Introduction
Benign prostate hyperplasia (BPH), also known as
prostate gland enlargement, is a genitourinary condition that is
most prevalent in aging males, usually starting at 50-61 years of
age (1), and causing lower urinary
tract symptoms (LUTS), such as urine flow blockage as a result of
the urethra being compressed by the enlarged gland. Other potential
complications may include bladder, urinary tract or kidney problems
(2). Most males have continued
prostate growth throughout their life (2) After the age of 30 years, males exhibit
a 1% drop in testosterone production per year and an increase in
the level of dihydrotestosterone, possibly due to the age-related
increase in 5α reductase (SRD5A2) activity. This rising level of
dihydrotestosterone appears to increase prostate cell longevity and
proliferation, leading to BPH (3).
BPH is a histological diagnosis that is specified by
non-malignant hyperplasia of the stromal and glandular epithelial
cells of the prostate, leading to an enlargement in its size
(4). Studies and meta-analyses have
revealed that BPH is associated with an increased risk of prostate
and bladder cancers (5) due to
their common pathophysiological driving factors (6). BPH arises mostly from the peripheral
zone (70%), followed by growth in the transition zone (20%) and in
the central zone (10%) of the gland. The public health burden of
BPH is high due to the increased associated morbidity and treatment
costs: As many as 33% of males older than 50 years, ~50% of those
>60 years, 70% of those >70 years and 90% of those >85
years develop BPH (7). The risk for
progressing into a cancerous state is small, but instead, the
entire prostate gland grows uniformly, with small smooth, elastic
and firm hyperplastic nodules. Common complications include urinary
tract infections, bladder stones and chronic kidney problems
(8). The symptoms maybe obstructive
(weakened urine stream, strained or prolonged voiding, urinary
hesitancy) or irritative (pain, nocturia, urge incontinence), or
may produce a constant sense of incomplete bladder emptying after
micturition that leads to the requirement of frequent urination
(9).
The primary risk factors for BPH include age, family
history, obesity and being of Afro-Caribbean descent. BPH is most
common in western countries and affects >1 billion males all
over the world (6). According to
GLOBOCAN estimates, 1.2 million novel cases of prostate cancer were
reported worldwide in 2018(10).
Diagnostic methods include physical and digital rectal
examinations, prostate-specific antigen (PSA) level measurements,
prostate biopsy, prostate ultrasound, urinalysis and urine culture
(11,12). PSA, a glycoprotein enzyme produced
by the epithelial cells of the prostate gland, is considered the
mainstay for BPH prognosis and diagnosis. However, it is difficult
to differentiate between the increase in PSA levels due to BPH and
prostate cancer. Furthermore, the test fails to discriminate
between low-risk and aggressive tumors (13). Major invasive and medical therapies
are available for BPH treatment (14). Invasive therapies include microwave
thermotherapy, prostate needle ablation, as well as surgical, laser
and transurethral therapies; medical therapies include a-adrenergic
blockers, 5a-reductase inhibitors, phosphodiesterase type-5
inhibitor therapy for BPH/LUTS and b-3-agonist therapy.
Pharmaceutical treatments include finasteride (5-alpha reductase
inhibitor) and alpha-1 antagonists. Finasteride shrinks the
prostate gland by inhibiting the conversion of testosterone into
dihydrotestosterone, resulting in urine flow obstruction relief
(15), and alpha-1 antagonists
(such as phenoxybenzamine) bind alpha-1 receptors of bladder-neck
smooth muscle, causing its relaxation and allowing urine to pass
(16).
The discovery of genomic mutations and development
of high-throughput screening and microarray technologies have
opened up possibilities for identifying gene biomarkers for the
diagnosis, prognosis and treatment of BPH (17). Genomic functional networks may help
reveal interactions between BPH-associated modules, genitourinary
diseases and hyperglycemia, and identify pathway-specific
interactions. Furthermore, as only a few drugs (with numerous side
effects) are available for treating BPH (18), alternative drugs are required.
Histopathology methods for BPH remain incomplete. Differentially
expressed genes (DEGs) in BPH and normal prostate tissues are
likely to reflect underlying pathogenic mechanisms involved in the
development of the disease. Complementary DNA microarray technology
may be used to identify genes associated with BPH. The present
study focused on a specific set of genes responsible for BPH and
performed protein-protein interaction analyses to disclose
functional networks. Potential prognostic biomarkers were
identified using in silico approaches, high-throughput
microarray data and comprehensive protein-protein interaction
analyses.
The objectives of the present study were to discover
genes that are differentially expressed in BPH and normal prostate
tissues, identify functional networks and look for potential
alternative BPH agents in a list of Chinese herbs.
Patients and methods
Datasets
The gene expression profiles of BPH patients from
the Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/) dataset with the
accession number GSE6099 were used. For the analysis, seven samples
were selected: A total of four human epithelium samples of BPH
nodules (EPI_BPH) and three samples from human epithelium of
individuals without a history of prostate disease (EPI_NOR). The
epithelium is one of the basic types of animal tissue that lines
the outer surface of the prostate gland. The pathological evidence
for prostate diseases confirmed the neoplastic changes of the
prostate epithelium.
Identification of DEGs
TheGEO2R web-based tool was used for basic
processing, analysis of gene expression datasets and identification
of DEGs in BPH. The GEO2R tool uses GEO query (19) and the R limma package (20) from the Bioconductor project
(https://www.bioconductor.org/). Fold
changes (FCs) were calculated as the ratio of the average
expression values of each probe across the samples under normal and
BPH conditions. Genes with logFC≥1.0 were considered as upregulated
and those with logFC≤-1.0 as downregulated (20,21).
Gene and functional annotation
clustering
Functional annotation clustering available in the
DAVID tool (https://david.ncifcrf.gov/) was performed.
Associations among the ‘annotation terms’ were measured based on
their co-association genes in order to cluster similar, redundant
and heterogeneous annotation content from the similar or different
resources into annotation groups, based on the protocol by Huang
et al (21) from 2009.
Protein-protein interaction study
The GeneMania database (https://genemania.org/) (22) was used to infer experimentally known
physical interactions between proteins to predict pathways, protein
functions and potential novel therapeutic targets.
RNA tissue specificity analysis
For the purpose of RNA tissue-specific analysis, the
Human Protein Atlas (HPA) (23) was
used. The HPA portal maps all the human proteins in cells, tissues
and organs using integrated omics technologies. In addition, the
distribution of proteins in the prostate was examined using the
Tissue Atlas (one of the three major HPA projects).
