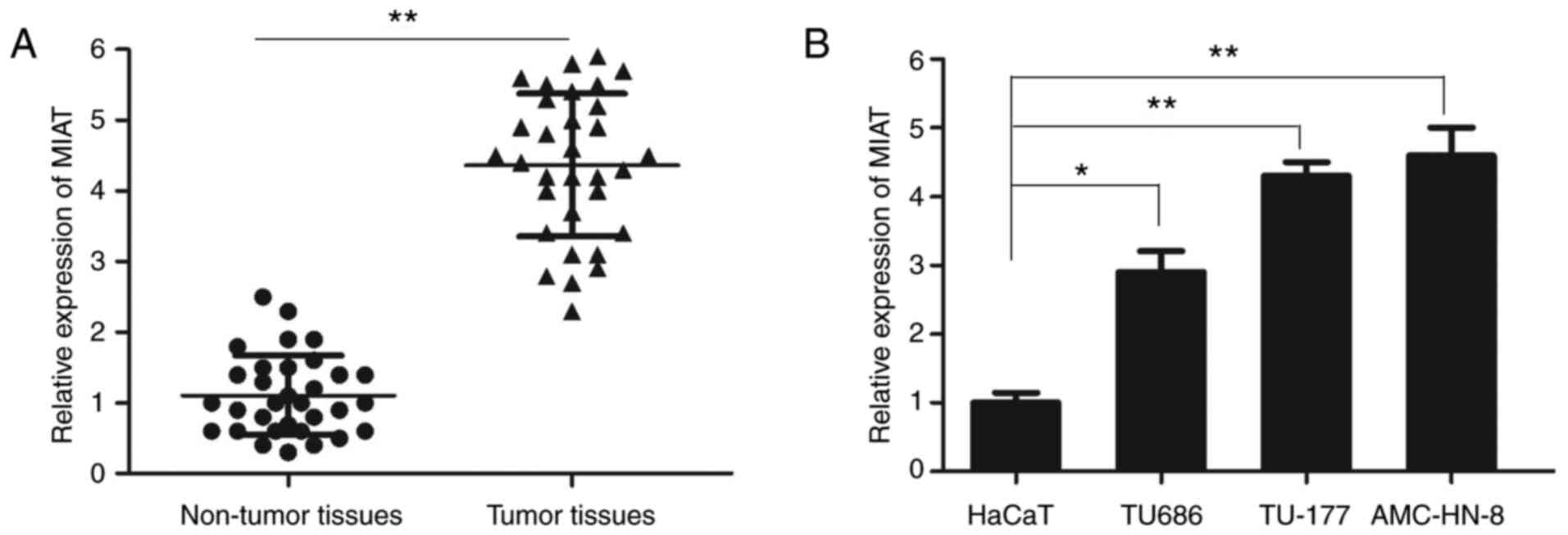
The expression of lncRNA MIAT in LSCC tissues and
cell lines was measured using RT-qPCR. The relative expression of
lncRNA MIAT in LSCC tissue samples was significantly increased
compared with that in the adjacent non-tumor tissues (Fig. 1A). In addition, the expression
levels of lncRNA MIAT were significantly higher in the LSCC cell
lines TU686, TU-177 and AMC-HN-8 compared with those in the control
HaCaT cell line (Fig. 1B). As the
relative level of MIAT was higher in TU-177 and AMC-HN-8 cells,
these cell lines were selected for further experimentation.
Knockdown of MIAT inhibits the
proliferation of LSCC cells
To investigate the effects of lncRNA MIAT on the
viability of LSCC cells further, small interfering RNA specifically
targeting MIAT (si-MIAT) or si-NC were transfected into the TU-177
and AMC-HN-8 cells. Transfection efficiency was confirmed by
RT-qPCR (Fig. 2A), where the
expression levels of MIAT were significantly decreased in the two
LSCC cell lines transfected with si-MIAT compared with those in the
si-NC group. The cell viability curves as determined by MTT assay
revealed that following MIAT knockdown, the viabilities of the
TU-177 and AMC-HN-8 cell lines were significantly inhibited at 72 h
(Fig. 2B). In addition, it was
found that knocking down MIAT expression significantly reduced the
number of colonies formed by the TU-177 and AMC-HN-8 cells
(Fig. 2C).
Knockdown of MIAT inhibits the
migration and invasion of LSCC cells
The migratory and invasive abilities of LSCC cells
was next examined using wound healing and Transwell invasion
assays, respectively. The migratory ability of TU-177 and AMC-HN-8
cells was significantly decreased following MIAT knockdown
(Fig. 3A). It was also found that
MIAT knockdown significantly reduced the number of invasive TU-177
and AMC-HN-8 cells (Fig. 3B).
