Introduction
Liver cancer was the sixth most common cancer and
the fourth leading cause of cancer deaths worldwide in 2018, which
resulted in 841,080 new cases and 781,265 deaths, accounting for
4.7% of all cancer cases and 8.2% of all cancer deaths (1). As the most common type of liver
cancer, hepatocellular carcinoma (HCC) comprises 75-85% of cases of
liver cancer (1). Surgical
resection is an optimal modality in patients with HCC who have
small solitary tumors and well-preserved liver function, and could
be subjected to this invasive operation and achieve satisfactory
efficacy (2). Patients who are
diagnosed in advanced stages of HCC, who account for the majority
of all cases, are often not suitable candidates for surgical
resection (2). Liver
transplantation, another curative treatment option, is most
suitable for patients with HCC who are not good candidates for
resection, although donor shortage and high medical costs limit its
application (2). Hence,
investigating the molecular mechanisms of HCC progression is
required for the detection of novel and effective therapeutic
targets to improve HCC prognosis.
Integrins, which are heterodimers, consist of α and
β subunits, which participate in a range of cellular processes
including cell survival, growth, invasion and migration (3). Integrin α7 (ITGA7), which belongs to
the integrin family of adhesion molecules, plays a role in
cell-cell and cell-extracellular matrix interactions in multiple
cellular processes (4). According
to previous studies, ITGA7 is oncogenic in the pathological
processes of several carcinomas including glioblastoma, esophageal
squamous cell carcinoma (OSCC) and lung cancer (4-6).
For instance, ITGA7 promotes OSCC cell migration and invasion,
while concurrently increasing E-cadherin and α-smooth muscle actin
(α-SMA) expression, which are common markers of
endothelial-mesenchymal transition (EMT), indicating that ITGA7 may
promote malignant cellular function and induce EMT in OSCC cells
(5). Hence, it was hypothesized
that ITGA7 may also have promotive effects on cellular function and
EMT regulation in HCC. However, to the best of our knowledge,
little is known about the role of ITGA7 in HCC. Thus, the present
study aimed to investigate the effects of ITGA7 on regulating HCC
progression and EMT.
Materials and methods
Cell culture
Human normal liver epithelial cells (THLE-3) and HCC
cell lines SKHEP1 and SNU449 were purchased from American Type
Culture Collection. HCC cell lines Li7 and Huh7 were purchased from
RIKEN BioResource Center. THLE-3 cells were cultured in 90%
bronchial epithelial growth medium (Lonza Group, Ltd.) and 10% FBS
(Gibco; Thermo Fisher Scientific, Inc.). Li7 cells and SNU449 cells
were cultured in 90% RPMI-1640 medium (Gibco; Thermo Fisher
Scientific, Inc.) and 10% FBS. Huh7 cells were cultured in 90% DMEM
(Gibco; Thermo Fisher Scientific, Inc.) and 10% FBS. SKHEP1 cells
were cultured in 90% Eagle's minimum essential medium (Gibco;
Thermo Fisher Scientific, Inc.) and 10% FBS. All medium was added
100 U/ml penicillin and 100 lg/ml treptomycin (Sigma-Aldrich; Merck
KGaA). Cells were maintained in a humid incubator at 37˚C.
ITGA7 small interfering RNA (siRNA)
construction and transfection
siRNA was used to knock down ITGA7 expression. ITGA7
siRNA and nonsense siRNA were designed and synthesized by Guangzhou
RiboBio Co., Ltd. ITGA7 siRNA (80 nM) and nonsense siRNA were
transfected into Huh7 cells and SNU449 cells using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer's instructions for
6 h at 37˚C. Subsequently, cells transfected with ITAG7 siRNA were
considered ITGA7-knockdown (KD) cells, and cells transfected with
nonsense siRNA were marked as control cells. At 24 h after
transfection, lTGA7 mRNA and protein expression levels were
determined by RT-qPCR and western blotting; cell apoptosis was
detected by an annexin V/propidium iodide (AV/PI) assay at 48 h,
expression of apoptosis-related protein cleaved caspase 3 was
detected by western blotting, and cell migration and invasion
abilities were assessed by wound scratch and Transwell assays,
respectively. Cell viability was assessed using a Cell Counting
Kit-8 (CCK-8) assay at 0, 24, 48 and 72 h. Additionally, whether
the mRNA and protein expression of E-cadherin, vimentin, N-cadherin
and α-SMA were regulated by ITGA7 was determined by RT-qPCR and
western blotting at 24 h after transfection. In addition, the
sequences of ITGA7 siRNA were as follows: Forward,
5'-GCAUCAAGAGCUUCGGCUATT-3' and reverse,
5'-UAGCCGAAGCUCUUGAUGCTT-3'.
RT-qPCR
ITGA7 mRNA expression was assessed in THLE-3, Li7,
Huh7, SKHEP1 and SNU449 cells. E-cadherin, vimentin, N-cadherin and
α-SMA mRNA expression was assessed in Huh7 and SNU449 cells.
Following cell dissociation using 0.25% trypsin (Gibco; Thermo
Fisher Scientific, Inc.), TRIzol® reagent (Invitrogen;
Thermo Fisher Scientific, Inc.) was used to extract total RNA.
