Introduction
Cardiovascular disease (CVD) is a leading cause of
mortality worldwide (1). One of the
main causes of CVD is atherosclerosis (AS) (2), and the pathological mechanisms
underlying AS have been the focus of a number of studies. AS is
recognized as a chronic arterial inflammatory disease that is
induced by oxidized low-density lipoprotein (ox-LDL) accumulation
and inflammation of the arterial intima under hypercholesterolemic
conditions (3). The results of
previous pharmacological studies have demonstrated that the main
risk factors of AS include dyslipidemia, hypertension, alcohol
consumption, smoking, diabetes, obesity and a lack of exercise
(4). Currently, dyslipidemia is
considered the most critical risk factor of AS (5). Ox-LDL is a critical diagnostic marker
of AS (6), and has the ability to
cause lipid metabolism disorders, promote the formation of foam
cells derived from vascular smooth muscle cells (VSMCs), and
regulate the proliferation, apoptosis, migration and
differentiation of VSMCs, which serve vital roles in the
development of AS (7).
The results of previous studies have revealed the
key signals and molecular pathways (e.g., MAPK/ERK, Nrf2)
underlying the formation and development of atherosclerotic plaques
(8,9). Moreover, the role of microRNAs
(miRNAs/miRs) in CVD have remained the focus of a number of
studies. miRNAs are a family of highly conserved, endogenous
non-coding small RNA molecules that regulate gene expression
through binding of the 3'-untranslated region (3'-UTR) of target
mRNAs (10-12).
miRNAs are involved in diverse cellular functions, including
differentiation, growth, proliferation, migration, senescence and
apoptosis in a number of cells including cancer cells, HUVECs and
H9c2(13). Moreover, A previous
study demonstrated that miRNAs serve essential roles in regulating
AS (14). Geng et al
(14) reported that ox-LDL
increased the expression levels of miR-129-5p in human aortic
endothelial cells, thereby decreasing cell proliferation. This led
to a decrease in the mRNA expression of Beclin-1, which decreased
endothelial cell autophagy in AS. Thus, miR-129-5p exerted
differing effects on a number of cells under the pathological
conditions of AS, which is dependent on the targeted cell type
(8). The results of previous
studies have provided novel molecular insights into the impact of
miRNAs in AS pathways, identifying them as novel therapeutic
targets. For example, miR-181b acts as an inhibitor of endothelial
inflammatory responses by targeting NF-κB binding in AS (15). The results of an study demonstrated
the role of miR-221/miR-222 in the regulation of platelet derived
growth factor-mediated VSMC proliferation (16). Moreover, miR-145 served a regulative
role in aberrant VSMC proliferation, which is a key pathological
process of AS (17). Liu et
al (18) demonstrated the role
of miR-21 as a key target for protective reagents against
ox-LDL-induced rat vascular endothelial cell injury, which may
serve critical roles in the development of AS (18). Furthermore, miR-129-5p has been
demonstrated to suppress carcinogenesis in a number of cancers
including gastric cancer and osteoscarcoma (19,20).
However, the specific role of miR-129-5p in the development of AS
is yet to be fully elucidated.
High-mobility group box 1 protein (HMGB1), a highly
conserved and widely expressed DNA-binding protein, is a key
mediator of cell migration and proliferation (21). Notably, the results of a previous
study demonstrated that HMGB1 was overexpressed in cancer cells and
promoted cell invasion and migration (22). HMGB1 has been reported to
participate in the regulation of AS progression. For example, Wu
et al (23) demonstrated
that miR-328 mitigated ox-LDL-induced endothelial cell injury by
targeting HMGB1 in AS. Moreno et al (24) also reported that HMGB1 was highly
expressed in atherosclerotic plaques.
In the present study, ox-LDL-induced A7r5 cells were
used to determine the role of miR-129-5p in cell migration and
proliferation, and to investigate the association of miR-129-5p
with HMGB1 and the PI3k/Akt signaling pathway.
Materials and methods
Materials
FBS, bovine serum albumin (BSA) and endothelial cell
growth supplement were purchased from Gibco; Thermo Fisher
Scientific, Inc. Ox-LDL was purchased from Unionbiol. Cell Counting
Kit-8 (CCK-8) was purchased from Dojindo Molecular Technologies,
Inc. (cat. no. CK04).
