Introduction
Periodontitis is one of the most common chronic
inflammatory diseases and is characterized by the progressive
destruction of the supporting tissues surrounding the tooth,
eventually leading to tooth loss (1-3).
Due to the lack of clear symptoms, the majority of patients
typically miss the early intervention period of this disease,
meaning that extensive damage has already been caused to the entire
dentition on presentation (4). A
review has previously revealed an association between periodontitis
and diabetes complications in the human body (5). Periodontitis is traditionally treated
using non-surgical methods, which mainly involve the subgingival
eradication of bacterial deposits on the dental surface (6). However, deeper understanding on the
etiopathogenesis of the disease is required to develop novelly
effective treatment options (1).
Homeobox containing 1 (HMBOX1) is a member of the
homeobox transcription factor family that has been identified to
serve a role in a large number of biological processes including
proliferation, inflammation and apoptosis (7-10).
A previous genomic and proteomic analysis found that the expression
levels of HMBOX1 were downregulated in gingival tissues from
patients with periodontitis compared with a control group without
periodontitis, which identified a potential association between
HMBOX1 expression levels and the pathogenesis of periodontitis
(11). In addition, mice deficient
in HMBOX1 exhibited enhanced levels of apoptosis in vascular
endothelial cells following challenge with lipopolysaccharide
(LPS), which promoted the inflammatory response in a mouse model of
acute lung injury (10,12). However, to the best of our
knowledge, no studies to date have determined the specific role of
HMBOX1 in the pathogenesis of periodontitis.
CXCL10 was first identified in 1985 and has been
found to be regulated by various cytokines, such as IFN-γ and TNF
(13-16).
It has been previously reported that the significantly upregulated
expression of CXCL10 in patients with periodontitis can serve as a
biomarker of periodontitis (17).
In addition, the NF-κB signaling pathway was found to upregulate
the expression of CXCL10 in COMM domain-containing protein
7-treated hepatocellular carcinoma cells (18). Aldo-keto reductase family 1 member
C2 was also discovered to exert potent anti-inflammatory and
antioxidant effects by inhibiting NF-κB activity downstream of
HMBOX1 activation in an atherosclerosis model of LPS-induced
EA.hy926 cells (9).
Therefore, it was hypothesized in the present study
that HMBOX1 may alleviate LPS-induced human periodontal ligament
stem cell (hPDLSC) injury by downregulating CXCL10 expression via
the NF-κB signaling pathway.
Materials and methods
Cell lines and culture
Human periodontal stem cells (hPDLSCs) were
purchased from YBIO (cat. no. YB-Cell-H002) and cultured in DMEM
(Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% FBS
(Gibco; Thermo Fisher Scientific, Inc.) in a humidified incubator
with 5% CO2 at 37˚C. Cells were treated with or without
1 µg/ml LPS (cat. no. L8880; Beijing Solarbio Science &
Technology Co., Ltd.) for 12 or 24 h at 37˚C to construct the in
vitro periodontitis model. The NF-κB activator phorbol
12-myristate 13-acetate (PMA) (19)
(20 ng/ml; Sigma-Aldrich; Merck KGaA) was used to pre-treat hPDLSCs
for 30 min at 37˚C on hPDLSCs before LPS treatment and then
transfected with the HMBOX1 overexpression (Ov-HMBOX1, pEGFP-N1;
vector) plasmid or its control plasmids (Ov-NC, empty plasmids) for
24 h.
Cell transfection
Ov-HMBOX1 (1 µg) and its negative control (Ov-NC;
empty plasmids. 1 µg), short interfering RNA (siRNA/si) for CXCL10
(30 pmol; si-CXCL10-1, 5'-GGUUAAUAAAGUAAUUAUAAC-3'; si-CXCL10-2,
5'-CGUGUUGAGAUCAUUGCUACA-3') and si-NC (30 pmol;
5'-GAUAAUUAUGGAUAAUAAUAC-3') were purchased from Shanghai
GenePharma Co., Ltd. Transfection was performed using
Lipofectamine® 3000 reagent (Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer's protocol.
Transfected cells were cultured for 24 h at 37˚C prior for use in
subsequent experiments.
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA was extracted from hPDLSCs using
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc.). Total RNA (2 µg) was reverse-transcribed into cDNA using a
PrimeScript™ RT kit (Takara Bio, Inc.) under a condition of 37˚C
for 1 h and 72˚C for 10 min. qPCR was subsequently performed using
a SYBR® Premix Ex Taq™ (cat. no. RR420A; Takara Bio,
Inc.) on a LightCycler 480 PCR system (Roche Diagnostics) according
to the manufacturer's protocol. The following thermocycling
conditions were used for the qPCR: Initial denaturation at 95˚C for
2 min; followed by 40 cycles of 95˚C for 20 sec, 60˚C for 20 sec
and 72˚C for 20 sec, followed by a final step at 78˚C for 5 min.