Subsequently, 1 µg RNA was reverse transcribed into cDNA using a
PrimeScript RT reagent kit (Takara Bio, Inc.) with following
thermocycling condition: 42˚C for 15 min and 85˚C for 5 sec. TB
Green™ Fast qPCR mix (Takara, Bio, Inc.) was used for qPCR. The
following thermocycling conditions were used: 95˚C for 5 min; 40
cycles of 95˚C for 5 sec and 61˚C for 30 sec. Gene expression was
calculated using the 2-∆∆Cq method (7). Primer sequences are shown in Table I. GAPDH was used as the internal
reference gene. In addition, the sequences of ITGA7 siRNA were as
follows: Forward, 5'-GCAUCAAGAGCUUCGGCUATT-3' and reverse,
5'-UAGCCGAAGCUCUUGAUGCTT-3'.
 | Table IPrimers used for reverse
transcription-quantitative PCR. |
Table I
Primers used for reverse
transcription-quantitative PCR.
| | Primer sequence
(5'-3') |
|---|
| Target gene | Forward | Reverse |
|---|
| ITGA7 |
GCCACTCTGCCTGTCCAATG |
GGAGGTGCTAAGGATGAGGTAGA |
| E-cadherin |
TGATTCTGCTGCTCTTGCTGTT |
CCTCTTCTCCGCCTCCTTCTT |
| α-SMA |
CATTCACGAGACCACCTACAACAG |
CGCCGATCCACACCGAGTAT |
| GAPDH |
GACCACAGTCCATGCCATCAC |
ACGCCTGCTTCACCACCTT |
Western blotting
Total protein was extracted using RIPA Lysis and
Extraction buffer (Thermo Fisher Scientific, Inc.). The Pierce™ BCA
Protein Assay kit (Thermo Fisher Scientific, Inc.) was used to
measure protein concentration. Subsequently, 20 µg protein sample
was fractionated using NuPAGE™ 4-20% Tris-Acetate Midi Protein Gels
(Thermo Fisher Scientific, Inc.), and transferred to PVDF
membranes. Membranes were blocked using 5% skim milk for 2 h at
room temperature and incubated with primary antibodies overnight at
4˚C. Membranes were incubated with a secondary antibody for 1 h at
room temperature. The chemiluminescence of blots was detected using
Pierce™ ECL Plus Western Blotting substrate (Invitrogen; Thermo
Fisher Scientific, Inc.) and then exposed to X-ray film (Kodak)
following treatment. GAPDH was used as the internal reference
protein. Antibodies used for western blotting are listed in
Table II.
 | Table IIAntibodies used for western
blotting. |
Table II
Antibodies used for western
blotting.
| Antibody | Manufacturer | Catalog number | Dilution |
|---|
| Primary
antibodies | | | |
|
ITGA7 mouse
mAb | Santa Cruz
Biotechnology, Inc. | sc-51576 | 1:1,000 |
|
E-cadherin
mouse mAb | Santa Cruz
Biotechnology, Inc. | sc-8426 | 1:1,000 |
|
Vimentin
mouse mAb | Santa Cruz
Biotechnology, Inc. | sc-6260 | 1:500 |
|
N-cadherin
mouse mAb | Santa Cruz
Biotechnology, Inc. | sc-393933 | 1:1,000 |
|
α-SMA mouse
mAb | Santa Cruz
Biotechnology, Inc. | sc-53142 | 1:1,000 |
|
Cleaved
caspase 3 mouse mAb | Cell Signaling
Technology, Inc. | 9664S | 1:1,000 |
|
GAPDH mouse
mAb | Santa Cruz
Biotechnology, Inc. | sc-47724 | 1:1,000 |
| Secondary
antibodies | | | |
|
Goat
anti-mouse IgG-HRP | Santa Cruz
Biotechnology, Inc. | sc-2005 | 1:5,000 |
CCK-8 assay
Cells were plated at a density of 3x104
the 96-well plates for 24 h. Following the addition of 10 µl CCK-8
solution (Dojindo Molecular Technologies, Inc.) and 90 µl RPMI-1640
medium to each plate, cells were incubated at 37˚C with 5%
CO2. Optical density values were detected using a
microplate reader (Biotek Instruments, Inc.).
AV/PI
Cells were digested with pancreatin and washed with
PBS. Following suspension in 100 µl binding buffer and addition of
5 µl AV and 5 µl PI, cells were incubated in the dark with a
Annexin V-FITC Apoptosis Detection kit according to the
manufacturer's protocol (Sigma-Aldrich; Merck KGaA).
Wound scratch assay
Cells, which were pre-culture in medium containing
1% FBS for 24 h, were cultured until 80% confluence and scraped
with a sterile pipette tip to create adherent cell gaps.
Subsequently, the cells were incubated and then observed at 0 and
24 h by inverted fluorescence microscopy (Nikon Corporation). The
migration rate was calculated as follows: Migration rate = (scraped
area- residual area)/scraped area.