Molecular docking of active components
of Chinese herbs to BPH receptor proteins
A total of 10 Chinese herbs that have been cited as
effective for treating BPH were selected (Table I). Using a systematic literature
review, the phytochemicals potentially contributing to the
effectiveness of these herbs were retrieved and listed in Table I.
 | Table IPhytochemical compounds occurring in
Chinese medicinal herbs used to treat benign prostate
hyperplasia. |
Table I
Phytochemical compounds occurring in
Chinese medicinal herbs used to treat benign prostate
hyperplasia.
| Chinese herb
name | Phytochemical
compound |
|---|
| Chaun Shan
Jia | N-butyl
tricosylamide, cyclo (L-seryl-L-tyrosyl) |
| E Shu |
Curdioneandgermacrone |
| Fu ling | Polyporenic acid
C |
| Huang
Bo | Berberine |
| Huang
Qi | Astragalosides |
| Rou Gui | Cinnamonitride |
| Shan Zhu
Yu | Cornuside |
| Shu Di
Huang | Rehmania
polysaccharide, Jionoside |
| Wang Bu Liu
Xing | Triterpene
saponins |
| Ze Xie | Alisol A
monoacetate |
In silico extraction of phytochemical
compounds
The structures of the phytochemical compounds were
obtained from various databases, such as ChEMBL (24), PubChem (25) and DrugBank (26).
Molecular docking
The structures of the relevant prostate receptor
proteins and the phytochemicals selected for the study
(cinnamonitride, astragaloside, cornuside, polyporenic acid C,
berberine and alisol A monoacetate) were first converted into pdbqt
files for docking. AutoDock Vina (27) was used with receptor proteins to
perform blind flexible dockings.
Interaction analysis
The Protein-Ligand Interaction Profiler (PLIP)
(28) was used to establish
interactions between the docked complexes.
Results and Discussion
The differentially expression genes (DEGs) in BPH
were identified using the methodology described in the
‘Identification of DEGs’ section. Out of a variety of existing
methods for identifying DEGs from microarray gene expression data,
such as the FC (29) or t-test
statistics (30), calculation of
the log(FC) was chosen as one of the simplest ad-hoc methods for
microarray analyses. The FC describes the change in expression of a
gene between two observed samples, i.e., between normal and BPH
tissues (31).
After selecting the gene expression datasets and
evaluating them for differential expression analysis, a boxplot of
the BPH and the normal samples was generated (Fig. 1). It was observed that the median of
the two sample types (i.e., BPH vs. normal) was close to zero.
However, there were significant variations in terms of their
minimum, first quartile, third quartile and maximum values between
BPH and normal groups, with BPH samples GSM141335 and GSM141337
showing lower values compared with those in normal samples
GSM141338 and GSM141339 (Fig.
1).
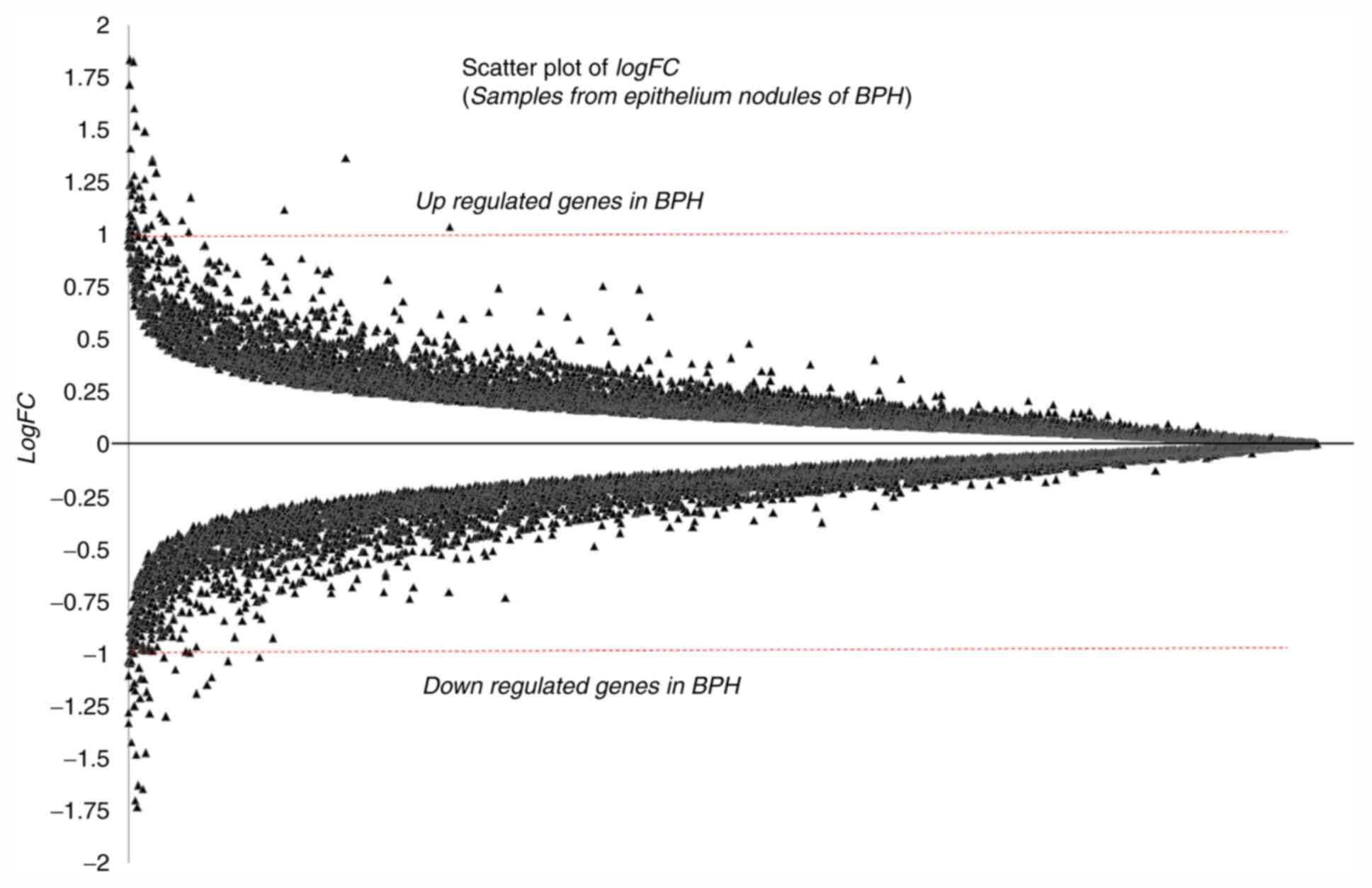
The logFC statistics for the entire genome were
computed. The logFC values of all the genes are presented in a
scatter plot in Fig. 2. Genes with
a logFC ≥1.0 were considered to be upregulated and those with logFC
≤-1.0 were considered to be downregulated in BPH tissues (32,33).