The primers sequences are as following: HMBOX1 forward,
5'-CTTCAGCGACTTCGGCGTA-3' and reverse, 5'-ATCATAACTGTTGCTAGGTG
ACG-3'; CXCL10 forward, 5'-GTGGCATTCAAGGAGTACCTC-3' and reverse,
5'-TGATGGCCTTCGATTCTGGATT-3' and GAPDH forward,
5'-GGAGCGAGATCCCTCCAAAAT-3' and reverse,
5'-GGCTGTTGTCATACTTCTCATGG-3' Relative mRNA expression levels were
quantified using the 2-ΔΔCq method and normalized to
GAPDH expression (20).
Western blotting
Total protein was extracted from hPDLSCs using RIPA
lysis buffer (Beyotime Institute of Biotechnology). Total protein
was quantified using a bicinchoninic acid assay kit (ProteinTech
Group, Inc.) and 50 µg protein/lane was separated by 12% SDS-PAGE.
The separated proteins were subsequently transferred onto
nitrocellulose membranes and non-specific binding was blocked with
5% skimmed milk diluted in TBS buffer for 3 h at room temperature.
The membranes were then incubated overnight at 4˚C with specific
primary antibodies against HMBOX1 (cat. no. 16123-1-AP; 1:1,000;
ProteinTech Group, Inc.), CXCL10 (cat. no. 10937-1-AP; 1:1,000;
Proteintech Group, Inc.), Ki67 (cat. no. ab16667; 1:1,000; Abcam),
proliferating cell nuclear antigen (PCNA; cat. no. ab92552;
1:1,000); IL-6 (cat. no. ab233551; 1:1,000; Abcam), TNF-α (cat. no.
ab183218; 1:1,000; Abcam), IL-1β (cat. no. ab216995; 1:1,000;
Abcam), Bcl-2 (cat. no. ab32124; 1:1,000; Abcam), Bax (cat. no.
ab32503; 1:1,000; Abcam), cleaved caspase-3 (cat. no. ab2302;
1:500; Abcam), caspase-3 (cat. no. ab184787; 1:2,000; Abcam),
phosphorylated (p-) p65 (cat. no. ab76302, 1:1,000; Abcam), p65
(cat. no. ab32536; 1:1,000; Abcam), IκBα (cat. no. sc-1643;
1:1,000; Santa Cruz Biotechnology, Inc.) p-IκBα (cat. no.
sc-101713; 1:500; Santa Cruz Biotechnology, Inc.) and GAPDH (cat.
no. ab9485; 1:1,000; Abcam). Following the primary antibody
incubation, the membranes were washed three times with TBS-Tween-20
(0.05% Tween-20) for 10 min each time and incubated with an
HRP-conjugated secondary antibody (cat. no. ab7090; 1:10,000;
Abcam) for 45 min at room temperature. Protein bands were
visualized using Immun-Star HRP Chemiluminescent Substrate kit
(cat. no. 1705040) on a detector (both from Bio-Rad Laboratories,
Inc.). The grey value was quantified with ImageJ software 1.46r
(National Institutes of Health).
Cell Counting Kit-8 (CCK-8) assay
Cells were seeded into 96-well plates at a density
of 2x104 cells/well and cultured for 24 h at 37˚C, after
which 10 µl CCK-8 reagent (Dojindo Molecular Laboratories, Inc.)
was added to each well and incubated for a further 4 h at 37˚C. The
absorbance was measured at a wavelength of 450 nm using a
microplate reader.
ELISA
The cell supernatant of transfected hPDLSCs was
collected and the levels of TNF-α (cat. no. EK0525), IL-6 (cat. no.
EK0410) andIL-1β (cat. no. EK0392) were determined using their
corresponding ELISA kits (Wuhan Boster Biological Technology Ltd.)
according to the manufacturer's protocols.
TUNEL assay
Apoptosis was conducted using an In Situ Cell Death
Detection kit (Roche Diagnostics) according to the manufacturer's
protocol. The cells were fixed with 4% paraformaldehyde for 30 min
at room temperature, followed by the permeation of 0.2% Triton
X-100 for 8 min at room temperature. TUNEL solution was dropped
into sections for incubation for 60 min at 37˚C, which were then
washed with PBS for three times, each for 5 min and the nuclei were
stained using DAPI for 8 min (0.4 µg/m) in the dark. Apoptotic
cells in four randomly selected views were visualized using an
inverted fluorescence microscope (IX73; Olympus Corporation;
magnification, x200). The apoptotic index (%) was calculated using
the following formula: Apoptotic cells (green fluorescence)/total
cells (blue fluorescence) x100%.