Transwell assay
After coating Matrigel basement membrane matrix (BD
Biosciences) on the upper Transwell chamber (Costar; Corning, Inc)
at 37˚C for 1 h. Cells (3x104) in FBS-free medium (DMEM
for Huh-7 cells and RPMI-1640 for SNU-449 cells) were seeded in the
upper chamber, and lower chamer was filled with 500 µl 10% FBS
containng-medim (DMEM for Huh-7 cells and RPMI-1640 for SNU-449
cells). Following incubation for 24 h at 37˚C and wiping of the
upper cells, cells in the lower chamber were fixed with
formaldehyde (Sigma-Aldrich; Merck KGaA). After staining with 0.5%
crystal violet (Sigma-Aldrich; Merck KGaA) for 15 min at room
temperature, the invasive cell count of each well was calculated by
the averaging the invasive cell count of five fields of view in
each well, which was observed using an inverted fluorescence
microscopy (Nikon Corporation) at a magnification of x200.
Statistical analysis
Statistical analysis and graph plotting were
performed using GraphPad Prism 7.02 (GraphPad Software, Inc.). All
assays were repeated in triplicate. Data are presented as the mean
± SD. Comparison between two groups were performed using unpaired
t-test, while multiple comparisons were performed using Dunnett's
t-test. P<0.05 was considered to indicate a statistically
significant difference.
Results
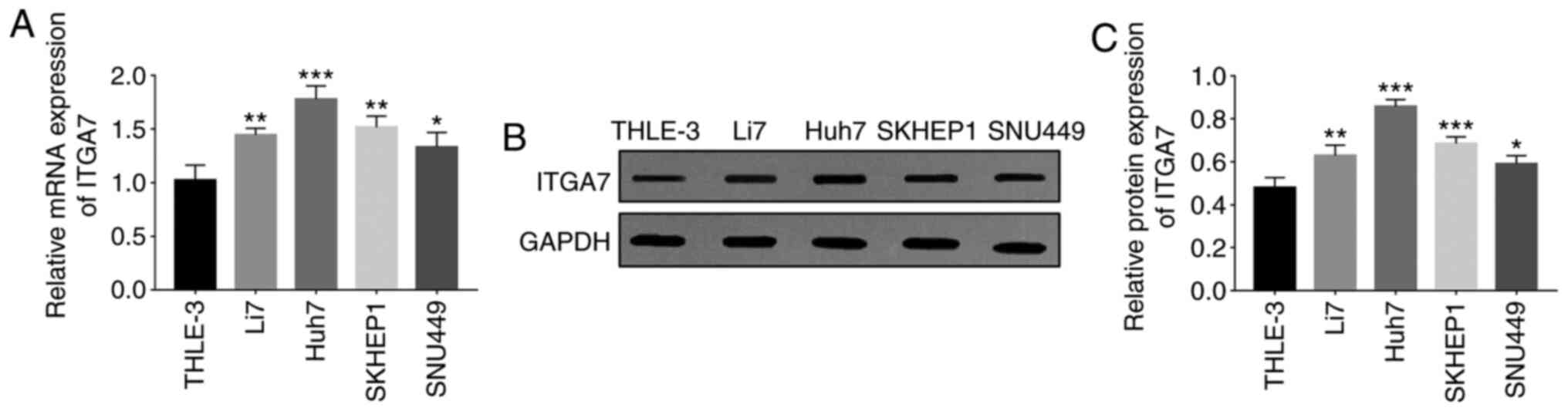
ITGA7 is highly expressed in HCC cell
lines compared with human normal liver epithelial cells
ITGA7 mRNA expression was higher in HCC cell lines
[including Li7 (P<0.01), Huh7 (P<0.001), SKHEP1 (P<0.01)
and SNU449 (P<0.05) cells] compared with human normal liver
epithelial cells (THLE-3 cells) (Fig.
1A). In addition, ITGA7 protein expression was elevated in HCC
cell lines [Li7 (P<0.01), Huh7 (P<0.001), SKHEP1 (P<0.001)
and SNU449 (P<0.05) cells] compared with human normal liver
epithelial cells (THLE-3 cells; Fig.
1B and C). ITGA7 expression was
the highest in Huh7 cells and the lowest in SNU449 cells. Hence,
Huh7 and SNU449 cells were selected for subsequent experiments to
assess the effects of ITGA7 knockdown on cell proliferation,
migration, invasion and EMT in HCC cells.
ITGA7 expression is attenuated in the
ITGA7-KD group compared with the control group after
transfection
ITGA7 mRNA (P<0.001; Fig. 2A) and protein (Fig. 2C) expression decreased in the
ITGA7-KD group compared with the control group in Huh7 cells. ITGA7
mRNA (P<0.001; Fig. 2B) and
protein (Fig. 2D) expression levels
were also lower in the ITGA7-KD group compared with the control
group in SNU449 cells.
ITGA7 knockdown decreases cell
proliferation but increases cell apoptosis
The effects of ITGA7 on regulating cell
proliferation and apoptosis in Huh7 and SNU449 cells were then
investigated. In Huh7 cells, cell proliferation decreased in the
ITGA7-KD group at 48 h (P<0.05) and 72 h (P<0.01) compared
with the control group (Fig. 3A).