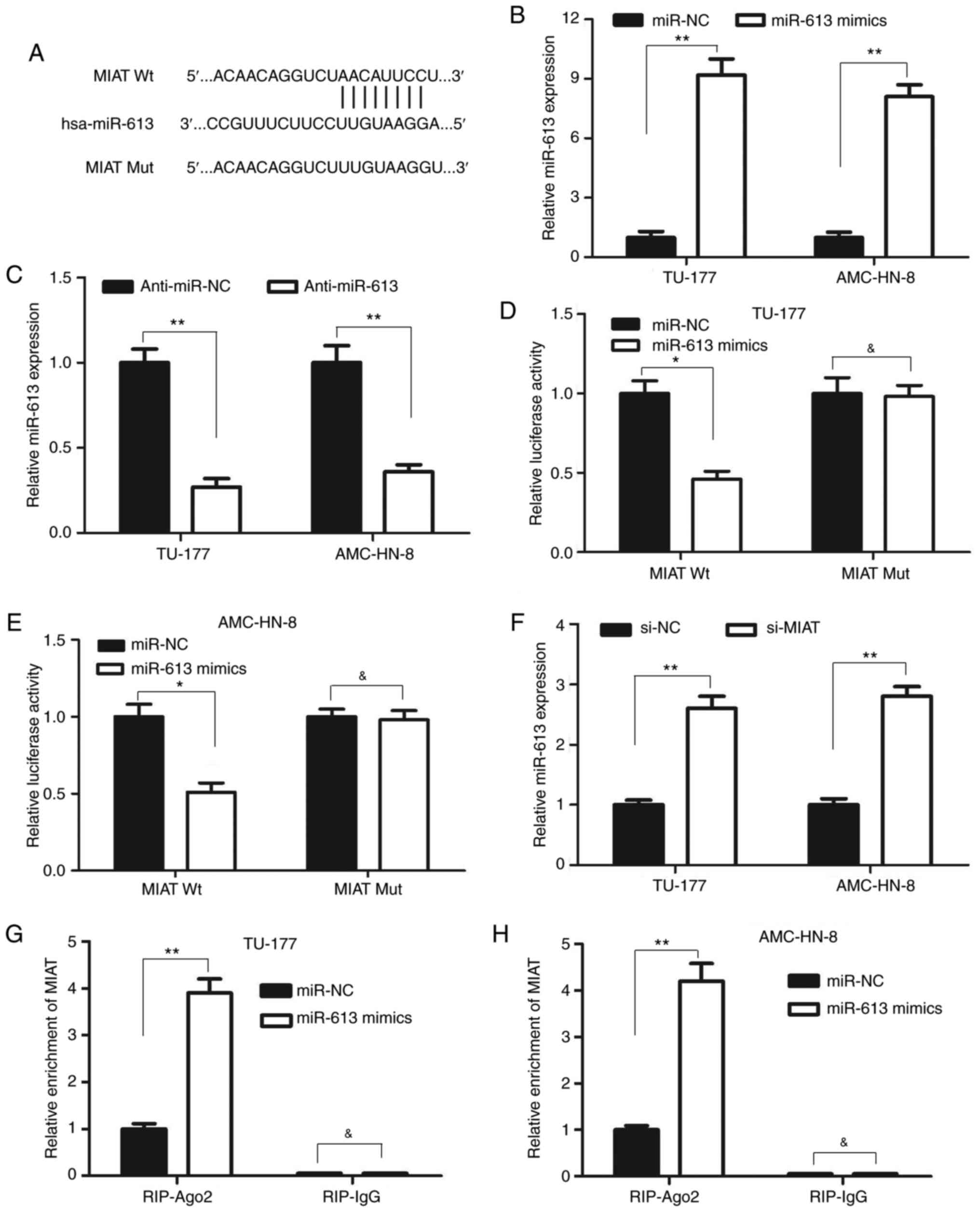
lncRNA MIAT directly binds miR-613 in
LSCC cells
To further elucidate the molecular mechanisms of
action of lncRNA MIAT in regulating the biological behaviors of
LSCC cells, the binding sites of MIAT were predicted using Starbase
v2.0 (https://starbase.sysu.edu.cn).
Previous studies have demonstrated that miR-613 is a tumor
suppressor that can inhibit cell proliferation and invasion in a
number of types of cancer, including LSCC (22-24).