Statistical analysis
Statistical analysis was performed using GraphPad
Prism version 6.0 software (GraphPad Software, Inc.) and data are
presented as the mean ± SD. Statistical differences between ≥3
groups were determined using one-way ANOVA followed by Tukey's
test. All experiments were repeated at least three times. P<0.05
was considered to indicate a statistically significant
difference.
Results
Overexpression of HMBOX1 promotes the
proliferation of LPS-induced hPDLSCs
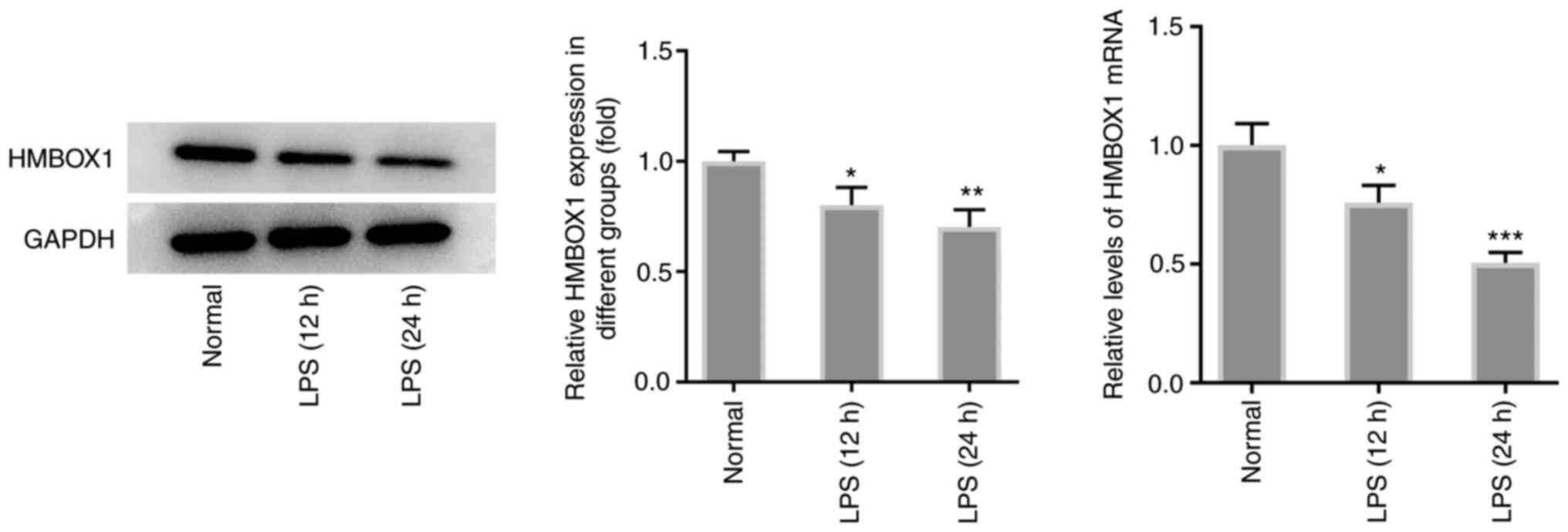
To determine the role of HMBOX1 in the pathogenesis
of periodontitis, the expression levels of HMBOX1 were first
measured in LPS-treated hPDLSCs. As shown in Fig. 1, exposure of hPDLSCs to LPS for 12
or 24 h significantly downregulated the mRNA and protein expression
levels of HMBOX1 compared with those in hPDLSCs without LPS
treatment.
Subsequently, transfection of hPDLSCs with Ov-HMBOX1
significantly increased the expression of HMBOX1 compared with that
in the control and Ov-NC groups (Fig.
2A). Furthermore, results from the CCK-8 and western blot
analyses demonstrated that LPS exposure significantly reduced the
viability of hPDLSCs and expression levels of the
proliferation-related proteins Ki-67 and PCNA (Fig. 2B. By contrast, Ov-HMBOX1
significantly reversed all aforementioned effects on cell viability
and Ki-67 and PCNA expression (Fig.
2B and C). These results
suggest that HMBOX1 overexpression can restore the proliferation of
LPS-induced hPDLSCs.