Meanwhile, the cell apoptosis rate at 48 h was increased in the
ITGA7-KD group compared with the control group (P<0.01; Fig. 3C and E), and cleaved caspase 3 protein
expression was increased in the ITGA7-KD group compared with the
control group (Fig. 3G). In SNU449
cells, cell proliferation was reduced in the ITGA7-KD group at 48 h
(P<0.05) and 72 h (P<0.05) compared with the control group
(Fig. 3B). The cell apoptosis rate
at 48 h was enhanced in the ITGA7-KD group compared with the
control group (P<0.01; Fig. 3D
and F), and cleaved caspase 3
protein expression increased in the ITGA7-KD group compared with
the control group (Fig. 3H).
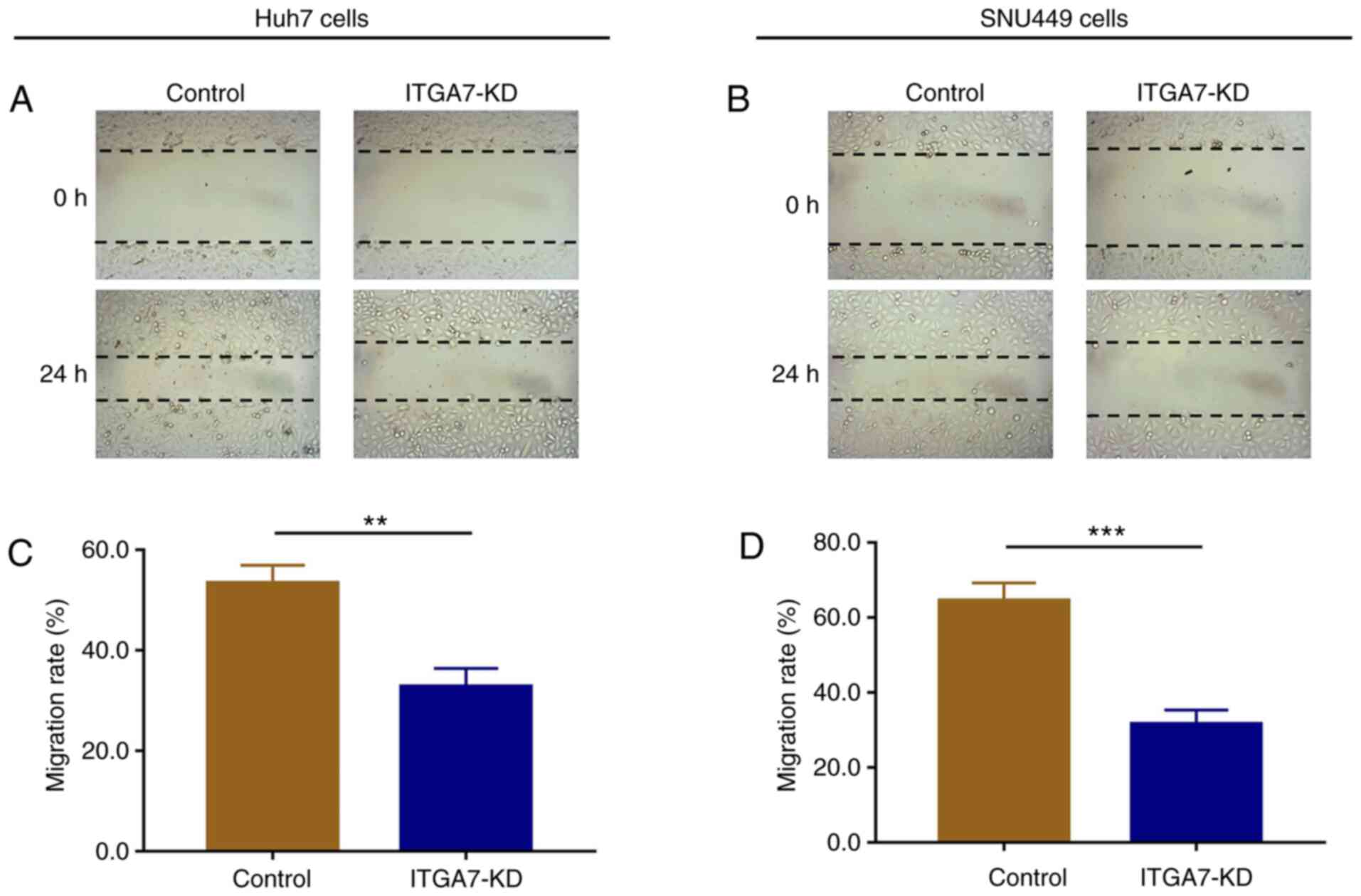
ITGA7 knockdown represses cell
migration
The effects of ITGA7 on regulating cell migration in
Huh7 and SNU449 cells were assessed. In Huh7 cells, the wound
scratch assay showed that the migration rate at 24 h after
transfection was lower in the ITGA7-KD group compared with the
control group (P<0.01; Fig. 4A
and C). In SNU449 cells, the
migration rate at 24 h after transfection was also attenuated in
the ITGA7-KD group compared with the control group (P<0.001;
Fig. 4B and D).