Following conventional rules, a threshold of a two-fold change in
gene expression (i.e., -1.0 ≤logFC ≥1.0), and P≤0.05 (5%
significance level) were used to short-list DEGs in BPH. Table II lists the identified DEGs with
their P-values and logFC values. Among the highest ranking
identified DEGs are Zinc finger proteins (ZNF3; P<0.0001,
logFC=3.0111), Acyl-CoA synthetase family member 3 (ACSF3;
P<0.0001, logFC=1.5768), Fibrinogen like-1 (FGL1; P=0.0001,
logFC=-1.4845), PMS1 homolog 1, mismatch repair system component
(PMS1; P=0.0001, logFC=-1.4611), Forkhead box P2 (FOXP2;
P<0.0001, logFC=-1.3491), anterior gradient 2 (AGR2;
P<0.0001, logFC=-1.3156) andRing finger protein 135 (RNF135;
P=0.0001, logFC=1.2748). In addition, profile graphs of identified
DEGs were plotted to obtain a graph of DEG expression across the
different samples, as presented in Fig.
3. The profile graphs point to differential expression
behaviours in the BPH and normal prostate samples.
 | Figure 3Profile graphs of differentially
expressed genes, namely ZNF3, ACSF3, FGL1, FOXP2, AGR2, RNF135,
BAZ2B, NCAPD3, PDK4, CDC45, FOLH1, CXXC4, PMS1, ARHGAP42, COL12A1,
HECTD4, IGFBP5, TF, ADAMTS1, TEAD1, EDRF1, KCTD13 and BACE2. BPH,
benign prostate hyperplasia; ZNF3, Zinc finger protein 3; ACSF3,
Acyl-CoA synthetase family member 3; FGL1, Fibrinogen like 1;
FOXP2, Forkhead box P2; AGR2, Anterior gradient 2; RNF135, Ring
finger protein 135; BAZ2B, Bromodomain adjacent to zinc finger
domain 2B; NCAPD3, Non-smooth muscle cell condensin II complex
subunit D3; PDK4, Pyruvate dehydrogenase kinase 4; CDC45, Cell
division cycle 45; FOLH1, Folate hydrolase (prostate-specific
membrane antigen) 1; CXXC4, CXXC finger protein 4; PMS1, PMS1
homolog 1, mismatch repair system component; ARHGAP42, Rho GTPase
activating protein 42; COL12A1, collagen type XII α1 chain; HECTD4,
HECT domain E3 ubiquitin protein ligase 4; IGFBP5, Insulin-like
growth factor binding protein 5; TF, Transferrin; ADAMTS1, ADAM
metallopeptidase with thrombospondin type 1 motif 1; TEAD1, TEA
domain transcription factor 1; EDRF1, Erythroid differentiation
regulatory factor 1; KCTD13, Potassium channel tetramerization
domain containing 13; BACE2, Beta-site APP-cleaving enzyme 2. |
 | Table IIList of differentially expressed
genes in BPH and predicted scores. |
Table II
List of differentially expressed
genes in BPH and predicted scores.
| ID | P-value | T | logFC | Gene symbol | Gene name | Gene function | Uniprot ID |
|---|
| Hs6-3-3-1 | <0.0001 | 11.0053 | 3.0111 | ZNF3 | Zinc finger protein
3 | Involved in cell
differentiation and/or proliferation. | https://www.uniprot.org/uniprot/P17036 |
| Hs6-11-20-9 | <0.0001 | 7.0481 | 1.5768 | ACSF3 | Acyl-CoA synthetase
family member 3 | Catalyzes the
initial reaction in intramitochondrial fatty acid synthesis into
respective CoA thioester, by activating malonate and
methylmalonate. | https://www.uniprot.org/uniprot/Q4G176 |
| Hs6-30-1-12 | 0.0001 | -6.6404 | -1.4845 | FGL1 | Fibrinogen like
1 | Upregulated in
various cancers, including lung cancer, prostate cancer, melanoma
and colorectal cancer. | https://www.uniprot.org/uniprot/Q08830 |
| Hs6-32-17-24 | 0.0001 | -6.2384 | -1.4611 | PMS1 | PMS1 homolog 1,
mismatch repair system component | Repair of DNA
mismatch. | https://www.uniprot.org/uniprot/P54277 |
| Hs6-18-1-13 | <0.0001 | -6.4221 | -1.3491 | FOXP2 | Forkhead box
P2 | A transcriptional
repressor that participate in the specification and differentiation
of the lung epithelium, development of neural, gastrointestinal and
cardiovascular tissues. | https://www.uniprot.org/uniprot/O15409 |
| Hs6-13-2-18 | <0.0001 | -7.5867 | -1.3156 | AGR2 | Anterior gradient
2, protein disulphide isomerase family member | Catalyze protein
folding and thiol-disulfide interchange reactions. | https://www.uniprot.org/uniprot/O95994 |
| Hs6-11-16-9 | 0.0001 | 5.9628 | 1.2748 | RNF135 | Ring finger protein
135 | Acts as an
E2-dependent E3 ubiquitin-protein ligase, involved in innate immune
defense against viruses. | https://www.uniprot.org/uniprot/Q8IUD6 |
| Hs6-20-10-21 | 0.0249 | 2.5918 | 1.2741 | HECTD4 | HECT domain E3
ubiquitinprotein ligase 4 | Transfers the
ubiquitin to targeted substrates directly. | https://www.uniprot.org/uniprot/Q9Y4D8 |
| Hs6-31-19-17 | 0.0001 | -6.0425 | -1.2109 | BAZ2B | Bromodomain
adjacent to zinc finger domain 2B | May play a role in
transcriptional regulation interacting with imitation SWI. | https://www.uniprot.org/uniprot/Q9UIF8 |
| Hs6-8-2-2 | 0.0018 | 3.8935 | 1.1828 | NCAPD3 | Non-smooth muscle
cellcondensin II complexsubunit D3 | Mitotic chromosome
assembly and segregation. | https://www.uniprot.org/uniprot/P42695 |
| Hs6-11-17-23 | 0.0112 | -2.9520 | -1.1392 | PDK4 | Pyruvate
dehydrogenase kinase 4 | A member of the PDK
protein kinase family that encodes a mitochondrial protein with a
histidine kinase domain. | https://www.uniprot.org/uniprot/Q16654 |
| Hs6-23-18-13 | 0.0087 | -3.0793 | -1.1176 | ADAMTS1 | ADAM
metallopeptidase with thrombospondin type 1 motif 1 | Angiogenic
inhibitor activity; has a critical role in follicular rupture;
associated with various inflammatory processes and development of
cancer cachexia. | https://www.uniprot.org/uniprot/Q9UHI8 |
| Hs6-20-19-25 | 0.0040 | 3.9720 | 1.0868 | CDC45 | Cell division cycle
45 | Required for
initiation of chromosomal DNA replication. | https://www.uniprot.org/uniprot/O75419 |
| Hs6-19-25-7 | 0.0024 | -3.7587 | -1.0450 | FOLH1 | Folate hydrolase
(prostate-specific membrane antigen) 1 | Folate hydrolase
and N-acetylated-alpha-linked-acidic dipeptidase activity;
associated with prostate tumor progression. | https://www.uniprot.org/uniprot/Q04609 |
| Hs6-28-5-13 | 0.0014 | -4.5094 | -1.0398 | CXXC4 | CXXC finger protein
4 | Represses Wnt
signaling pathway via interaction with DVL1; binds to DNA
containing CpG dinucleotides over CpH (H=A, T, and C),
hemimethylated-CpG and hemimethylated-hydroxymethyl-CpG. | https://www.uniprot.org/uniprot/Q9H2H0 |
| Hs6-10-24-14 | 0.0012 | 4.6071 | 1.0299 | KCTD13 | Potassium channel
tetramerization domain containing 13 | Substrate-specific
adapter of a BCR E3 ubiquitin-protein ligase complex required for
synaptic transmission; mediates the ubiquitination of RhoA, leading
to its degradation by the proteasome. | https://www.uniprot.org/uniprot/Q8WZ19 |