Overexpression of HMBOX1 inhibits
inflammation and apoptosis in LPS-induced hPDLSCs
Since periodontitis is one of the most prevalent
inflammatory diseases (21), the
effects of HMBOX1 on the inflammation and apoptosis of LPS-induced
hPDLSCs were analyzed. LPS stimulation significantly increased the
production and expression of inflammatory cytokines IL-6, TNF-α and
IL-1β, which was significantly reversed by Ov-HMBOX1 transfection
(Fig. 3A and B). LPS stimulation also significantly
increased the apoptosis of hPDLSCs, whilst significantly
downregulating the expression levels of the anti-apoptotic proteins
Bcl-2. However, expression of the proapoptotic proteins Bax and
cleaved caspase-3 were significantly increased y LPS treatment
(Fig. 3C and D). Conversely, these observations
aforementioned were significantly reversed after these LPS-induced
hPDLSCs were transfected with Ov-HMBOX1 (Fig. 3C and D). These findings suggest that HMBOX1
overexpression can inhibit the inflammation and apoptosis of
LPS-induced hPDLSCs.
 | Figure 3Overexpression of HMBOX1 blunts the
inflammation and apoptosis of LPS-induced hPDLSCs. (A) Expression
of inflammation-related proteins IL-6, TNF-α and IL-1β measured by
western blotting. (B) Secretion of inflammation-related proteins
IL-6, TNF-α and IL-1β measured by ELISA. (C) Apoptosis was measured
and quantified by TUNEL. (D) Expression of apoptosis-related
proteins Bcl-1, Bax and caspase 3 was measured by western blotting.
***P<0.001 vs. control; #P<0.05,
##P<0.01, ###P<0.001 vs. LPS+Ov-NC.
hPDLSCs, human periodontal ligament stem cells; HMBOX1, Homeobox
containing 1; LPS, lipopolysaccharide; Ov, overexpression; NC,
negative control. |
HMBOX1 modulates the expression of
CXCL10 by regulating the NF-κB signaling pathway
To investigate the effects of HMBOX1 on CXCL10
expression and the NF-κB signaling pathway in LPS-induced hPDLSCs,
western blotting and RT-qPCR were used to measure CXCL10 expression
levels following the overexpression of HMBOX1. The results revealed
that Ov-HMBOX1 transfection significantly downregulated the
expression levels of CXCL10 compared with those in cells
transfected with Ov-NC (Fig. 4A).
Furthermore, LPS-induced phosphorylation of p65 and IκBα was
significantly abrogated by Ov-HMBOX1 transfection (Fig. 4B), suggesting a possible association
between HMBOX1 and the NF-κB signaling pathway. Therefore, a NF-κB
agonist, PMA, was used to assess the potential effects of NF-κB on
the regulation of CXCL10 expression. The significant suppressive
effects of Ov-HMBOX1 on the expression of CXCL10 were significantly
reversed by PMA (Fig. 4C). These
findings suggest that HMBOX1 may modulate the expression of CXCL10
by regulating the NF-κB signaling pathway.
 | Figure 4HMBOX1 modulates the expression of
CXCL10 through the NF-κB signaling pathway. (A) The expression of
CXCL10 in LPS-induced hPDLSCs overexpressing HMBOX1 was measured by
western blotting and reverse transcription-quantitative PCR. (B)
Phosphorylation of NF-κB signaling pathway-related proteins IκBα
and p65 in LPS-induced hPDLSCs overexpressing HMBOX1 was measured
by western blotting. (C) After PMA treatment, the expression of
CXCL10 in LPS-induced hPDLSCs overexpressing CXCL10 was measured.
**P<0.01, ***P<0.001 vs. control;
#P<0.05, ##P<0.01,
###P<0.001 vs. LPS+Ov-NC; ∆∆∆P<0.001
vs. LPS+Ov-HMBOX1. hPDLSCs, human periodontal ligament stem cells;
HMBOX1, Homeobox containing 1; LPS, lipopolysaccharide; Ov,
overexpression; NC, negative control; CXCL10, C-X-C motif chemokine
ligand 10; PMA, phorbol 12-myristate 13-acetate. |
HMBOX1 enhances the proliferation
whilst inhibiting the inflammation and apoptosis of LPS-induced
hPDLSCs through the NF-κB/CXCL10 axis
Based on the aforementioned findings, it was
hypothesized that HMBOX1 may influence the physiology of
LPS-induced hPDLSCs through the NF-κB/CXCL10 signaling axis. The
expression of CXCL10 was knocked down using si-CXCL10-1 and
si-CXCL10-2. si-CXCL10-2 was selected for use in subsequent
experiments as it was able to significantly downregulate CXCL10
expression, with a markedly greater magnitude compared with that
mediated by si-CXCL10-1 (Fig. 5A).