Cell culture
A7r5 cells were purchased from The American Type
Culture Collection and maintained in DMEM (Gibco; Thermo Fisher
Scientific, Inc.) containing 10% FBS (Gibco; Thermo Fisher
Scientific, Inc.), 1 mmol/l sodium pyruvate, 4 mmol/l L-glutamine,
4.5 g/l glucose, 1.5 g/l sodium bicarbonate, 100 mg/ml streptomycin
and 100 U/ml penicillin at 37˚C in a humidified atmosphere with 5%
CO2.
Cell transfection
A7r5 cells were transfected with 20 nM miR-129-5p
mimics (5'CUUUUUGCGGUCUGGGCUUGC3') or the corresponding negative
control (Shanghai GenePharma Co., Ltd.) using
Lipofectamine® 2000 reagent (Invitrogen; Thermo Fisher
Scientific, Inc.) for 48 h at 37˚C. For HMGB1 overexpression, the
recombinant sense expression vector plasmid Cytomegalovirus
promoter DNA 3.1 for HMGB1 (1 µg/µl pcDNA3.1-HMGB1; Invitrogen;
Thermo Fisher Scientific, Inc.) was constructed by subcloning the
cDNA fragment of HMGB1 containing the complete coding sequence
between KpnI and BamHI. Cells were transfected using
Lipofectamine® 2000 according to the manufacturer's
protocol. Cells in the control group were transfected with
pcDNA3.1-NC (empty vector). Following transfection, cells were
incubated with fresh DMEM for 24 h, the medium was replaced with
fresh and cells were incubated for a further 24 h at 37˚C. Cells
were collected for subsequent experiments.
Wound healing
A7r5 cells were cultured in six-well culture plates
(1x106 cells/well) for 48 h at 37˚C. The wound healing
assay was conducted as previously described (25). The cell monolayers on the surface of
the six-well plate were scratched with a 200 µl micropipette tip.
Cells were subsequently incubated at 37˚C for 24 h in DMEM
containing 2% FBS (26).
Non-adherent cells were washed with PBS and the remaining cells
were treated with ox-LDL (0, 10, 20 and 40 µg/ml) based on
preliminary experiments. Images were captured using light
microscope (magnification, x40), and images of linear wounds were
obtained from nine fields/well at 0 and 48 h after injury. Three
independent repeats were carried out.
CCK-8 assays for the detection of cell
viability
Cell viability was assessed using a CCK-8 assay kit.
A7r5 cells were cultured in 96-well plates overnight at a density
of 104 cells/well at 37˚C, and subsequently transfected
with miR-129-5p mimics or an inhibitor as previously described. At
48 h after transfection, 10 µl CCK-8 solution was added to each
well for 1 h and absorbance readings at 450 nm were obtained in
triplicate using a spectrophotometric plate reader. Three wells
were measured for each data point and three independent repeats
were conducted.
Transwell migration assay
A total of 1x105 A7r5 cells were
collected and seeded in a six-well plate. After the cells had
attached, they were treated with ox-LDL at 0, 10, 20, 40 µg/ml for
48 h. The cells were collected and 5,000 cells/well in 200 µl/well
were seeded into the upper chambers filled DMEM without FBS that
had been pre-coated with 150 mg Matrigel (BD Biosciences) 24 h at
37˚C for invasion assays. The lower chambers were filled with DMEM
containing 10% FBS. Following incubation at 37˚C for 48 h, cells
remaining on the upper surface of the membrane were removed. Cells
on the lower surface of the membrane were fixed with 4%
paraformaldehyde and stained with 0.1% crystal violet for 15 min at
room temperature. Stained cells were subsequently observed and
counted under a light microscope (magnification, x100). Three
independent experiments were performed.
Colony formation assay
A7r5 cells were treated with ox-LDL (0, 10, 20 and
40 µg/ml), seeded in 60-mm plates at 1,000 cells/well and the
culture was terminated when colonies became visible to the naked
eye and contained >50 cells. The cells were washed with 1X PBS,
fixed in 4% paraformaldehyde for 15 min and stained with 0.1% (w/v)
Giemsa at room temperature. Colonies were subsequently viewed and
counted in 10 randomly selected fields under a light microscope
magnification, x100, Nikon Corporation), and images were captured
using a digital camera (Canon Inc.). The percentage of colony
formation for each group was calculated using the following
equation: Percentage of colony formation (%)=the number of
colonies/1,000 x100. Three independent experiments were
performed.