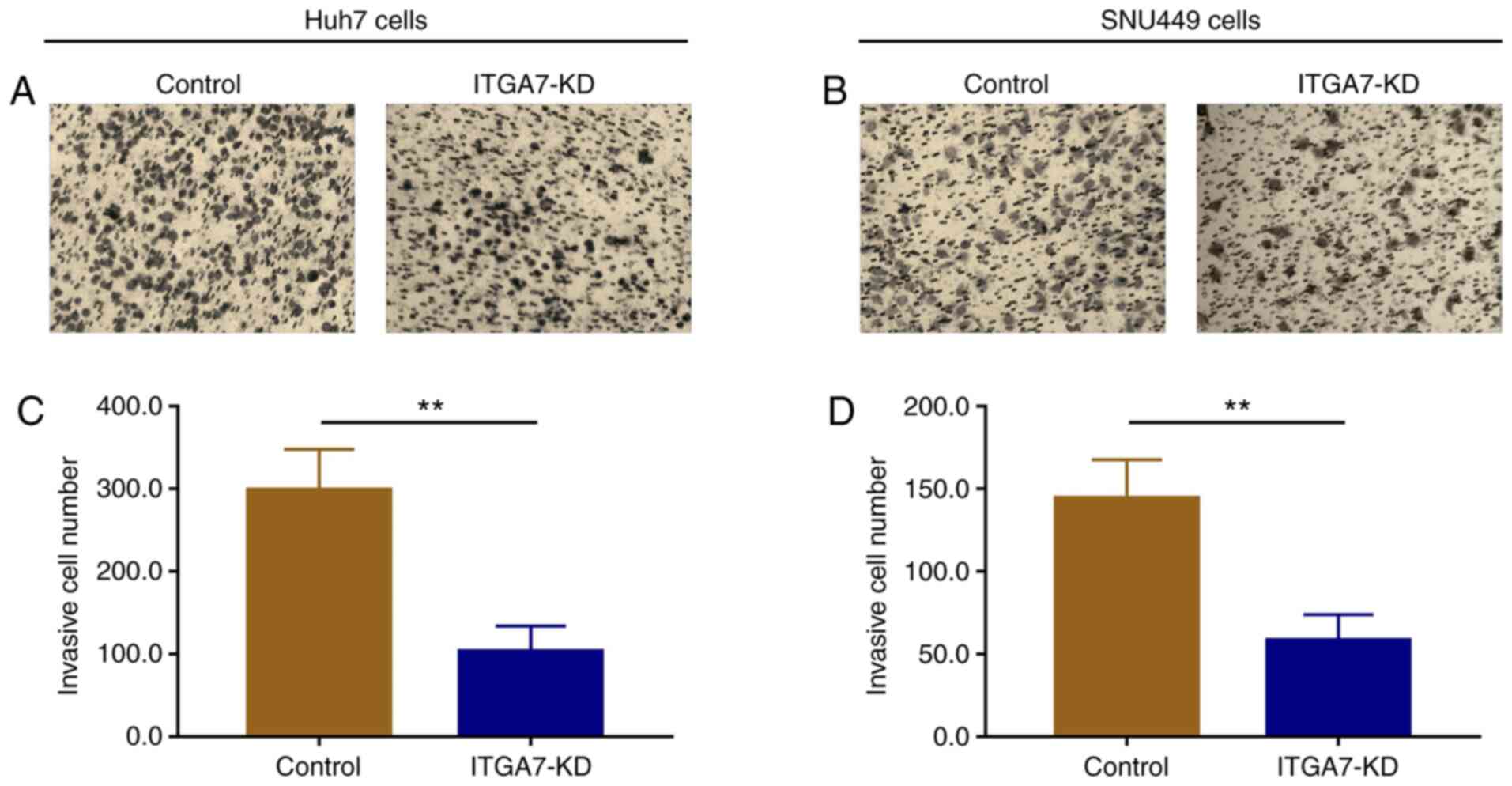
ITGA7 knockdown suppresses cell
invasion
Transwell assays were performed to investigate the
effects of ITGA7 on regulating Huh7 and SNU449 cell invasion. The
number of invasive Huh7 cells was decreased in the ITGA7-KD group
compared with the control group (P<0.01) at 24 h (Fig. 5A and C). The number of invasive SNU449 cells was
also lower in the ITGA7-KD group compared with the control group
(P<0.01) at 24 h (Fig. 5B and
D).