PMA treatment inhibited the proliferation of hPDLSCs in
LPS+Ov-HMBOX1+PMA+si-NC group compared with LPS+Ov-HMBOX1+si-NC
group, which was significantly abolished by co-transfection with
si-CXCL10 (Fig. 5B). Furthermore,
the protein levels of proliferation markers, Ki67 and PCNA, were
significantly decreased by PMA treatment compared with that in the
co-treatment of LPS and Ov-HMBOX1 transfection, but were able to be
markedly reversed by CXCL10 silencing (Fig. 5C).
 | Figure 5HMBOX1 enhances the proliferation of
LPS-induced hPDLSCs through the NF-κB/CXCL10 axis. (A) The
expression level of CXCL10 after knocking down its expression by
si-CXCL10-1 and si-CXCL10-2 was measured by western blotting and
reverse transcription-quantitative PCR. After PMA treatment, (B)
cell viability and (C) the expression of proliferation-related
proteins Ki-67 and PCNA in LPS-induced hPDLSCs co-transfected with
Ov-HMBOX1 and si-CXCL10 were measured. ***P<0.001 vs.
control; ##P<0.01, ###P<0.001 vs.
LPS+Ov-NC+si-NC; ∆P<0.05, ∆∆P<0.01 vs.
LPS+Ov-HMBOX1+si-NC; @@P<0.01,
@@@P<0.001 vs. LPS+Ov-HMBOX1+PMA+si-NC. hPDLSCs,
human periodontal ligament stem cells; HMBOX1, Homeobox containing
1; LPS, lipopolysaccharide; Ov, overexpression; NC, negative
control; CXCL10, C-X-C motif chemokine ligand 10; PMA, phorbol
12-myristate 13-acetate; PCNA, proliferating cell nuclear antigen;
si, small interfering. |
The inflammation and apoptosis of hPDLSCs in
LPS+Ov-HMBOX1+PMA+si-NC group were enhanced by PMA as relative to
LPS+Ov-HMBOX1 group, as evidenced by the increased production and
secretion of inflammatory cytokines IL-6, TNF-α and IL-1β (Fig. 6A and B) and the increased apoptotic rate of
these cells. In addition, increased expression levels of Bax and
cleaved caspase-3, and a decrease in Bcl-2 expression were observed
after addition of PMA compared with the LPS+Ov-HMBOX1 group
(Fig. 6C and D). By contrast, all of these effects
aforementioned were reversed following co-transfection with
si-CXCL10 (Fig. 6). Taken together,
these data suggest that HMBOX1 may enhance the proliferation and
inhibit the inflammation and apoptosis of LPS-induced hPDLSCs
through the NF-κB/CXCL10 signaling axis.
 | Figure 6HMBOX1 blunts the inflammation and
apoptosis of LPS-induced hPDLSCs through the NF-κB/CXCL10 axis.
After PMA treatment, (A) the expression of inflammatory factors
IL-6, TNF-α and IL-1β was measured by western blotting, whilst (B)
the secretion of inflammatory factors IL-6, TNF-α and IL-1β was
measured by ELISA. After PMA treatment, (C) apoptosis was measured
using TUNEL, whereas (D) the expression of apoptosis-related
proteins Bcl-2, Bax and caspase 3 in LPS-induced hPDLSCs
transfected with Ov-HMBOX1 and si-CXCL10 was measured by western
blotting. ***P<0.001 vs. control;
##P<0.01, ###P<0.001 vs.
LPS+Ov-NC+si-NC; ∆P<0.05, ∆∆P<0.01,
∆∆∆P<0.001 vs. LPS+Ov-HMBOX1+si-NC;
@@P<0.01, @@@P<0.001 vs.