Bioinformatics analysis
Bioinformatics analysis was performed to predict the
downstream miRNA that would interact with HMGB1, to further
investigate the regulatory mechanism of miR129-5p. TargetScan
(http://www.targetscan.org/vert_72/)
and miRBase (https://www.mirbase.org/) were used
for the gene prediction, according to the online software operating
instructions.
RNA isolation and reverse
transcription-quantitative (RT-q)PCR for miR-129-5p
A total of 1 µg RNA was extracted from A7r5 cells
using TRIzol® reagent (Thermo Fisher Scientific, Inc.).
Reverse transcription was performed using RT Reagent kit (cat. no.
RR037A; Takara, Bio, Inc.). qPCR (cat. no. RR820A; Takara, Bio,
Inc.) was performed using SYBR Green mix (Takara Bio, Inc.) with
primers specific to miR-129-5p (Guangzhou RiboBio Co., Ltd.). The
PCR conditions were as follows: 95˚C for 10 min, followed by 40
cycles at 95˚C for 30 sec, 60˚C for 30 sec and 72˚C for 1 min.
Relative quantification of the miRNA expression was calculated
using the 2-ΔΔCq method (27). The sequence of miR-129-5p was:
5'CUUUUUGCGGUCUGGGCUUGC3'. HMGB1, Forward
5'ATCCTGGCTTATCCATTGGTGAT3'; Reverse 5'CTCGTCGTCTTCCTCTTCCTTCT3'.
The corresponding PCR primers were: miR-129-5p, Forward
5'-ACACTCCAGCTGGGTCCCTGAGACCCTTAA-3' and Reverse
CTCAACTGGTGTGGAGT-; U6, Forward: 5'-CTCGCTTCGGCAGCACA-3' and
Reverse 5'-AACGCTTCACGAAYYYGCGT-3'. U6 was selected as the
housekeeping gene to normalize the expression of miR-129-5p.
Dual-luciferase reporter assay
Wild-type (WT) or mutant (MUT) versions of
miR-129-5p were subcloned into the pGL3 Basic vector (Promega
Corporation). A total of 20 nM miR-129-5p mimics
(5'CUUUUUGCGUCUGGGCUUGC3') (Guangzhou RiboBio Co., Ltd.) were
co-transfected with pLUC-WT-HMGB1 (5'UACCACUCUGUAAUUGCAAAAAA) or
pLUC-MUT-HMGB1 (5'UACCACUCUGUAAUUCCUAUAUA) (500 ng) into
3x104 A7r5 cells. Cells were transfected using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer's protocol. After
48 h incubation at 37˚C, the cells were lysed by lysis buffer as
supplied by the Dual-Luciferase Detection kit (Beyotime Institute
of Biotechnology) on ice and luciferase activity was tested using
the Dual-Luciferase Reporter assay system (Promega Corporation).
Luciferase activity was normalized to Renilla luciferase
activity. After transfection for 48 h, relative luminescence was
tested using luminometry according to the manufacturer's
instructions.
Western blotting assay
Cell lysates from ox-LDL treated A7r5 cells were
placed on ice in 1X RIPA lysis buffer (Sigma-Aldrich; Merck KGaA)
containing protease and phosphatase inhibitors (Thermo Fisher
Scientific, Inc.). The concentration of the protein in cells
lysates was detected using a BCA kit (Beijing Solarbio Science
& Technology Co., Ltd.). The proteins in cell lysates (10 µg)
were separated using SDS-PAGE on a 10% gel. The separated proteins
were subsequently transferred onto a nitrocellulose membrane and
incubated with 5% non-fat milk solution or 5% BSA for 1 h at room
temperature. The membranes were incubated with primary antibodies
against HMGB1, focal adhesion kinase (FAK; cat. no. 3285, 1:1,000;
Cell Signaling Technology, Inc.), Akt (cat. no. 9272, 1:1,000; Cell
Signaling Technology, Inc.), PI3k (cat. no. 4255, 1:1,000; Cell
Signaling Technology, Inc.), phosphorylated (p)-FAK (cat. no. 3281,
1:1,000; Cell Signaling Technology, Inc.), p-Akt (cat. no. 9275,
1:1,000; Cell Signaling Technology, Inc.), and GAPDH (cat. no.