| Hs6-15-23-4 | 0.0049 | -3.3798 | -1.0297 | ARHGAP42 | Rho GTPase
activating protein 42 | May influence blood
pressure by functioning as a GTPase-activating protein for RhoAin
vascular smooth muscle. | https://www.uniprot.org/uniprot/A6NI28 |
| Hs6-11-11-1 | 0.0052 | -3.3483 | -1.0290 | TEAD1 | TEA domain
transcription factor 1 | Plays a key role in
the Hippo signaling pathway, a pathway involved in organ size
control, and tumor suppression by restricting proliferation and
promoting apoptosis; cardiac development; binds to the M-CAT
motif. | https://www.uniprot.org/uniprot/P28347 |
| Hs6-28-7-19 | 0.0015 | 4.0068 | 1.0283 | COL12A1 | Collagen type XII
alpha 1 chain | Interacts with type
I collagen-containing fibrils, the COL1 domain could be associated
with the surface of the fibrils, and the COL2 and NC3 domains may
be localized in perifibrillar matrix. | https://www.uniprot.org/uniprot/Q99715 |
| Hs6-16-23-19 | 0.0014 | -4.5432 | -1.0194 | EDRF1 | Erythroid
differentiation regulatory factor 1 | A transcription
factor involved in erythroid differentiation; transcriptional
activation of the globin gene. | https://www.uniprot.org/uniprot/Q3B7T1 |
| Hs6-32-13-1 | 0.0003 | 4.7861 | 1.0155 | IGFBP5 | Insulin-like growth
factor binding protein 5 | IGF-binding
proteins prolong the half-life of the IGFs and either inhibit or
stimulate the growth-promoting effects of the IGFs in cell culture;
alter the interaction of IGFs with their cell surface
receptors. | https://www.uniprot.org/uniprot/P24593 |
| Hs6-20-10-20 | 0.0014 | -4.5268 | -1.0133 | BACE2 | Beta-site
APP-cleaving enzyme 2 | Proteolytic
processing of the APP; cleaves APP, between residues 690 and 691,
leading to the generation and extracellular release of beta-cleaved
soluble APP, and a corresponding cell-associated C-terminal
fragment. | https://www.uniprot.org/uniprot/Q9Y5Z0 |
| Hs6-27-5-1 | 0.0298 | -2.4615 | -1.0002 | TF | Transferrin | Iron binding
transport proteins, bind two Fe3+ ions in association
with the binding of an anion, usually bicarbonate; responsible for
the transport of iron from sites of absorption and heme degradation
to those of storage and utilization; stimulating. cell
proliferation. | https://www.uniprot.org/uniprot/P02787 |
The list of identified DEGs was validated against
the published literature in order to find evidence for their
involvement in BPH or other prostatic conditions. It was revealed
that, for instance, the association of the genes ADAM
metallopeptidase with thrombospondin type 1 motif 1 (ADAMTS1)
(34), folate hydrolase 1 (FOLH1)
or Prostate-specific membrane antigen (35) and insulin like growth factor binding
protein 5 (IGFBP5) (36) with BPH
was listed in the DisGeNET database (37). Modified ADAMTS1 expression results
in markedly changed blood vessel morphology and altered
thrombospondin-1(TSP1) levels in tumors. Loss of ADAMTS1 is
associated with small-diameter vessels that are consistent with
more aggressive prostate tumors (38). These results suggest that ADAMTS1 is
an important regulatory factor of tumor growth and angiogenesis
during prostate cancer progression. According to the Human Protein
Atlas (https://www.proteinatlas.org/),
ADAMTS1RNA expression is significantly enhanced in prostate tissue.
The DisGeNET database reports that PMS1 is a biomarker of malignant
prostate neoplasms (39,40), and anterior gradient 2(AGR2) is
highly associated with prostate neoplasms (41-44)
and prostate carcinoma (44). Zinc
finger proteins (ZNF) ZNF91(45),
ZFX (46), ZNF185(47), ZNF132(48) and myc associated zinc finger protein
(49), as the family of ZNF3, have
been associated with prostate pathology, prostate cancer
progression and prostate cancer pathogenesis. The association of
zinc finger proteins with BPH and prostate cancer was reviewed in
Rahman (50) in 2016. Fibrinogen
like-1 (FGL1) has been associated with prostate cancer and
high-grade prostatic intraepithelial neoplasia (HGPIN) (51). Collagen type XII α1 chain (COL12A1)
is upregulated in BPH (52-54).
Similarly, other identified DEGs are involved in different prostate
diseases. One of the key biomarkers involved in BPH, but not in
prostate cancer, is Steroid 5α-reductase 2 (SRD5A2) (55-58).
It was not identified in the differential expression analysis of
the present study, but it was considered for drug interaction
studies.
Another gene identified in this analysis was FOLH1,
also known as prostate-specific membrane antigen. It encodes a type
II transmembrane glycoprotein expressed in a number of tissues,
including the prostate. In the prostate, FOLH1 is upregulated in
cancerous cells, has been used as a diagnostic and prognostic
marker for prostate cancer (59)
and was also proposed as a possible marker for neurological
disorders such as Alzheimer's and Huntington's disease (60). According to GeneCards, the Human
Gene Database (https://www.genecards.org), FOLH1 is involved in
prostate tumor progression (61,62).
Finally, insulin-like growth factor binding proteins (IGFBPs)
exhibit abnormalities in prostatic stromal cells in BPH (36,63).
Gene and functional annotation
clustering
Clustered annotations make the functional analyses
more clear and focused. Clustering algorithms rely on the
hypothesis that similar annotations should have similar gene
members (64). The functional
annotation clustering of the present study used Kappa statistics to
estimate the degree of the common genes between two annotations,
and fuzzy heuristic clustering was used to classify the groups of
similar annotations based on kappa values. Hence, common gene
annotations have a high chance of being grouped together. This
eases the biological analysis and interpretation at the group
level.
After gene clustering and functional annotation
clustering, only two gene clusters were obtained with the ‘lowest’
classification stringency. The first cluster contained five genes,
namely bromo domain adjacent to zinc finger domain 2B, TEA domain
transcription factor 1 (TEAD1), erythroid differentiation
regulatory factor 1, ZNF3 and forkhead box P2with an enrichment
score of 0.68 and the second cluster contained only three genes,
namelyβ-secretase 2, ADAMTS1 and FOLH1 with an enrichment score of
0.47. Fig. 4 presents a 2D view of
clustered genes with their associated gene terms.