LPS+Ov-HMBOX1+PMA+si-NC. hPDLSCs, human periodontal ligament stem
cells; HMBOX1, Homeobox containing 1; LPS, lipopolysaccharide; Ov,
overexpression; NC, negative control; CXCL10, C-X-C motif chemokine
ligand 10; PMA, phorbol 12-myristate 13-acetate; si, small
interfering. |
Discussion
The homeobox family of protein contains ~300 gene
members, most of which have been implicated in the regulation of
embryonic development (22). In
particular, HMBOX1 is ubiquitously expressed in human tissues and
has been identified to be a novel transcription factor (6). HMBOX1 consists of a homeobox domain in
the N-terminal region and an hepatocyte nuclear factor 1 domain at
the C-terminus (7). In the present
study, the expression levels of HMBOX1 were found to be
downregulated in LPS-induced hPDLSCs. Therefore, the potential
effects of HMBOX1 on the physiology of LPS-treated hPDLSCs were
investigated by constructing an Ov-HMBOX1 plasmid and transfecting
it into hPDLSCs. The cytotoxicity of LPS-induced hPDLSCs was
mitigated following Ov-HMBOX1 transfection, which was accompanied
by enhanced cell proliferation. In addition, the inflammation and
apoptosis of LPS-induced hPDLSCs were alleviated following
Ov-HMBOX1 transfection. The present study found also that increased
HMBOX1 inhibited NF-κB activation in hPDLSCs in response to LPS. In
addition to the potential involvement of NF-κB in the regulatory
role of HMBOX1 in response to LPS, a previous study also showed
that 5'AMP-activated protein kinase mediates the regulation of
HMBOX1, which is associated with inflammation after EA.hy926 cells
were subjected to LPS stimulation (9). Additionally, HMBOX1 has been reported
to be involved in cancer progression in an AKT-dependent manner,
including osteosarcoma, lung cancer and liver cancer (23-25).
However, whether MAPKs and AKT serve a role in the regulation of
HMBOX1 upstream of inflammation and apoptosis in response to LPS
require further study.
CXCL10 is an inflammatory cytokine that can be
secreted by numerous cell types, including monocytes, endothelial
cells, fibroblasts and mesenchymal cells (17). Previous studies reported that CXCL10
can regulate cell recruitment, migration and invasion (26,27).
This subsequently attracted the attention of researchers, who later
proposed that the expression profile of CXCL10 can be used as a
novel type of biomarker for acute lung injury (28). CXCL10 was also discovered to serve a
regulatory role during the inflammatory response against hepatic
ischemia and reperfusion injury, suggesting its potential for use
in a novel therapeutic approach for this disease (29). In addition, a previous study
reported a close association between CXCL10 expression and
periodontitis (30).
The NF-κB signaling pathway is an extensively
studied signaling pathway that can modulate the secretion of
cytokines, chemokines and adhesion molecules (20). Activation of the NF-κB signaling
pathway by osteopontin was found to increase bone destruction
during periapical periodontitis (31). In addition, another study reported
that NF-κB serve an important role in the pathogenesis of
periodontitis (32). It was also
previously reported that NF-κB signaling was activated in highly
inflamed white adipose tissues in obese rats with periodontitis
compared with that in obese rats without periodontitis, suggesting
a role for NF-κB in the pathogenesis of periodontitis (33). In the present study, following the
treatment of PMA, an activator of the NF-κB signaling pathway,
CXCL10 expression were reduced. In addition, hPDLSCs treated with
PMA showed decreased cell viability and increased apoptosis, and
increased levels of proinflammatory factors TNF-α, IL-6 and IL-1β
compared with those in cells treated with LPS and transfected with
Ov-HMBOX1 but without PMA treatment. However, the effects of
inhibiting the NF-κB signaling pathway on CXCL10 expression require
further study, which is a limitation of the present study. The
NF-κB signaling pathway has also been implicated in the regulation
of CXCL10. For example, the modulation of NF-κB was found to be
mediated by COMM domain-containing 7 during anti-hepatocellular
carcinoma therapy via CXCL10 upregulation (18). Furthermore, NF-κB signaling was
revealed to regulate CXCL10 production in 4T1 breast cancer cells
(34). Findings of the present
study revealed that LPS-induced upregulation of CXCL10 expression
was downregulated by Ov-HMBOX1 transfection, which was subsequently
reversed by PMA treatment. These results support the hypothesis
that HMBOX1 may regulate the physiology of LPS-induced hPDLSCs
through the NF-κB/CXCL10 signaling axis. Furthermore, CXCL10
knockdown reversed the PMA-induced inhibition of cell proliferation
whilst reducing the PMA-induced production of inflammatory
cytokines and apoptosis in these cells.
In conclusion, to the best of our knowledge, the
present study is the first to provide evidence to suggest that
HMBOX1 overexpression can attenuate LPS-induced hPDLSC injury by
downregulating CXCL10 expression through the NF-κB signaling
pathway. This may provide a novel insight into the development of
targeted treatment options for periodontitis in the future.
Acknowledgements
No applicable.