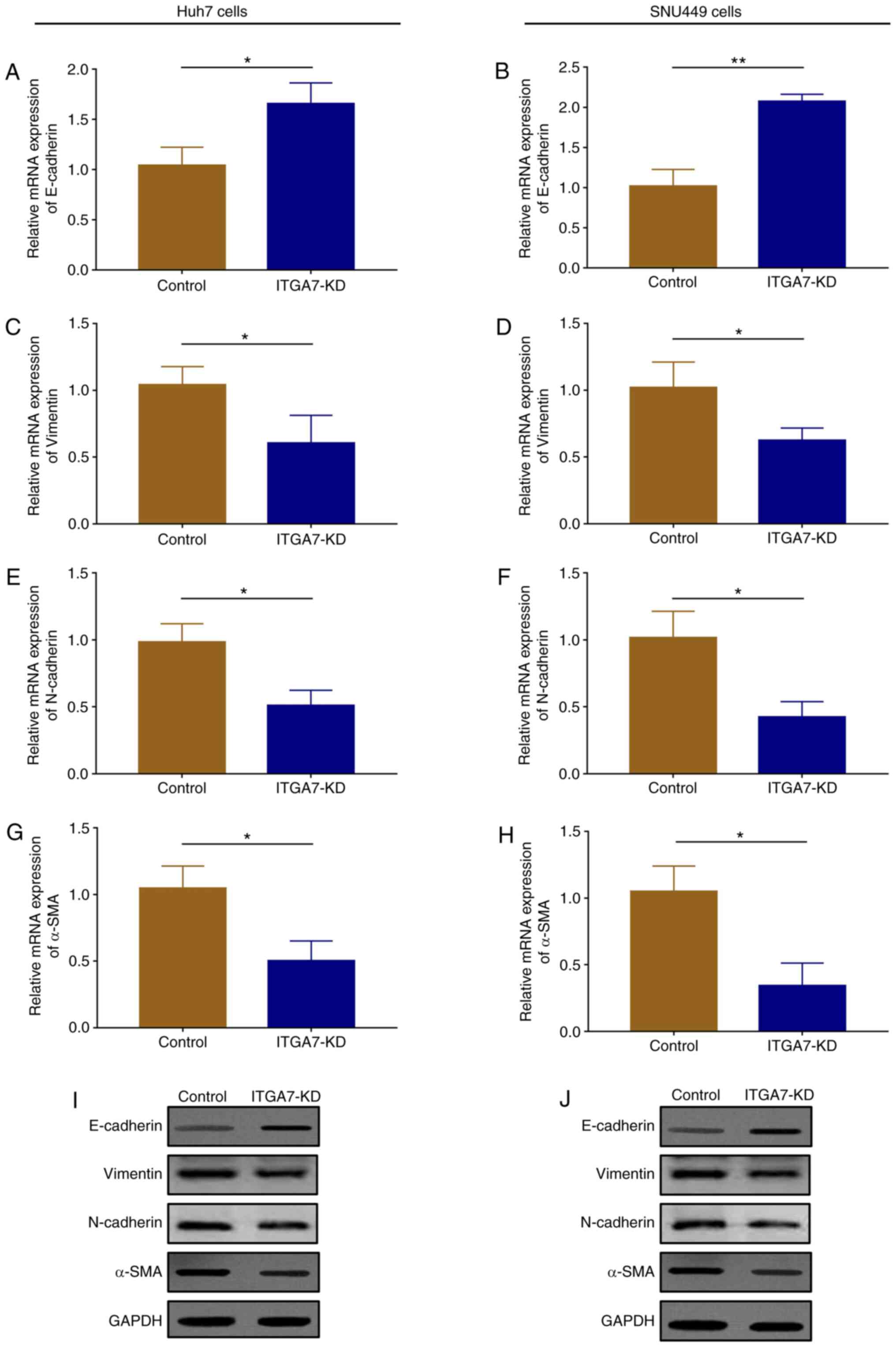
ITGA7 knockdown represses EMT
To assess the effects of ITGA7 on EMT underlying HCC
pathogenesis, the expression levels of EMT markers, including
E-cadherin, vimentin, N-cadherin and α-SMA, were detected in Huh7
and SNU449 cells after transfection. In Huh7 cells, mRNA
(P<0.05) and protein expression of E-cadherin increased
(Fig. 6A and I); however, mRNA (all P<0.05) and
protein expression levels of vimentin (Fig. 6C and I), N-cadherin (Fig. 6E and I) and α-SMA (Fig. 6G and I) were decreased in the ITGA7-KD group
compared with the control group at 24 h. In SNU449 cells, mRNA
(P<0.01) and protein expression levels of E-cadherin (Fig. 6B and J) increased, while mRNA (all P<0.05)
and protein expression levels of vimentin (Fig. 6D and J), N-cadherin (Fig. 6F and J) and α-SMA (Fig. 6H and J) were decreased in the ITGA7-KD group
compared with the control group at 24 h.
Discussion
Integrins are transmembrane protein receptors that
attach cells to the extracellular matrix and bind ligands secreted
by other cells (8,9). As one of the integrins, ITGA7 is
proposed to serve as a key regulator in tumor propagation and
cancer stem cell properties (5,10).
Previous studies revealed that ITGA7 is highly expressed in various
cancer cells, including OSCC and mesothelioma (5,11).
Although these previous studies detected an upregulation of ITGA7
in various cancer cells, to the best of our knowledge, its role in
HCC is still unclear. The present study revealed that ITGA7 was
overexpressed in HCC cell lines, including Li7, Huh7, SKHEP1 and
SNU449 cells, compared with human normal liver epithelial THLE-3
cells. ITGA7 regulates several genes and pathways, including the
focal adhesion kinase (FAK)/AKT-zinc finger E-box Binding Homeobox
1 (ZEB1) signaling pathway, to promote cell proliferation and
metastasis, subsequently contributing to the malignant
transformation of HCC (5,12). Thus, ITGA7 was overexpressed in HCC
cells compared with healthy control cells.
Previous studies indicated that ITGA7 is involved in
the pathological progression of different carcinomas through
affecting cell activities such as cell migration and invasion
(5,6,9,13-15).