5174; 1:1,000, Cell signal Technology, Inc.) at 4˚C overnight. The
membrane was washed with TBS-Tween-20 (20%) 3 times (8 min) and
subsequently incubated with anti-rabbit IgG (TA354196, 1:10,000) or
anti-mouse IgG antibodies (TA35266, 1:10,000; OriGene Technologies,
Inc.) conjugated to horseradish peroxidase for 1 h at room
temperature. Bands were visualized using an ECL detection kit
(Thermo Fisher Scientific, Inc.). Then band intensities were
determined using ImageJ software v. 1.4 (National Institutes of
Health), and normalized to GAPDH.
Statistical analysis
Each experiment was carried out at least three
times, and all values are presented as the mean ± SD. SPSS 22.0
software (IBM Corp.) was used to analyze the data. Comparisons
between two means were evaluated using a paired Student's t-test.
Comparisons among multiple groups were evaluated using one-way
ANOVA followed by Bonferroni's post hoc test. P<0.05 was
considered to indicate a statistically significant difference.
Results
Ox-LDL increases the viability and
migration of A7r5 cells
The viability of A7r5 cells induced by 0, 10, 20 and
40 µg/ml ox-LDL was investigated using CCK-8 and clone formation
assays. Following ox-LDL treatment, the results of the CCK-8 assay
demonstrated that compared with control group, the absorbance of
A7r5 cells significantly increased with the increase of ox-LDL
concentration, indicating that ox-LDL increased viability in a
dose-dependent manner (Fig. 1A). As
presented in Fig. 1, A7r5 cells
induced by 40 µg/ml ox-LDL demonstrated the highest level of
viability. As exhibited in Fig. 1B,
the clone formation of A7r5 cells was significantly increased with
the increase of ox-LDL concentration compared with control group,
further indicating that ox-LDL increased viability in a
dose-dependent manner, and that A7r5 cells induced by 40 µg/ml
ox-LDL demonstrated the highest level of viability. Moreover, the
results of wound healing and Transwell assays revealed a high level
of A7r5 cell migration induced by 40 µg/ml ox-LDL compared with
control group (Fig. 1C and D).
miR-129-5p expression levels decrease
and HMGB1 expression levels increase in A7r5 cells following
treatment with ox-LDL
The results of RT-qPCR demonstrated that the level
of miR-129-5p decreased with the increase of ox-LDL in A7r5 cells
compared with control group. Notably, the expression level of
miR-129-5p was the lowest in A7r5 cells following treatment with 40
µg/ml ox-LDL, compared with the control group (Fig. 2A). Furthermore, HMGB1 mRNA and
protein expression levels in A7r5 cells induced by 0, 10, 20 and 40
µg/ml ox-LDL were evaluated using western blot analysis and
RT-qPCR. As demonstrated in Fig.
2B-D, both HMGB1 protein and mRNA expression levels were the
highest in A7r5 cells induced by 40 µg/ml ox-LDL compared with the
control group. Thus, the expression levels of HMGB1 were increased,
and the expression levels of miR-129-5p were decreased in A7r5
cells following treatment with ox-LDL.