Funding
Funding: No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
MN, HL, PL and PD conceived and designed the study,
performed the experiment, collected, analyzed and interpreted the
data and revised the manuscript. MN wrote the manuscript. All
authors read and approved the final manuscript. MN and PD confirm
the authenticity of all the raw data.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Krishna R and De Stefano JA: Ultrasonic
vs. hand instrumentation in periodontal therapy: Clinical outcomes.
Periodontol 2000. 71:113–127. 2016.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Van Dyke TE: The management of
inflammation in periodontal disease. J Periodontol. 79 (Suppl
8):S1601–S1608. 2008.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Papapanou PN, Sanz M, Buduneli N, Dietrich
T, Feres M, Fine DH, Flemmig TF, Garcia R, Giannobile WV, Graziani
F, et al: Periodontitis: Consensus report of workgroup 2 of the
2017 World Workshop on the classification of periodontal and
Peri-implant diseases and conditions. J Periodontol. 89 (Suppl
1):S173–S182. 2018.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Slots J: Periodontitis: Facts, fallacies
and the future. Periodontol 2000. 75:7–23. 2017.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Borgnakke WS, Ylostalo PV, Taylor GW and
Genco RJ: Effect of periodontal disease on diabetes: Systematic
review of epidemiologic observational evidence. J Periodontol. 40
(Suppl 14):S135–S152. 2013.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Manresa C, Sanz-Miralles EC, Twigg J and
Bravo M: Supportive periodontal therapy (SPT) for maintaining the
dentition in adults treated for periodontitis. Cochrane Database
Syst Rev. 1(CD009376)2018.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Chen S, Saiyin H, Zeng X, Xi J, Liu X, Li
X and Yu L: Isolation and functional analysis of human HMBOX1, a
homeobox containing protein with transcriptional repressor
activity. Cytogenet Genome Res. 114:131–136. 2006.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Diao N, Li Y, Yang J, Jin C, Meng X, Jiao
W, Feng J, Liu Z and Lu N: High expression of HMBOX1 contributes to
poor prognosis of gastric cancer by promoting cell proliferation
and migration. Biomed Pharmacother. 115(108867)2019.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Yuan HX, Feng XE, Liu EL, Ge R, Zhang YL,
Xiao BG and Li QS: 5,2'-dibromo-2,4',5'-trihydroxydiphenylmethanone
attenuates LPS-induced inflammation and ROS production in EA.hy926
cells via HMBOX1 induction. J Cell Mol Med. 23:453–463.
2019.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Ma H, Su L, He X and Miao J: Loss of
HMBOX1 promotes LPS-induced apoptosis and inhibits LPS-induced
autophagy of vascular endothelial cells in mouse. Apoptosis.
24:946–957. 2019.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Guzeldemir-Akcakanat E, Alkan B,
Sunnetci-Akkoyunlu D, Gurel B, Balta VM, Kan B, Akgun E, Yilmaz EB,
Baykal AT, Cine N, et al: Molecular signatures of chronic
periodontitis in gingiva: A genomic and proteomic analysis. J
Periodontol. 90:663–673. 2019.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Zhao H, Han Q, Lu N, Xu D, Tian Z and
Zhang J: HMBOX1 in hepatocytes attenuates LPS/D-GalN-induced liver
injury by inhibiting macrophage infiltration and activation. Mol
Immunol. 101:303–311. 2018.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Zhang F, Mears JR, Shakib L, Beynor JI,
Shanaj S, Korsunsky I and Nathan A: Accelerating Medicines
Partnership Rheumatoid Arthritis and Systemic Lupus Erythematosus
(AMP RA/SLE) Consortium. Donlin LT and Raychaudhuri S: IFN-γ and
TNF-α drive a CXCL10+ CCL2+ macrophage
phenotype expanded in severe COVID-19 and other diseases with
tissue inflammation. bioRxiv: Aug 5, 2020 (Epub ahead of print).
doi: 10.1101/2020.08.05.238360.
|
|
14
|
Luster AD, Unkeless JC and Ravetch JV:
Gamma-interferon transcriptionally regulates an early-response gene
containing homology to platelet proteins. Nature. 315:672–676.
1985.PubMed/NCBI View
Article : Google Scholar
|
|
15
|
Groom JR and Luster AD: CXCR3 ligands:
Redundant, collaborative and antagonistic functions. Immunol Cell
Biol. 89:207–215. 2011.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Gao J, Wu L, Wang S and Chen X: Role of
chemokine (C-X-C Motif) ligand 10 (CXCl10) in renal diseases.
Mediators Inflamm. 2020(6194864)2020.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Aldahlawi S, Youssef AR and Shahabuddin S:
Evaluation of chemokine CXCL10 in human gingival crevicular fluid,
saliva, and serum as periodontitis biomarker. J Inflamm Res.