Therefore, the present study focused on miR-613 among these miRNAs
that were predicted to interact with MIAT. lncRNA MIAT was found to
contain a complimentary binding site of miR-613 (Fig. 4A). The expression level of miR-613
was also significantly upregulated in both TU-177 and AMC-HN-8
cells after transfection with miR-613 mimics compared with that
after transfection with miR-NC (Fig.
4B). By contrast, the level of miR-613 expression was
significantly downregulated after transfection with anti-miR-613 in
both TU-177 and AMC-HN-8 cells mimics compared with that after
transfection with anti-miR-NC (Fig.
4C). Subsequently, the targeted effects of miR-613 on MIAT were
validated using luciferase reporter assay. The results revealed
that co-transfection with the MIAT wild-type luciferase reporter
and miR-613 mimics significantly reduced the luciferase activity
compared with that after co-transfection with MIAT wild-type
luciferase reporter and miR-NC (Fig.
4D and E). However, no marked
changes were observed in the luciferase activity of the MIAT mutant
luciferase reporter group in the TU-177 and AMC-HN-8 cells after
co-transfection with miR-613 (Fig.
4D and E). These data strongly
suggest that miR-613 specifically binds to MIAT. Subsequently, it
was found that the knockdown of MIAT significantly increased
miR-613 expression in the TU-177 and AMC-HN-8 cells (Fig. 4F). Therefore, RIP assay was
performed to elucidate the possible endogenous association between
MIAT and miR-613. Transfection with the miR-613 mimics led to a
significant enrichment of MIAT in lysates precipitated using the
anti-Ago2 antibody in the TU-177 and AMC-HN-8 cells compared with
that after transfection with the miR-NC (Fig. 4G and H). Taken together, these results suggested
that lncRNA MIAT can directly bind to miR-613 and function as a
sponge of miR-613 in LSCC.
miR-613 expression is downregulated in
human LSCC tissues and cell lines and inversely correlates with
MIAT expression
The expression levels of miR-613 in the three LSCC
cell lines TU686, TU-177 and AMC-HN-8 and the HaCaT cell line, in
addition to those in the LSCC tissues and the corresponding
adjacent non-tumor tissues were also measured using RT-qPCR. The
expression level of miR-613 was significantly lower in the three
LSCC cells compared with that in HaCaT cells (Fig. 5A). Additionally, it was found that
the expression of miR-613 was also significantly lower in LSCC
tissues compared with that in adjacent non-tumor tissues (Fig. 5B). Pearson's correlation analysis
was subsequently performed to examine the correlation between MIAT
and miR-613 expression in LSCC tissues. The expression level of
miR-613 correlated inversely with that of MIAT expression in LSCC
tissue samples (Fig. 5C).
Overexpression of miR-613 inhibits the
progression of LSCC
To investigate the role of miR-613 in the
proliferative, migratory and invasive ability of LSCC cells,
miR-613 mimics or miR-NC was transfected into TU-177 and AMC-HN-8
cells. The cell viability at 72 h and colony formation ability of
TU-177 and AMC-HN-8 cells in the miR-613 mimics group were
significantly reduced compared with those in the miR-NC group
(Fig. 6A-C). In addition, the
migratory ability of the TU-177 and AMC-HN-8 cells was
significantly decreased following miR-613 overexpression compared
with that after miR-NC transfection (Fig. 6D). It was also found that
overexpression of miR-613 significantly reduced the number of
invasive TU-177 and AMC-HN-8 cells compared with that after miR-NC
transfection (Fig. 6E).
Inhibition of miR-613 reverses the
effects of lncRNA MIAT knockdown on the proliferation and invasion
of LSCC cells
To further investigate whether lncRNA MIAT promotes
the proliferation, migratory and invasive abilities of LSCC cells
by negatively regulating miR-613 expression, anti-miR-613 was
transfected into TU-177 and AMC-HN-8 cells where MIAT expression
was also knocked down. Downregulation of miR-613 markedly reversed
the inhibitory effects of si-MIAT on the viability of TU-177 and
AMC-HN-8 cells at 72 h (Fig. 7A and
B). In addition, the colony forming
efficiency of the TU-177 and AMC-HN-8 cells was significantly
increased in the si-MIAT + anti-miR-613 group compared with that in
the si-MIAT + anti-miR-NC group (Fig.
7C and D). It was also found
that the downregulation of miR-613 significantly reversed the
inhibitory effects of si-MIAT on the migration and invasion of
TU-177 and AMC-HN-8 cells (Fig.