For the functional annotation clustering analysis, a
‘high’ classification stringency was selected and 4 annotation
clusters were obtained, as presented in Table III and Fig. 5. Out of the 4 clusters (Table III and Fig. 5), the first 3 were significantly
enriched (enrichment scores of 0.95, 0.70 and 0.52). The P-values
of functional Gene Ontology terms of these 3 significantly enriched
clusters are also reasonably acceptable. Each of the four terms
within cluster 1 was associated with both overlapping as well as
differing genes (Fig. 5B). The
terms of clusters 2 and 4 were only associated with overlapping
genes.
 | Table IIIResults of functional annotation
clustering analysis. |
Table III
Results of functional annotation
clustering analysis.
| A, Annotation
cluster 1 (enrichment score: 0.95) |
|---|
| Category | Term | Count | P-value | Genes | Benjamini | FDR |
|---|
| UP_KEYWORDS | Secreted | 6 | 0.0526 | TF, COL12A1,
ADAMTS1, FGL1, AGR2, IGFBP5 | 0.9020 | 44.1446 |
| UP_KEYWORDS | Signal | 8 | 0.1386 | TF, CDC45, BACE2,
COL12A1, ADAMTS1, FGL1, AGR2, IGFBP5 | 0.8822 | 79.9935 |
| UP_SEQ_FEATURE | Signal peptide | 7 | 0.1472 | TF, BACE2, COL12A1,
ADAMTS1, FGL1, AGR2, IGFBP5 | 1.0000 | 85.0691 |
| UP_KEYWORDS | Disulfide bond | 7 | 0.1475 | TF, BACE2, COL12A1,
ADAMTS1, FGL1, AGR2, IGFBP5 | 0.8592 | 82.1005 |
| B, Annotation
cluster 2 (enrichment score: 0.70) |
| Category | Term | Count | P-value | Genes | Benjamini | FDR |
| UP_KEYWORDS | Protease | 3 | 0.1139 | FOLH1, BACE2,
ADAMTS1 | 0.9257 | 72.8568 |
|
GOTERM_BP_DIRECT | GO:0006508~
proteolysis | 3 | 0.1283 | FOLH1, BACE2,
ADAMTS1 | 0.9987 | 80.3993 |
| UP_KEYWORDS | Hydrolase | 3 | 0.5431 | FOLH1, BACE2,
ADAMTS1 | 0.9810 | 99.9785 |
| C, Annotation
cluster 3 (enrichment score: 0.52) |
| Category | Term | Count | P-value | Genes | Benjamini | FDR |
| UP_KEYWORDS | Transcription
regulation | 5 | 0.2338 | EDRF1, TEAD1,
BAZ2B, ZNF3, FOXP2 | 0.8282 | 94.3394 |
| UP_KEYWORDS | Transcription | 5 | 0.2496 | EDRF1, TEAD1,
BAZ2B, ZNF3, FOXP2 | 0.8287 | 95.4801 |
|
GOTERM_BP_DIRECT | GO:0006351~
transcription, DNA-templated | 4 | 0.4491 | EDRF1, BAZ2B, ZNF3,
FOXP2 | 1.0000 | 99.9156 |
| D, Annotation
cluster 4 (enrichment score: 0.01) |
| Category | Term | Count | P-value | Genes | Benjamini | FDR |
|
GOTERM_CC_DIRECT | GO:0016021~
integral component of membrane | 4 | 0.9615 | FOLH1, BACE2,
HECTD4, ADAMTS1 | 1.0000 | 100 |
| UP_KEYWORDS | Transmembrane
helix | 4 | 0.9631 | FOLH1, BACE2,
HECTD4, ADAMTS1 | 1.0000 | 100 |
| UP_KEYWORDS | Transmembrane | 4 | 0.9637 | FOLH1, BACE2,
HECTD4, ADAMTS1 | 1.0000 | 100 |
| UP_KEYWORDS | Membrane | 4 | 0.9958 | FOLH1, BACE2,
HECTD4, ADAMTS1 | 1.0000 | 100 |
Protein interaction analysis
Protein-protein interactions (PPIs) have a crucial
role in cells and control essential cellular and biological
processes. Any disease-causing mutations affecting PPIs may lead to
disruption of protein-DNA interactions, protein misfolding and new
undesirable interactions (65). A
better understanding of possible PPIs allows for the prediction of
pathways, protein functions and potential novel therapeutic
targets. In the present study, gene interactions were predicted
using the GeneMania tool with customized gene-gene interaction
parameters, such as physical interactions and gene co-expression
interactions only, as presented in Fig.
6. The physical interactions are depicted in pink, while
co-expression interactions are displayed in purple. The maximum
resultant genes and maximum resultant attributes were set to
default, i.e., 20 and 10, respectively. The genes that do not
interact with any other genes under these parameter settings were
removed from the network (Fig. 6).
This way, 3 different interaction networks were obtained (Fig. 6). The size of the nodes represents
the gene score, i.e., the degree with which GeneMania predicted the
gene-gene association. Similarly, the thickness of the edges
represents the strength of the interaction. The topological
analysis of gene-gene interactions suggested that the genes IGFBP5,
TEAD1 and transferrin are hub genes that have direct or indirect
interactions with other DEGs identified. Most of these interactions
are physical interactions with good strengths (pink color). This
suggests that these DEGs may be used for the rapeutic strategies
and as drug targets (66,67).
RNA tissue specificity analysis
Genes have unique expression patterns that are
broadly classified as tissue-specific or housekeeping. In a
multicellular organism, knowledge of the tissue-specificity of a
gene contributes to a better understanding of its function
(68). In the present study, tissue
specificity was measured by counting the number of tissues each
gene was expressed in.
Both disease and trait phenotypes are under dynamic
tissue-specific regulation. The major purpose of performing RNA
tissue specificity analyses was to better understand how the
expression of genes and its regulatory processes may be affected by
disease or other biological factors. The ADAMTS1 gene is expressed
in numerous tissues, including the ovary, adipose tissue,
gallbladder and placenta. Fig. 7
presents the results of the tissue specificity analysis of certain
DEGs in BPH identified in the present study. It was observed that
the prostate tissue specificity score of all of the DEGs identified
was low (normalized expression, <25), and the differential
expression of these genes in BPH samples may be due to genetic
variations leading to BPH.
Molecular docking and interaction
analysis Extraction of phytochemical compounds
In the present study, only six compounds (namely
cinnamonitride, astragaloside, cornuside, polyporenic acid C,
berberine and alisol A monoacetate) were considered, since they
have been used for the treatment of BPH and its phytochemical
compound structures are available.
Docking
By docking ligands (phytochemical compounds) to
receptor proteins, it was indicated that polyporenic acid C and
alisol A were notable to bind to the proteins under any of the
tested conditions. Dockings were possible only between the
prostatic receptors and cinnamonitride, astragaloside, berberine
and cornuside. Fig. 8 presents the
best binding positions for each receptor-ligand complex
docking.