 | Figure 2miR-129-5p is downregulated and HMGB1
is upregulated in ox-LDL-induced A7r5 cells. (A) RT-qPCR assays
were used to determine the expression levels of miR-129-5p in A7r5
cells following treatment with 0, 10, 20 and 40 µg/ml ox-LDL. (B)
RT-qPCR assays were used to determine the expression levels of
HMGB1 in A7r5 cells following treatment with 0, 10, 20 and 40 µg/ml
ox-LDL. (C) Western blot assays were used to determine the protein
expression levels of HMGB1 in A7r5 cells following treatment with
0, 10, 20 and 40 µg/ml ox-LDL. (D) Quantification of HMGB1 protein
expression levels in A7r5 cells. Experiments were repeated three
times. **P<0.01 and ***P<0.001 vs. 0
µg/ml ox-LDL. miR, microRNA; HMGB1, high-mobility group box 1
protein; ox-LDL, oxidized low-density lipoprotein; RT-qPCR, reverse
transcription-quantitative PCR. |
miR-129-5p regulates HMGB1 expression
by binding to the HMGB1 3'UTR
Results generated from TargetScan (http://www.targetscan.org/vert_72/) and miRBase
(https://www.mirbase.org/) databases suggested
that the 3'UTR of HMGB1 contained miR-129-5p seed sites (Fig. 3A). In order to determine whether
miR-129-5p directly targeted HMGB1, a dual-luciferase reporter
assay was performed after A7r5 cells were co-transfected with
HMGB1-WT or HMGB1-MUT reporter plasmids, and either miR-129-5p or
miR-NC. The results of the present study demonstrated that
miR-129-5p significantly downregulated the luciferase activity of
HMGB1-WT plasmid (Fig. 3B), but
exerted no notable effects on the activity of the HMGB1-MUT
plasmid, indicating that miR129-5p directly targeted HMGB1. RT-qPCR
results revealed that the expression levels of miR-129-5p in the
miR-129-5p mimics group were significantly increased compared with
the mimics control group (Fig. 3C).
Moreover, the expression levels of miR-129-5p in the miR-129-5p
inhibitor group were significantly reduced compared with the
inhibitor control group. Thus, the results of the RT-qPCR analysis
confirmed transfection efficiency. In addition, western blotting
and RT-qPCR results revealed differences in the expression levels
of HMGB1 in miR-129-5p mimics group. As demonstrated in Fig. 3D and E, miR-129-5p downregulated both the HMGB1
mRNA and protein expression levels in A7r5 cells.
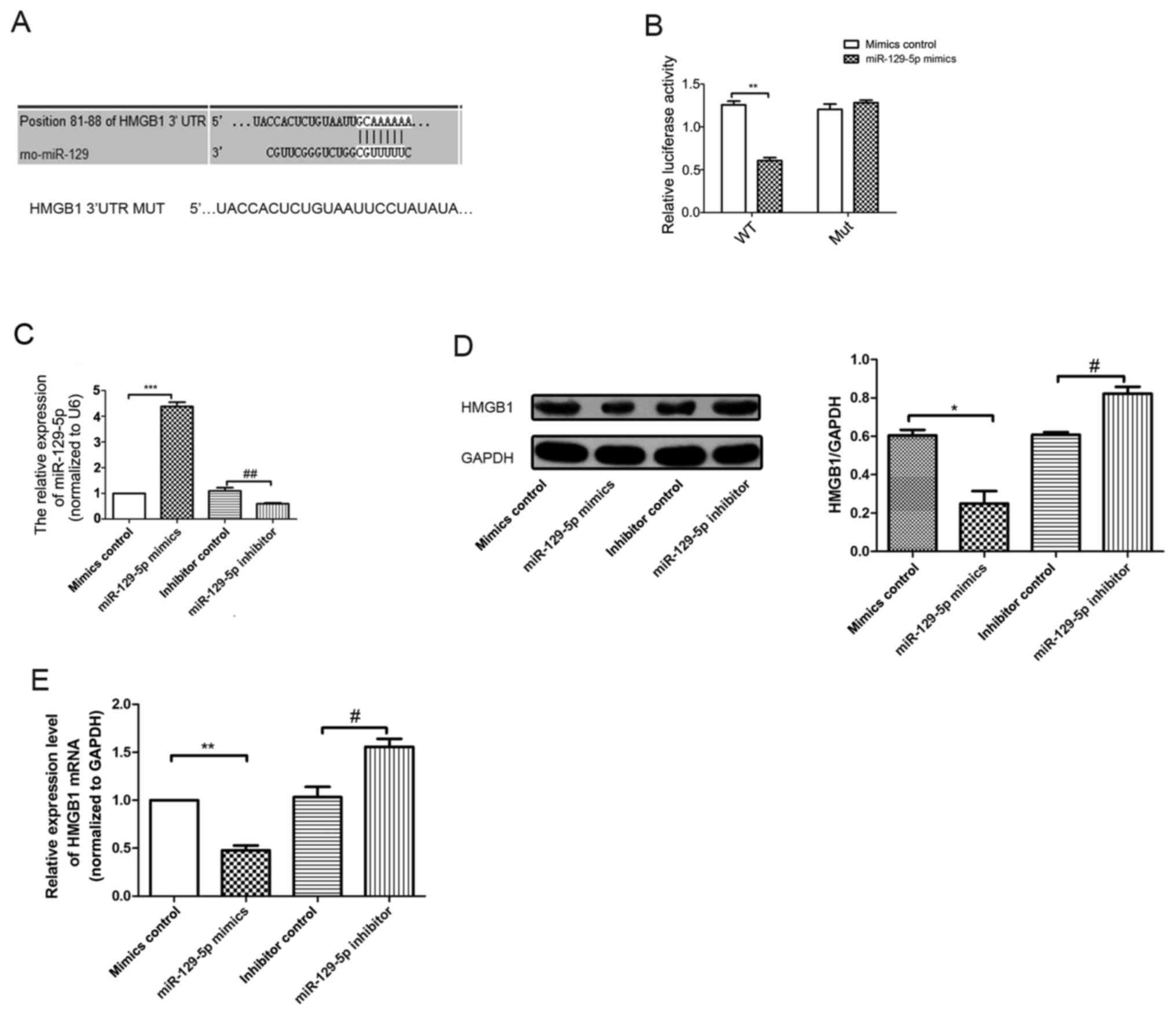 | Figure 3miR-129-5p directly targets HMGB1 in
A7r5 cells. (A) Bioinformatics analysis demonstrated that
miR-129-5p targets the 3'UTR of HMGB1 mRNA at the specific binding