7E-G).
 | Figure 7lncRNA MIAT promotes LSCC development
by regulating miR-613 expression. The effects of co-transfection of
si-MIAT and anti-miR-613 on the viability of (A) TU-177 and (B)
AMC-HN-8 cells were detected by MTT assay. The effects of
co-transfection of si-MIAT and anti-miR-613 on the proliferation of
(C) TU-177 and (D) AMC-HN-8 cells were detected by colony formation
assay. The effects of co-transfection of si-MIAT and anti-miR-613
on the migratory ability of (E) TU-177 and (F) AMC-HN-8 cells were
detected using wound healing assay (magnification, x100). (G)
Transwell assay were used to examine TU-177 and AMC-HN-8 cell
invasion after si-MIAT and anti-miR-613 co-transfection
(magnification, x100). *P<0.05,
**P<0.01 and &P>0.05, si-MIAT +
anti-miR-NC group vs. si-NC + anti-miR-NC group;
#P<0.05, ##P<0.01 and
&P>0.05, si-MIAT + anti-miR-613 group vs. si-MIAT
+ anti-miR-NC group. MIAT, myocardial infarction associated
transcript; OD, optical density; NC, negative control; miR,
microRNA. |
Discussion
LSCC accounts for >90% of all laryngeal cancer
(LCa) cases, which is one of the most commonly observed
malignancies of the head and neck (25). Risk factors for LSCC include alcohol
consumption, a history of smoking, older age and male sex (26). Surgery is one of the main treatment
methods for this disease, but the surgical resection margin is
generally insufficient for treating patients presenting with
perineural invasion (27).
Mesolella et al (28)
previously reported that it is important to complement the surgical
therapeutic treatment with adjuvant radiotherapy. In spite of
synthetic serial treatments, including surgical intervention,
chemotherapy and radiation therapy, long-term prognosis of patients
with LSCC remains largely unsatisfactory (29). Therefore, understanding of the
precise molecular mechanisms underlying the pathogenesis of LSCC
and identifying novel targeted treatments for LSCC remain important
issues that need to be addressed.
The occurrence and progression of LSCC require
complex biological processes involving multiple genetic and
epigenetic changes. Recent studies have highlighted that multiple
types of RNA, including lncRNAs, circular RNAs and microRNAs, can
all regulate the expression levels of mRNA to serve important roles
in LSCC (30-32).
Takeuchi et al (33)
recently reported that a number of microRNAs, including miR-9,
miR-10b, miR-21 and miR-132 are aberrantly upregulated in LCa,
where they can contribute to the aggressiveness of the LCa. In
addition, miR-449a upregulation was found to inhibit cell invasion
and motility, suppressed cell proliferation and induced the
downregulation of Notch1 and Notch2 in the LCa cell line
HEP-2(34). At present, there are
no valid prognostic factors that can systematically drive the
choice of nodal treatment for LCa. Ricciardiello et al
(35) previously identified 20
miRNAs specific for LCa and a tissue-specific miRNA signature that
is predictive of lymph node metastases. In laryngeal carcinoma
characterized by 11 miRNAs, seven of which were found to be
upregulated and four were downregulated (36). lncRNAs are a novel class of RNAs
that do not encode proteins and contribute to the development and
progression of malignant tumors (7). An increasing number of studies have
suggested that lncRNA dysregulation is involved in epigenetic
alterations to serve important roles in the tumorigenesis and
metastasis of various types of tumors, including breast cancer
(37), osteosarcoma (38), NSCLC (39) and colorectal cancer (40). Of note, lncRNAs, including
NEAT1(10), H19(41), taurine upregulated-1(42) and plasmacytoma variant translocation
1 (PVT1) (43), function as
oncogenes to promote the aggressive phenotypes of LSCC, whilst
RP11-169D4.1(44), neighboring
enhancer of FOXA2(45) and
maternally expressed 3(46)
function as tumor suppressors in LSCC. In addition, lncRNAs have
been reported to be important regulators of gene expression and
represent an innovative pharmacological target as molecular
biomarkers of LSCC (30).
Currently, an increasing number of studies have
reported that lncRNA MIAT serves an important role in the
initiation and development of malignant tumors. In particular, MIAT
has been shown to enhance papillary thyroid cancer growth and
metastasis by binding miR-324-3p and upregulating LIM and SH3
protein 1(47). lncRNA MIAT also
serves as a key factor to promote cell invasion, migration and
proliferation through the PI3K/AKT signaling pathway in melanoma
(48). In addition, it promotes
cell migration and invasion in NCSLC by sponging miR-1246(49). In the present study, one major
finding was that lncRNA MIAT was significantly upregulated in LSCC
tissues and cell lines (TU686, TU-177 and AMC-HN-8), where the MIAT
expression level was higher in TU-177 and AMC-HN-8. Therefore,
TU-177 and AMC-HN-8 were chosen to evaluate the effects of MIAT on
LSCC cells. The results revealed that the knockdown of MIAT
expression exerted inhibitory effects on LSCC cell proliferation,
migration and invasion. This suggest that MIAT promotes the
tumorigenesis of LSCC and may function as an oncogene. However, it
is a limitation for the lack of parallel experiments on the TU686
cell line in the present study. Additionally, further investigation
is required to elucidate the role of MIAT in the progression of
LSCC in vivo in future studies.