Interaction analysis
After comparing the dockings, it was revealed that
berberine had the higher binding affinity for BPH target receptors.
Therefore, the best-bound complexes with berberine were selected
and subjected to a PLIP interaction analysis. Fig. 9 demonstrates interactions formed
between berberine and the selected target receptors, including
hydrophobic interactions, hydrogen bonds and salt bridges. Out of
the four complexes subjected to the interaction analysis, the
interactions formed between AGR2 (protein databank ID, 2LNT) and
berberine were the most stable due to a balance between the number
of hydrogen and hydrophobic bonds. The complex between berberine
and AGR2 had 5 hydrogen bonds stabilizing the complex (the more
hydrogen bonds in a complex, the more stable the complex).
Conclusions
In the present study, genes with differential
expression between BPH and normal prostatic tissues were discovered
and interaction analyses associated with BPH phenotypes were
performed. A general framework for mapping complex interactions
from genome-wide genotype data was established and interactions
with Chinese herbal drugs were identified.
The recent discovery of novel genomic mutations and
the availability of high-throughput screening and microarray
technologies have facilitated the uncovering of gene biomarkers for
the diagnosis, prognosis and treatment of various diseases, such as
BPH (31-33).
With the help of in silico approaches, high-throughput
microarray data were analyzed to identify DEGs as biomarkers for
BPH; furthermore, PPI, gene clustering and tissue specificity
analyses were performed to associate their expression to BPH
phenotypes. In addition, molecular docking studies of certain
short-listed gene biomarkers [AGR2 (2LNT), SRD5A2 (6OQX), ZNF3
(5T00) and COL12A1 (1U5M)] were performed to identify alternative
Chinese herbal drugs for the treatment of BPH. The results suggest
that the AGR2 receptor (2LNT) and berberine (Huang Bo) form a
stable complex that maybe examined in further pharmacological
studies. Further experimental studies are required to confirm the
present computational results.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets generated and/or analyzed during the
current study are available in the NCBI-GEO repository (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE6099).
Authors' contributions
MG conceived and designed the study; MG and ZC
collected and analyzed the data for this study; ZC wrote the
manuscript; and MG reviewed and edited the manuscript. All authors
read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Lorenzo G, Hughes TJR, Dominguez-Frojan P,
Reali A and Gomez H: Computer simulations suggest that prostate
enlargement due to benign prostatic hyperplasia mechanically
impedes prostate cancer growth. Proc Natl Acad Sci USA.
116:1152–1161. 2019.PubMed/NCBI View Article : Google Scholar
|
|
2
|
NIH-NIDDK. Prostate Enlargement (Benign
Prostatic Hyperplasia)|NIDDK [Internet]. National Institute of
Diabetes and Digestive and Kidney Diseases. 2019 [cited 2020 Jan
16]. Available from: https://www.niddk.nih.gov/health-information/urologic-diseases/prostate-problems/prostate-enlargement-benign-prostatic-hyperplasia.
|
|
3
|
Nicholson TM and Ricke WA: Androgens and
estrogens in benign prostatic hyperplasia: Past, present and
future. Differentiation. 82:184–199. 2011.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Hata J, Satoh Y, Akaihata H, Hiraki H,
Ogawa S, Haga N, Ishibashi K, Aikawa K and Kojima Y: Molecular
classification of benign prostatic hyperplasia: A gene expression
profiling study in a rat model. Int J Urol. 23:599–612.
2016.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Dai X, Fang X, Ma Y and Xianyu J: Benign
prostatic hyperplasia and the risk of prostate cancer and bladder
cancer: A meta-analysis of observational studies. Medicine
(Baltimore). 95(e3493)2016.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Miah S and Catto J: BPH and prostate
cancer risk. Indian J Urol. 30:214–218. 2014.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Chughtai B, Forde JC, Thomas DD, Laor L,
Hossack T, Woo HH, Te AE and Kaplan SA: Benign prostatic
hyperplasia. Nat Rev Dis Primers. 2(16031)2016.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Kim EH, Larson JA and Andriole GL:
Management of benign prostatic hyperplasia. Annu Rev Med.
67:137–151. 2016.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Barry MJ, Fowler FJ, O'Leary MP,
Bruskewitz RC, Holtgrewe HL and Mebust WK: Measuring
disease-specific health status in men with benign prostatic
hyperplasia. Measurement committee of the American urological
association. Med Care. 33 (Suppl 4):AS145–AS155. 1995.PubMed/NCBI
|
|
10
|
Rawla P: Epidemiology of prostate cancer.
World J Oncol. 10:63–89. 2019.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Levitt JM and Slawin KM: Prostate-specific
antigen and prostate-specific antigen derivatives as predictors of
benign prostatic hyperplasia progression. Curr Urol Rep. 8:269–274.
2007.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Nickel JC, Aaron L, Barkin J, Elterman D,
Nachabé M and Zorn KC: Canadian urological association guideline on
male lower urinary tract symptoms/benign prostatic hyperplasia
(MLUTS/BPH): 2018 update. Can Urol Assoc J. 12:303–312.
2018.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Chen J, Zhang D, Yan W, Yang D and Shen B:
Translational bioinformatics for diagnostic and prognostic
prediction of prostate cancer in the next-generation sequencing
era. Biomed Res Int. 2013(901578)2013.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Mobley RE and Bizzarro MJ: Central
line-associated bloodstream infections in the NICU: Successes and
controversies in the quest for zero. Semin Perinatol. 41:166–174.
2017.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Tacklind J, Fink HA, Macdonald R, Rutks I
and Wilt TJ: Finasteride for benign prostatic hyperplasia. Cochrane
Database Syst Rev. 6(CD006015)2010.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Michel MC and Vrydag W: Alpha1-, alpha2-
and beta-adrenoceptors in the urinary bladder, urethra and
prostate. Br J Pharmacol. 147 (Suppl 2):S88–S119. 2006.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Yadav SS, Li J, Lavery HJ, Yadav KK and
Tewari AK: Next-generation sequencing technology in prostate cancer
diagnosis, prognosis, and personalized treatment. Urol Oncol.
33:267.e1–e13. 2015.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Yu ZJ, Yan HL, Xu FH, Chao HC, Deng LH, Xu
XD, Huang JB and Zeng T: Efficacy and side effects of drugs
commonly used for the treatment of lower urinary tract symptoms
associated with benign prostatic hyperplasia. Front Pharmacol.
11(658)2020.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Davis S and Meltzer PS: GEOquery: A bridge
between the gene expression omnibus (GEO) and BioConductor.
Bioinformatics. 23:1846–1847. 2007.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Smyth GK: Limma: Linear models for
microarray data. In: Gentleman R, Carey VJ, Huber W, Irizarry RA
and Dudoit S (eds): Bioinformatics and Computational Biology
Solutions Using R and Bioconductor. Statistics for Biology and
Health, Springer, New York, NY, pp397-420, 2005.
|
|
21
|
Huang DW, Sherman BT and Lempicki RA:
Bioinformatics enrichment tools: Paths toward the comprehensive
functional analysis of large gene lists. Nucleic Acids Res.