11:389–396. 2018.PubMed/NCBI View Article : Google Scholar
|
|
18
|
You N, Li J, Huang X, Wu K, Tang Y, Wang
L, Li H, Mi N and Zheng L: COMMD7 activates CXCl10 production by
regulating NF-κB and the production of reactive oxygen species. Mol
Med Rep. 17:6784–6788. 2018.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Chen H, Lin W, Lin P, Zheng M, Lai Y, Chen
M, Zhang Y, Chen J, Lin X, Lin L, et al: IL-10 produces a dual
effect on OGD-induced neuronal apoptosis of cultured cortical
neurons via the NF-κB pathway. Aging (Albany NY). 11:10796–10813.
2019.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Zhou W, Su L, Duan X, Chen X, Hays A,
Upadhyayula S, Shivde J, Wang H, Li Y, Huang D and Liang S:
MicroRNA-21 down-regulates inflammation and inhibits periodontitis.
Mol Immunol. 101:608–614. 2018.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Holland PW, Booth HA and Bruford EA:
Classification and nomenclature of all human homeobox genes. BMC
Biol. 5(47)2007.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Chen S, Li Y, Zhi S, Ding Z, Wang W, Peng
Y, Huang Y, Zheng R, Yu H, Wang J, et al: WTAP promotes
osteosarcoma tumorigenesis by repressing HMBOX1 expression in an
m6A-dependent manner. Cell Death Dis.
11(659)2020.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Li J, Zhao Y and Wang J: Extracellular
vesicle-associated microRNA-221-3p secreted by drug-resistant lung
cancer cells targets HMBOX1 to promote the progression of lung
cancer. Cancer Gene Ther. 28:679–692. 2021.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Zhao H, Jia H, Han Q and Zhang J: Homeobox
containing 1 inhibits liver cancer progression by promoting
autophagy as well as inhibiting stemness and immune escape. Oncol
Rep. 40:1657–1665. 2018.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Tsai CF, Chen JH and Yeh WL: Pulmonary
fibroblasts-secreted CXCL10 polarizes alveolar macrophages under
pro-inflammatory stimuli. Toxicol Appl Pharmacol.
380(114698)2019.PubMed/NCBI View Article : Google Scholar
|
|
27
|
McGrath-Morrow SA, Lee S, Gibbs K, Lopez
A, Collaco JM, Neptune E, Soloski MJ, Scott A and D'Alessio F:
Immune response to intrapharyngeal LPS in neonatal and juvenile
mice. Am J Respir Cell Mol Biol. 52:323–331. 2015.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Chalin A, Lefevre B, Devisme C, Pronier C,
Carrière V, Thibault V, Amiot L and Samson M: Serum CXCL10, CXCL11,
CXCL12, and CXCL14 chemokine patterns in patients with acute liver
injury. Cytokine. 111:500–504. 2018.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Zhai Y, Shen XD, Gao F, Zhao A, Freitas
MC, Lassman C, Luster AD, Busuttil RW and Kupiec-Weglinski JW:
CXCL10 regulates liver innate immune response against ischemia and
reperfusion injury. Hepatology. 47:207–214. 2008.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Shimada Y, Tabeta K, Sugita N and Yoshie
H: Profiling biomarkers in gingival crevicular fluid using
multiplex bead immunoassay. Arch Oral Biol. 58:724–730.
2013.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Dong M, Yu X, Chen W, Guo Z, Sui L, Xu Y,
Shang Y, Niu W and Kong Y: Osteopontin promotes bone destruction in
periapical periodontitis by activating the NF-κB pathway. Cell
Physiol Biochem. 49:884–898. 2018.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Hiyari S, Wong RL, Yaghsezian A, Naghibi
A, Tetradis S, Camargo PM and Pirih FQ: Ligature-induced
peri-implantitis and periodontitis in mice. J Clin Periodontol.
45:89–99. 2018.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Huang Y, Zeng J, Chen G, Xie X, Guo W and
Tian W: Periodontitis contributes to adipose tissue inflammation
through the NF-B, JNK and ERK pathways to promote insulin
resistance in a rat model. Microbes Infect. 18:804–812.
2016.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Jin WJ, Kim B, Kim D, Park Choo HY, Kim
HH, Ha H and Lee ZH: NF-κB signaling regulates cell-autonomous
regulation of CXCL10 in breast cancer 4T1 cells. Exp Mol Med.
49(e295)2017.PubMed/NCBI View Article : Google Scholar
|