Recent studies have demonstrated that lncRNAs serve
a role in cancer development as ceRNAs that can sponge miRNAs
(50-52).
lncRNA PVT1 has been previously shown to function as an oncogene in
the development of LSCC, partly by sponging miR-519d-3p and
inhibiting its expression (43). In
the present study, bioinformatics analysis predicted that several
potential complementary sites existed between MIAT and miR-613,
which was verified further using luciferase reporter assay in
TU-177 and AMC-HN-8 cells. Notably, it was found that the knockdown
of MIAT increased miR-613 expression, where MIAT acted as a miR-613
sponge in LSCC as shown by results from RIP assays. In addition, it
was found that miR-613 was expressed at lower levels in LSCC
tissues, where its expression negatively correlated with MIAT
expression. According to these results, it can be suggested that a
reciprocal relationship exists between miR-613 and MIAT expression
in LSCC. Previous studies have revealed that miR-613 acts as either
an oncogene or a tumor suppressor (22,23,53-56).
miR-613 expression was documented to be significantly upregulated
in cervical cancer (CC) tissues and cells, which promotes the cell
proliferation, invasion and migration of CC by targeting protein
tyrosine phosphatase non-receptor type 9(53). Similarly, miR-613 functions as an
oncogene in colon cancer, where it promotes cell proliferation,
invasion and migration by targeting protein atonal homolog 1,
likely by activating the JNK1 pathway (54). Furthermore, previous studies have
also indicated that miR-613 inhibits the progression of NSCLC
(22), pancreatic cancer (23), breast cancer (55) and gastric cancer (56). Wang et al (57) reported that miR-613 serves as a
tumor suppressor gene in LSCC partly by inhibiting
phosphoinositide-dependent kinase-1 expression. In the present
study, the expression of miR-613 was markedly downregulated in LSCC
cell lines, such that the overexpression of miR-613 inhibited the
proliferation, migration and invasion of LSCC cells. To investigate
whether lncRNA MIAT promotes the progression of LSCC by regulating
miR-613, anti-miR-613 was transfected into TU-177 and AMC-HN-8
cells, where MIAT expression was also knocked down. The data
revealed that the knockdown of miR-613 restored the proliferative,
migratory and invasive activities in LSCC cells following MIAT
silencing, suggesting that MIAT may promote the malignant
progression of LSCC by sponging miR-613.
In conclusion, the present study, to the best of our
knowledge, was the first to demonstrate that MIAT functions as a
novel tumor-promoting lncRNA in LSCC. In addition, MIAT functions
as a ceRNA to negatively regulate the expression of miR-613 by
competitive binding, thereby promoting the tumorigenesis and
development of LSCC.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
FS and YY designed the study and performed the
experiments. JL performed statistical analysis and revised the
manuscript. FS, YY and JL confirmed the authenticity of all the raw
data. All authors read and approved the final version of the
manuscript.
Ethics approval and consent to
participate
All patients signed informed consent prior to the
use of their tissues for the present study according to the
principles of the Declaration of Helsinki. The present study was
approved by the Ethics Committee of Tianjin Union Medical Center
(Tianjin, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2015. CA Cancer J Clin. 65:5–29. 2015.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Marur S and Forastiere AA: Head and neck
squamous cell carcinoma: Update on epidemiology, diagnosis, and
treatment. Mayo Clin Proc. 91:386–396. 2016.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Almadori G, Bussu F, Cadoni G, Galli J,
Paludetti G and Maurizi M: Molecular markers in laryngeal squamous
cell carcinoma: Towards an integrated clinicobiological approach.
Eur J Cancer. 41:683–693. 2005.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Yang L, Froberg JE and Lee JT: Long
noncoding RNAs: Fresh perspectives into the RNA world. Trends
Biochem Sci. 39:35–43. 2014.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Choudhry H, Harris AL and McIntyre A: The
tumour hypoxia induced non-coding transcriptome. Mol Aspects Med.
47-48:35–53. 2016.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Peng WX, Koirala P and Mo YY:
lncRNA-mediated regulation of cell signaling in cancer. Oncogene.