37:1–13. 2009.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Warde-Farley D, Donaldson SL, Comes O,
Zuberi K, Badrawi R, Chao P, Franz M, Grouios C, Kazi F, Lopes CT,
et al: The GeneMANIA prediction server: Biological network
integration for gene prioritization and predicting gene function.
Nucleic Acids Res. 38:W214–W220. 2010.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Uhlén M, Fagerberg L, Hallström BM,
Lindskog C, Oksvold P, Mardinoglu A, Sivertsson Å, Kampf C,
Sjöstedt E, Asplund A, et al: Proteomics. Tissue-based map of the
human proteome. Science. 347(1260419)2015.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Gaulton A, Bellis LJ, Bento AP, Chambers
J, Davies M, Hersey A, Light Y, McGlinchey S, Michalovich D,
Al-Lazikani B and Overington JP: ChEMBL: A large-scale bioactivity
database for drug discovery. Nucleic Acids Res. 40:D1100–D1107.
2012.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Kim S, Thiessen PA, Bolton EE, Chen J, Fu
G, Gindulyte A, Han L, He J, He S, Shoemaker BA, et al: PubChem
substance and compound databases. Nucleic Acids Res.
44:D1202–D1213. 2016.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Wishart DS, Knox C, Guo AC, Cheng D,
Shrivastava S, Tzur D, Gautam B and Hassanali M: DrugBank: A
knowledgebase for drugs, drug actions and drug targets. Nucleic
Acids Res. 36:D901–D906. 2008.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Trott O and Olson AJ: AutoDock Vina:
Improving the speed and accuracy of docking with a new scoring
function, efficient optimization, and multithreading. J Comput
Chem. 31:455–461. 2010.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Salentin S, Schreiber S, Haupt VJ, Adasme
MF and Schroeder M: PLIP: Fully automated protein-ligand
interaction profiler. Nucleic Acids Res. 43:W443–W447.
2015.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Schena M, Shalon D, Davis RW and Brown PO:
Quantitative monitoring of gene expression patterns with a
complementary DNA microarray. Science. 270:467–470. 1995.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Draghici S: Data Analysis Tools for DNA
Microarrays. Chapman and Hall/CRC Press, Boca Raton (FL), 2003.
Available from: https://trove.nla.gov.au/version/46530985.
|
|
31
|
Raza K: Reconstruction, topological and
gene ontology enrichment analysis of cancerous gene regulatory
network modules. Curr Bioinform. 11:243–258. 2016.
|
|
32
|
Raza K and Hasan AN: A comprehensive
evaluation of machine learning techniques for cancer class
prediction based on microarray data. Int J Bioinform Res Appl.
11:397–416. 2015.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Raza K: Analysis of microarray data using
artificial intelligence based techniques. In: Handbook of Research
on Computational Intelligence Applications in Bioinformatics,
pp216-239, 2016.
|
|
34
|
Cross NA, Chandrasekharan S, Jokonya N,
Fowles A, Hamdy FC, Buttle DJ and Eaton CL: The expression and
regulation of ADAMTS-1, -4, -5, -9, and -15, and TIMP-3 by TGFbeta1
in prostate cells: Relevance to the accumulation of versican.
Prostate. 63:269–275. 2005.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Burger MJ, Tebay MA, Keith PA, Samaratunga
HM, Clements J, Lavin MF and Gardiner RA: Expression analysis of
delta-catenin and prostate-specific membrane antigen: Their
potential as diagnostic markers for prostate cancer. Int J Cancer.
100:228–237. 2002.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Cohen P, Peehl DM, Baker B, Liu F, Hintz
RL and Rosenfeld RG: Insulin-like growth factor axis abnormalities
in prostatic stromal cells from patients with benign prostatic
hyperplasia. J Clin Endocrinol Metab. 79:1410–1415. 1994.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Piñero J, Ramírez-Anguita JM,
Saüch-Pitarch J, Ronzano F, Centeno E, Sanz F and Furlong LI: The
DisGeNET knowledge platform for disease genomics: 2019 update.
Nucleic Acids Res. 48:D845–D855. 2020.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Gustavsson H, Tesan T, Jennbacken K, Kuno
K, Damber JE and Welén K: ADAMTS1 alters blood vessel morphology
and TSP1 levels in LNCaP and LNCaP-19 prostate tumors. BMC Cancer.
10(288)2010.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Chen Y, Wang J, Fraig MM, Metcalf J,
Turner WR, Bissada NK, Watson DK and Schweinfest CW: Defects of DNA
mismatch repair in human prostate cancer. Cancer Res. 61:4112–4121.
2001.PubMed/NCBI
|
|
40
|
Armenia J, Wankowicz SAM, Liu D, Gao J,
Kundra R, Reznik E, Chatila WK, Chakravarty D, Han GC, Coleman I,
et al: The long tail of oncogenic drivers in prostate cancer. Nat
Genet. 50:645–651. 2018.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Morgenbesser SD, McLaren RP, Richards B,
Zhang M, Akmaev VR, Winter SF, Mineva ND, Kaplan-Lefko PJ, Foster
BA, Cook BP, et al: Identification of genes potentially involved in
the acquisition of androgen-independent and metastatic tumor growth
in an autochthonous genetically engineered mouse prostate cancer
model. Prostate. 67:83–106. 2007.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Maresh EL, Mah V, Alavi M, Horvath S,
Bagryanova L, Liebeskind ES, Knutzen LA, Zhou Y, Chia D, Liu AY and
Goodglick L: Differential expression of anterior gradient gene AGR2
in prostate cancer. BMC Cancer. 10(680)2010.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Bu H, Bormann S, Schäfer G, Horninger W,
Massoner P, Neeb A, Lakshmanan VK, Maddalo D, Nestl A, Sültmann H,
et al: The anterior gradient 2 (AGR2) gene is overexpressed in
prostate cancer and may be useful as a urine sediment marker for
prostate cancer detection. Prostate. 71:575–587. 2011.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Neeb A, Hefele S, Bormann S, Parson W,
Adams F, Wolf P, Miernik A, Schoenthaler M, Kroenig M, Wilhelm K,
et al: Splice variant transcripts of the anterior gradient 2 gene
as a marker of prostate cancer. Oncotarget. 5:8681–8689.
2014.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Paschke L, Rucinski M, Ziolkowska A,
Zemleduch T, Malendowicz W, Kwias Z and Malendowicz LK: ZFP91-a
newly described gene potentially involved in prostate pathology.
Pathol Oncol Res. 20:453–459. 2014.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Jiang H, Zhang L, Liu J, Chen Z, Na R,
Ding G, Zhang H and Ding Q: Knockdown of zinc finger protein
X-linked inhibits prostate cancer cell proliferation and induces
apoptosis by activating caspase-3 and caspase-9. Cancer Gene Ther.