For example, the interaction between ITGA7 and laminin-induced
outside-in signaling contributed to glioblastoma stem-like cell
growth and invasion (12).
Furthermore, the interaction between ITGA7 and S100P activated the
FAK/AKT-ZEB1 signaling pathway, which induced lung cancer cell
migration and invasion (6).
Furthermore, ITGA7 is associated with cancer cell stemness. In
another study, ITGA7 promoted the stemness of OSCC cells via
FAK/MAPK/ERK signaling, which subsequently induced the
tumorigenicity and metastasis of OSCC (5). In addition, ITGB7 knockdown enhanced
cell apoptosis but inhibited cell proliferation and invasion in
breast cancer (14). Although a few
studies have been performed to explore the role of ITGA7 in
different types of carcinoma, there remain certain contradictions.
Several lines of evidence revealed the role of ITGA7 as a tumor
suppressor in various malignancies. For example, ITGA7 appears to
activate cyclin-dependent kinase inhibitor 3 (CDKN3) and Rac
GTPase-activating protein 1 (RACGAP1) expression to inhibit cell
motility and metastasis of HCC cells (16). Another study revealed that ITGA7 may
be a tumor suppressor that impedes tumor growth and inhibits
migration in prostate cancer (9).
Additionally, ITGB7 interacts with high temperature requirement A2
to promote prostate cancer cell death (13). To the best of our knowledge, little
is known about the role of ITGA7 in HCC. The present study
investigated the effect of ITGA7 on regulating HCC cell activities.
It was found that ITGA7 knockdown decreased cell proliferation,
migration and invasion, but increased apoptosis of HCC cells, which
suggested that ITGA7 knockdown might suppress the function of HCC
cells. There are a few possible explanations for this. Similar to
its cancerogenic effect on tumor progression in lung cancer, ITGA7
might interact with S100P to trigger FAK/AKT-ZEB1 signaling to
enhance HCC cell proliferation, migration and invasion, thereby
contributing to HCC tumor progression (6). Similar to the promotive effects of
ITGA7 on tumor progression in glioblastoma, ITGA7 might accelerate
HCC cell growth and invasion via interacting with laminin-induced
outside-in signaling, thereby leading to tumor progression of HCC
(12). ITGA7 may attach cells to
the extracellular matrix and interact with ligands secreted by
other cells to activate HCC cell invasion and migration, which
subsequently promotes tumor progression of HCC (8,9). ITGA7
may also regulate CDKN3, which dephosphorylates tyrosine residues
of different cyclin-dependent kinases and represses cell cycle
progression in yeast and mammalian cells, to increase HCC cell
invasion and motility, subsequently accelerating tumor progression
in HCC (16-18).
ITGA7 also may modulate RACGAP1 to increase cell growth, enhance
cell motility and promote tumor metastasis. Taken together, it may
be hypothesized that ITGA7 knockdown suppresses tumor progression
of HCC (16,19). In addition, the discrepancies in
results between the present study and previous studies might result
from differences between the malignances studied. The present study
focused on HCC, while the majority of previous studies focused on
other types of cancer. Due to the complexity of malignant
pathological processes, different malignances might be distinctive
in terms of pathological features. Thus, the effects of ITGA7 on
cellular function and its underlying mechanisms in other
malignances may differ. Besides, different cell lines, different
assay operation times and experimental procedures may have also
contributed to distinctive results among different studies.
To the best of our knowledge, limited information is
available regarding the role of ITGA7 in EMT and tumor metastasis
(5,20,21).
One previous study reported that ITGA7 promoted OSCC cell migration
and invasion and induced EMT (5).
EMT is not only a well-coordinated process controlled by multiple
signaling pathways during embryonic development, but also a
pathological characteristic in neoplasia and fibrosis (22-25).
EMT has been considered as an essential regulator linked to tumor
progression and tumor metastasis through accelerating cancer cell
invasion and dissemination to distant organs (22-25).
To assess the effects of ITGA7 on regulating EMT in HCC,
E-cadherin, α-SMA, vimentin and N-cadherin levels were detected.
ITGA7 knockdown increased E-cadherin expression and decreased α-SMA
expression in HCC cells. To summarize, ITGA7 knockdown may repress
EMT in HCC. However, how ITGA7 knockdown suppressed cell
proliferation, migration, invasion and ETM in HCC remains unclear.
Further experiments, such as RNA sequencing, bioinformatics and
subsequent validation by RT-qPCR are required.
In conclusion, ITGA7 knockdown suppressed HCC cell
proliferation, migration, invasion and EMT, and promoted apoptosis.
These data indicated that ITGA7 might be a novel and effective
treatment target for HCC.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
ZWu and XK made substantial contributions to the
design of the present study, ZWu, XK and ZW were responsible for
data acquisition and interpretation. All authors read and approved
the final manuscript. All authors agree to be accountable for all
aspects of the work in ensuring that questions related to the
accuracy or integrity of the work are appropriately investigated
and resolved.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Forner A, Llovet JM and Bruix J:
Hepatocellular carcinoma. Lancet. 379:1245–1255. 2012.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Desgrosellier JS and Cheresh DA: Integrins
in cancer: Biological implications and therapeutic opportunities.