36:5661–5667. 2017.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Shen Z, Hao W, Zhou C, Deng H, Ye D, Li Q,
Lin L, Cao B and Guo J: Long non-coding RNA AC026166.2-001 inhibits
cell proliferation and migration in laryngeal squamous cell
carcinoma by regulating the miR-24-3p/p27 axis. Sci Rep.
8(3375)2018.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Wang R, Ma Z, Feng L, Yang Y, Tan C, Shi
Q, Lian M, He S, Ma H and Fang J: lncRNA MIR31HG targets HIF1A and
P21 to facilitate head and neck cancer cell proliferation and
tumorigenesis by promoting cell-cycle progression. Mol Cancer.
17(162)2018.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Wang P, Wu T, Zhou H, Jin Q, He G, Yu H,
Xuan L, Wang X, Tian L, Sun Y, et al: Long noncoding RNA NEAT1
promotes laryngeal squamous cell cancer through regulating
miR-107/CDK6 pathway. J Exp Clin Cancer Res. 35(22)2016.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Hao YR, Zhang DJ, Fu ZM, Guo YY and Guan
GF: Long non-coding RNA ANRIL promotes proliferation,
clonogenicity, invasion and migration of laryngeal squamous cell
carcinoma by regulating miR-181a/Snai2 axis. Regen Ther.
11:282–289. 2019.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Ishii N, Ozaki K, Sato H, Mizuno H, Saito
S, Takahashi A, Miyamoto Y, Ikegawa S, Kamatani N, Hori M, et al:
Identification of a novel non-coding RNA, MIAT, that confers risk
of myocardial infarction. J Hum Genet. 51:1087–1099.
2006.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Sha M, Lin M, Wang J, Ye J, Xu J, Xu N and
Huang J: Long non-coding RNA MIAT promotes gastric cancer growth
and metastasis through regulation of miR-141/DDX5 pathway. J Exp
Clin Cancer Res. 37(58)2018.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Zhang HY, Zheng FS, Yang W and Lu JB: The
long non-coding RNA MIAT regulates zinc finger E-box binding
homeobox 1 expression by sponging miR-150 and promoting cell
invasion in non-small cell lung cancer. Gene. 633:61–65.
2017.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Qu Y, Xiao H, Xiao W, Xiong Z, Hu W, Gao
Y, Ru Z, Wang C, Bao L, Wang K, et al: Upregulation of MIAT
regulates LOXL2 expression by competitively binding miR-29c in
clear cell renal cell carcinoma. Cell Physiol Biochem.
48:1075–1087. 2018.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Liu Z, Wang H, Cai H, Hong Y, Li Y, Su D
and Fan Z: Long non-coding RNA MIAT promotes growth and metastasis
of colorectal cancer cells through regulation of miR-132/Derlin-1
pathway. Cancer Cell Int. 18(59)2018.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Barnes L, Eveson JW, Reichart P and
Sidransky D: WHO pathology and genetic classification of tumors of
head and neck tumours. 3rd edition, Lyon, IARC Press, pp107-162,
2005.
|
|
18
|
Li Y, Tao C, Dai L, Cui C, Chen C, Wu H,
Wei Q and Zhou X: MicroRNA-625 inhibits cell invasion and
epithelial-mesenchymal transition by targeting SOX4 in laryngeal
squamous cell carcinoma. Biosci Rep. 39(BSR20181882)2019.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Nowinska K, Ciesielska U, Piotrowska A,
Jablonska K, Partynska A, Paprocka M, Zatonski T, Podhorska-Okolow
M and Dziegiel P: MCM5 expression is associated with the grade of
malignancy and Ki-67 antigen in LSCC. Anticancer Res. 39:2325–2335.
2019.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Giordano C, Barone I, Vircillo V, Panza S,
Malivindi R, Gelsomino L, Pellegrino M, Rago V, Mauro L, Lanzino M,
et al: Activated FXR inhibits leptin signaling and counteracts
tumor-promoting activities of cancer-associated fibroblasts in
breast malignancy. Sci Rep. 6(21782)2016.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Li D, Li DQ, Liu D and Tang XJ: miR-613
induces cell cycle arrest by targeting CDK4 in non-small cell lung
cancer. Cell Oncol (Dordr). 39:139–147. 2016.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Cai H, Yao J, An Y, Chen X, Chen W, Wu D,
Luo B, Yang Y, Jiang Y, Sun D and He X: lncRNA HOTAIR acts a
competing endogenous RNA to control the expression of notch3 via
sponging miR-613 in pancreatic cancer. Oncotarget. 8:32905–32917.