19:684–689. 2012.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Vanaja DK, Cheville JC, Iturria SJ and
Young CY: Transcriptional silencing of zinc finger protein 185
identified by expression profiling is associated with prostate
cancer progression. Cancer Res. 63:3877–3882. 2003.PubMed/NCBI
|
|
48
|
Abildgaard MO, Borre M, Mortensen MM,
Ulhøi BP, Tørring N, Wild P, Kristensen H, Mansilla F, Ottosen PD,
Dyrskjøt L, et al: Downregulation of zinc finger protein 132 in
prostate cancer is associated with aberrant promoter
hypermethylation and poor prognosis. Int J Cancer. 130:885–895.
2012.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Jiao L, Li Y, Shen D, Xu C, Wang L, Huang
G, Chen L, Yang Y, Yang C, Yu Y and Sun Y: The prostate
cancer-up-regulated Myc-associated zinc-finger protein (MAZ)
modulates proliferation and metastasis through reciprocal
regulation of androgen receptor. Med Oncol. 30(570)2013.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Rahman MT: Zinc and benign prostatic
hyperplasia (BPH) & prostate cancer (PCa) association. Med Res
Arch. 4:1–16. 2016.
|
|
51
|
Dunn TA, Fedor HL, De Marzo AM and Luo J:
Molecular profiling of indolent human prostate cancer: Tackling
technical challenges to achieve high-fidelity genome-wide data.
Asian J Androl. 14:385–392. 2012.PubMed/NCBI View Article : Google Scholar
|
|
52
|
O'Malley KJ, Eisermann K, Pascal LE,
Parwani AV, Majima T, Graham L, Hrebinko K, Acquafondata M, Stewart
NA, Nelson JB, et al: Proteomic analysis of patient tissue reveals
PSA protein in the stroma of benign prostatic hyperplasia.
Prostate. 74:892–900. 2014.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Adeola HA, Calder B, Soares NC, Kaestner
L, Blackburn JM and Zerbini LF: In silico verification and parallel
reaction monitoring prevalidation of potential prostate cancer
biomarkers. Future Oncol. 12:43–57. 2015.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Sacca PA, Mazza ON, Scorticati C,
Vitagliano G, Casas G and Calvo JC: Human periprostatic adipose
tissue: Secretome from patients with prostate cancer or benign
prostate hyperplasia. Cancer Genomics Proteomics. 16:29–58.
2019.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Hsing AW, Chen C, Chokkalingam AP, Gao YT,
Dightman DA, Nguyen HT, Deng J, Cheng J, Sesterhenn IA, Mostofi FK,
et al: Polymorphic markers in the SRD5A2 gene and prostate cancer
risk: A population-based case-control study. Cancer Epidemiol
Biomarkers Prev. 10:1077–102. 2001.PubMed/NCBI
|
|
56
|
Choi SY, Kim HJ, Cheong HS and Myung SC:
The association of 5-alpha reductase type 2 (SRD5A2) gene
polymorphisms with prostate cancer in a Korean population. Korean J
Urol. 56:19–30. 2015.PubMed/NCBI View Article : Google Scholar
|
|
57
|
Choubey VK, Sankhwar SN, Carlus SJ, Singh
AN, Dalela D, Thangaraj K and Rajender S: SRD5A2 gene polymorphisms
and the risk of benign prostatic hyperplasia but not prostate
cancer. Asian Pac J Cancer Prev. 16:1033–1036. 2015.PubMed/NCBI View Article : Google Scholar
|
|
58
|
Zeng XT, Su XJ, Li S, Weng H, Liu TZ and
Wang XH: Association between SRD5A2 rs523349 and rs9282858
polymorphisms and risk of benign prostatic hyperplasia: A
meta-analysis. Front Physiol. 8(688)2017.PubMed/NCBI View Article : Google Scholar
|
|
59
|
Lodewijk L, Willems SM, Dreijerink KMA, de
Keizer B, van Diest PJ, Schepers A, Morreau H, Bonenkamp HJ, Van
Engen-van Grunsven IACH, Kruijff S, et al: The theranostic target
prostate-specific membrane antigen is expressed in medullary
thyroid cancer. Hum Pathol. 81:245–254. 2018.PubMed/NCBI View Article : Google Scholar
|
|
60
|
Sácha P, Zámecník J, Barinka C, Hlouchová
K, Vícha A, Mlcochová P, Hilgert I, Eckschlager T and Konvalinka J:
Expression of glutamate carboxypeptidase II in human brain.
Neuroscience. 144:1361–1372. 2007.PubMed/NCBI View Article : Google Scholar
|
|
61
|
O'Keefe DS, Bacich DJ and Heston WD:
Comparative analysis of prostate-specific membrane antigen (PSMA)
versus a prostate-specific membrane antigen-like gene. Prostate.
58:200–210. 2004.PubMed/NCBI View Article : Google Scholar
|
|
62
|
Kinoshita Y, Kuratsukuri K, Landas S,
Imaida K, Rovito PM Jr, Wang CY and Haas GP: Expression of
prostate-specific membrane antigen in normal and malignant human
tissues. World J Surg. 30:628–636. 2006.PubMed/NCBI View Article : Google Scholar
|
|
63
|
Kumar R, Verma V, Sarswat A, Maikhuri JP,
Jain A, Jain RK, Sharma VL, Dalela D and Gupta G: Selective
estrogen receptor modulators regulate stromal proliferation in
human benign prostatic hyperplasia by multiple beneficial
mechanisms-action of two new agents. Invest New Drugs. 30:582–593.
2012.PubMed/NCBI View Article : Google Scholar
|
|
64
|
Huang DW, Sherman BT, Tan Q, Collins JR,
Alvord WG, Roayaei J, Stephens R, Baseler MW, Lane HC and Lempicki
RA: The DAVID gene functional classification tool: A novel
biological module-centric algorithm to functionally analyze large
gene lists. Genome Biol. 8(R183)2007.PubMed/NCBI View Article : Google Scholar
|
|
65
|
Raza K: Protein features identification
for machine learning-based prediction of protein-protein
interactions. In: International Conference on Information,
Communication and Computing Technology, pp305-317, 2017.
|
|
66
|
Raza K and Parveen R: Soft computing
approach for modeling genetic regulatory networks. In: Advances in
Computing and Information Technology. Advances in Intelligent
Systems and Computing. Meghanathan N, Nagamalai D and Chaki N
(eds). Springer, Berlin, Heidelberg, pp1-11, 2013.
|
|
67
|
Raza K and Parveen R: Reconstruction of
gene regulatory network of colon cancer using information theoretic
approach. In: Confluence 2013: The Next Generation Information
Technology Summit (4th International Conference), pp461-466,
2013.
|
|
68
|
Kryuchkova-Mostacci N and Robinson-Rechavi
M: A benchmark of gene expression tissue-specificity metrics. Brief
Bioinform. 18:205–214. 2017.PubMed/NCBI View Article : Google Scholar
|