Nat Rev Cancer. 10:9–22. 2010.PubMed/NCBI View
Article : Google Scholar
|
|
4
|
Carrasco-Garcia E, Auzmendi-Iriarte J and
Matheu A: Integrin α7: A novel promising target in glioblastoma
stem cells. Stem Cell Investig. 5(2)2018.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Ming XY, Fu L, Zhang LY, Qin YR, Cao TT,
Chan KW, Ma S, Xie D and Guan XY: Integrin α7 is a functional
cancer stem cell surface marker in oesophageal squamous cell
carcinoma. Nat Commun. 7(13568)2016.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Hsu YL, Hung JY, Liang YY, Lin YS, Tsai
MJ, Chou SH, Lu CY and Kuo PL: S100P interacts with integrin α7 and
increases cancer cell migration and invasion in lung cancer.
Oncotarget. 6:29585–29598. 2015.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) Method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Hynes RO: Integrins: Bidirectional,
allosteric signaling machines. Cell. 110:673–687. 2002.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Tan LZ, Song Y, Nelson J, Yu YP and Luo
JH: Integrin α7 binds tissue inhibitor of metalloproteinase 3 to
suppress growth of prostate cancer cells. Am J Pathol. 183:831–840.
2013.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Nunes AM, Barraza-Flores P, Smith CR and
Burkin DJ: Integrin α7: A major driver and therapeutic target for
glioblastoma malignancy. Stem Cell Investig. 4(97)2017.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Burkin DJ and Fontelonga TM: Mesothelioma
cells breaking bad: Loss of integrin α7 promotes cell motility and
poor clinical outcomes in patients. J Pathol. 237:282–284.
2015.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Haas TL, Sciuto MR, Brunetto L, Valvo C,
Signore M, Fiori ME, di Martino S, Giannetti S, Morgante L, Boe A,
et al: Integrin α7 Is a Functional Marker and Potential Therapeutic
Target in Glioblastoma. Cell Stem Cell. 21:35–50.e9.
2017.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Zhu ZH, Yu YP, Zheng ZL, Song Y, Xiang GS,
Nelson J, Michalopoulos G and Luo JH: Integrin alpha 7 interacts
with high temperature requirement A2 (HtrA2) to induce prostate
cancer cell death. Am J Pathol. 177:1176–1186. 2010.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Bai X, Gao C, Zhang L and Yang S: Integrin
α7 high expression correlates with deteriorative tumor features and
worse overall survival, and its knockdown inhibits cell
proliferation and invasion but increases apoptosis in breast
cancer. J Clin Lab Anal. 33(e22979)2019.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Su Y, Guan XQ, Liu FQ and Wang YL: The
effects of MIBG on the invasive properties of HepG2 hepatocellular
carcinoma cells. Int J Mol Med. 34:842–848. 2014.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Ren B, Yu YP, Tseng GC, Wu C, Chen K, Rao
UN, Nelson J, Michalopoulos GK and Luo JH: Analysis of integrin
alpha7 mutations in prostate cancer, liver cancer, glioblastoma
multiforme, and leiomyosarcoma. J Natl Cancer Inst. 99:868–880.
2007.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Gyuris J, Golemis E, Chertkov H and Brent
R: Cdi1, a human G1 and S phase protein phosphatase that associates
with Cdk2. Cell. 75:791–803. 1993.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Hannon GJ, Casso D and Beach D: KAP: A
dual specificity phosphatase that interacts with cyclin-dependent
kinases. Proc Natl Acad Sci USA. 91:1731–1735. 1994.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Kawashima T, Hirose K, Satoh T, Kaneko A,
Ikeda Y, Kaziro Y, Nosaka T and Kitamura T: MgcRacGAP is involved
in the control of growth and differentiation of hematopoietic
cells. Blood. 96:2116–2124. 2000.PubMed/NCBI
|
|
20
|
Brabletz T: EMT and MET in metastasis:
Where are the cancer stem cells? Cancer Cell. 22:699–701.
2012.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan
A, Zhou AY, Brooks M, Reinhard F, Zhang CC, Shipitsin M, et al: The
epithelial-mesenchymal transition generates cells with properties
of stem cells. Cell. 133:704–715. 2008.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Zou J, Li H, Huang Q, Liu X, Qi X, Wang Y,
Lu L and Liu Z: Dopamine-induced SULT1A3/4 promotes EMT and cancer
stemness in hepatocellular carcinoma. Tumour Biol.
39(1010428317719272)2017.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Santamaria PG, Moreno-Bueno G, Portillo F
and Cano A: EMT: Present and future in clinical oncology. Mol
Oncol. 11:718–738. 2017.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Nistico P: Bissell MJandRadisky DC:
Epithelial-mesenchymal transition: general principles and
pathological relevance with special emphasis on the role of matrix
metalloproteinases. Cold Spring Harb Perspect Biol.
4(a011908)2012.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Gaianigo N: Melisi D and Carbone C: EMT
and Treatment Resistance in Pancreatic Cancer. Cancers (Basel).
9(122)2017.PubMed/NCBI View Article : Google Scholar
|