2017.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Xiong H, Yan T, Zhang W, Shi F, Jiang X,
Wang X, Li S, Chen Y, Chen C and Zhu Y: miR-613 inhibits cell
migration and invasion by downregulating Daam1 in triple-negative
breast cancer. Cell Signal. 44:33–42. 2018.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Marioni G, Marchese-Ragona R, Cartei G,
Marchese F and Staffieri A: Current opinion in diagnosis and
treatment of laryngeal carcinoma. Cancer Treat Rev. 32:504–515.
2006.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Menach P, Oburra HO and Patel A: Cigarette
smoking and alcohol ingestion as risk factors for laryngeal
squamous cell carcinoma at Kenyatta National Hospital, Kenya. Clin
Med Insights Ear Nose Throat. 5:17–24. 2012.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Nix P, Cawkwell L, Patmore H, Greenman J
and Stafford N: Bcl-2 expression predicts radiotherapy failure in
laryngeal cancer. Br J Cancer. 92:2185–2189. 2005.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Mesolella M, Iorio B, Misso G, Luce A,
Cimmino M, Iengo M, Landi M, Sperlongano P, Caraglia M and
Ricciardiello F: Role of perineural invasion as a prognostic factor
in laryngeal cancer. Oncol Lett. 11:2595–2598. 2016.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Chu EA and Kim YJ: Laryngeal cancer:
Diagnosis and preoperative work-up. Otolaryngol Clin North Am.
41:673–695. 2008.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Cossu AM, Mosca L, Zappavigna S, Misso G,
Bocchetti M, De Micco F, Quagliuolo L, Porcelli M, Caraglia M and
Boccellino M: Long non-coding RNAs as important biomarkers in
laryngeal cancer and other head and neck tumours. Int J Mol Sci.
20(3444)2019.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Ekmekci CG, Coskunpinar E, Avci H, Farooqi
AA, Orhan KS and Akbas F: Integrative analysis of mRNA and microRNA
expression profiles in laryngeal squamous cell carcinoma. J Cell
Biochem. 120:3415–3422. 2019.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Zhao X, Zhang W and Ji W: miR-196b is a
prognostic factor of human laryngeal squamous cell carcinoma and
promotes tumor progression by targeting SOCS2. Biochem Biophys Res
Commun. 501:584–592. 2018.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Takeuchi T, Kawasaki H, Luce A, Cossu AM,
Misso G, Scrima M, Bocchetti M, Ricciardiello F, Caraglia M and
Zappavigna S: Insight toward the microRNA profiling of laryngeal
cancers: Biological role and clinical impact. Int J Mol Sci.
21(3693)2020.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Kawasaki H, Takeuchi T, Ricciardiello F,
Lombardi A, Biganzoli E, Fornili M, De Bortoli D, Mesolella M,
Cossu AM, Scrima M, et al: Definition of miRNA signatures of nodal
metastasis in LCa: miR-449a targets notch genes and suppresses cell
migration and invasion. Mol Ther Nucleic Acids. 20:711–724.
2020.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Ricciardiello F, Capasso R, Kawasaki H,
Abate T, Oliva F, Lombardi A, Misso G, Ingrosso D, Leone CA, Iengo
M and Caraglia M: A miRNA signature suggestive of nodal metastases
from laryngeal carcinoma. Acta Otorhinolaryngol Ital. 37:467–474.
2017.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Lian Y, Li Z, Fan Y, Huang Q, Chen J, Liu
W, Xiao C and Xu H: The lncRNA-HOXA-AS2/EZH2/LSD1 oncogene complex
promotes cell proliferation in pancreatic cancer. Am J Transl Res.
9:5496–5506. 2017.PubMed/NCBI
|
|
37
|
Liu XM, Yang B and Han J: Increased long
noncoding RNA LINP1 expression and its prognostic significance in
human breast cancer. Eur Rev Med Pharmacol Sci. 22:8749–8754.
2018.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Li H, Tian G, Tian F and Shao L: Long
non-coding RNA TUG1 promotes osteosarcoma cell proliferation and
invasion through inhibition of microRNA-212-3p expression. Exp Ther
Med. 16:779–787. 2018.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Zhang G, An X and Zhao H, Zhang Q and Zhao
H: Long non-coding RNA HNF1A-AS1 promotes cell proliferation and
invasion via regulating miR-17-5p in non-small cell lung cancer.
Biomed Pharmacother. 98:594–599. 2018.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Liu Y, Zhou J, Wang S, Song Y, Zhou J and
Ren F: Long non-coding RNA SNHG12 promotes proliferation and
invasion of colorectal cancer cells by acting as a molecular sponge
of microRNA-16. Exp Ther Med. 18:1212–1220. 2019.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Guan GF, Zhang DJ, Wen LJ, Xin D, Liu Y,
Yu DJ, Su K, Zhu L, Guo YY and Wang K: Overexpression of lncRNA
H19/miR-675 promotes tumorigenesis in head and neck squamous cell
carcinoma. Int J Med Sci. 13:914–922. 2016.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Zhang Z, Wang X, Cao S, Han X, Wang Z,
Zhao X, Liu X, Li G, Pan X and Lei D: The long noncoding RNA TUG1
promotes laryngeal cancer proliferation and migration. Cell Physiol
Biochem. 49:2511–2520. 2018.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Zheng X, Zhao K, Liu T, Liu L, Zhou C and
Xu M: Long noncoding RNA PVT1 promotes laryngeal squamous cell
carcinoma development by acting as a molecular sponge to regulate
miR-519d-3p. J Cell Biochem. 120:3911–3921. 2019.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Zhao J, Lv K, Li ZH, Wu J, Gao W, Wong TS,
Luo J, Qin H, Wang B, Fu Q and Lei WB: Functional significance of
the long non-coding RNA RP11-169D4.1 as a metastasis suppressor in
laryngeal squamous cell carcinoma by regulating CDH1. Oncol Rep.
38:211–220. 2017.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Cui X, Fang N, Cui Y, Xiao D and Wang X:
Long non-coding RNA NEF inhibits proliferation and promotes
apoptosis of laryngeal squamous cell carcinoma cells by inhibiting
Wnt/β-catenin signaling. Oncol Lett. 17:4928–4934. 2019.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Zhang X, Wu N, Wang J and Li Z: lncRNA
MEG3 inhibits cell proliferation and induces apoptosis in laryngeal
cancer via miR-23a/APAF-1 axis. J Cell Mol Med. 23:6708–6719.
2019.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Liu W, Wang Z, Wang C and Ai Z: Long
non-coding RNA MIAT promotes papillary thyroid cancer progression
through upregulating LASP1. Cancer Cell Int. 19(194)2019.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Yang Y, Zhang Z, Wu Z, Lin W and Yu M:
Downregulation of the expression of the lncRNA MIAT inhibits
melanoma migration and invasion through the PI3K/AKT signaling
pathway. Cancer Biomark. 24:203–211. 2019.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Lin D, Xu HP, Lin JH, Hu HH, Wang Q and
Zhang J: Long non-coding RNA MIAT promotes non-small cell lung
cancer progression by sponging miR-1246. Eur Rev Med Pharmacol Sci.
23:5795–5801. 2019.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Kartha RV and Subramanian S: Competing
endogenous RNAs (ceRNAs): New entrants to the intricacies of gene
regulation. Front Genet. 5(8)2014.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Russo F, Fiscon G, Conte F, Rizzo M, Paci
P and Pellegrini M: Interplay between long noncoding RNAs and
microRNAs in cancer. Methods Mol Biol. 1819:75–92. 2018.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Denzler R, McGeary SE, Title AC, Agarwal
V, Bartel DP and Stoffel M: Impact of microRNA levels, target-site
complementarity, and cooperativity on competing endogenous
RNA-regulated gene expression. Mol Cell. 64:565–579.
2016.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Li WT, Wang BL, Yang CS, Lang BC and Lin
YZ: miR-613 promotes cell proliferation and invasion in cervical
cancer via targeting PTPN9. Eur Rev Med Pharmacol Sci.
22:4107–4114. 2018.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Yang X, Zhang L, Song X, He W, Zhang D, Lu
Q, Wu J, Wu C and Jiang J: MicroRNA-613 promotes colon cancer cell
proliferation, invasion and migration by targeting ATOH1. Biochem
Biophys Res Commun. 504:827–833. 2018.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Wu J, Yuan P, Mao Q, Lu P, Xie T, Yang H
and Wang C: miR-613 inhibits proliferation and invasion of breast
cancer cell via VEGFA. Biochem Biophys Res Commun. 478:274–278.
2016.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Lu Y, Tang L, Zhang Q, Zhang Z and Wei W:
MicroRNA-613 inhibits the progression of gastric cancer by
targeting CDK9. Artif Cells Nanomed Biotechnol. 46:980–984.
2018.PubMed/NCBI View Article : Google Scholar
|
|
57
|
Wang J, Yang S, Ge W, Wang Y, Han C and Li
M: miR-613 suppressed the laryngeal squamous cell carcinoma
progression through regulating PDK1. J Cell Biochem. 119:5118–5125.
2018.PubMed/NCBI View Article : Google Scholar
|