site at 81-88 base pairs. (B) The mutant reporter of HMGB1 was
constructed, and A7r5 cells were co-transfected with either
miR-129-5p mimics or mimics control. The relative luciferase
activity was detected after 48 h. (C) RT-qPCR analysis was used to
detect the expression levels of miR-129-5p in different groups
(mimics control, miR-129-5p mimics, inhibitor control and
miR-129-5p inhibitor). (D) Western blotting analysis was used to
detect HMGB1 protein levels following miR-129-5p overexpression in
A7r5 cells. (E) mRNA expression levels of HMGB1 in A7r5 cells were
detected using RT-qPCR analysis. Experiments were repeated three
times. *P<0.05, **P<0.01 and
***P<0.001 vs. mimics control group;
#P<0.05 and ##P<0.01 vs. inhibitor
control group. miR, microRNA; HMGB1,high-mobility group box 1
protein; UTR, untranslated region; RT-qPCR, reverse
transcription-quantitative PCR; MUT, mutant; WT, wild-type. |
miR-129-5p inhibits the viability and
migration of A7r5 cells induced by ox-LDL
RT-qPCR was conducted to detect the mRNA levels of
miR-129-5p in A7r5 cells induced by 0, 10, 20 and 40 µg/ml ox-LDL.
The expression levels of miR-129-5p in A7r5 cells induced by 40
µg/ml ox-LDL was significantly reduced compared with the control
group (Fig. 4A). Moreover, the
expression levels of miR-129-5p were significantly increased in the
miR-129-5p mimics group, compared with the ox-LDL group (Fig. 4A). A CCK-8 assay was conducted to
analyze the viability of A7r5 cells induced by 40 µg/ml ox-LDL.
Notably, the viability of A7r5 cells induced by 40 µg/ml ox-LDL was
increased compared with the corresponding control group. However,
the viability of A7r5 cells was reduced following transfection with
the miR-129-5p mimics and treatment with 40 µg/ml ox-LDL compared
with mimics control + 40 g/ml ox-LDL group (Fig. 4B). Furthermore, colony formation and
Transwell assays were performed to determine the viability and
migration rate of A7r5 cells induced by ox-LDL. The results of the
present study demonstrated that the viability and migration of A7r5
cells induced by 40 µg/ml ox-LDL were increased compared with
control group. However, the viability and migration of the A7r5
cells were significantly decreased following transfection with the
miR-129-5p mimics and treatment with 40 µg/ml ox-LDL compared with
40 µg/ml ox-LDL + mimics control group (Fig. 4C and D).
Effects of miR-129-5p on the viability
and migration of A7r5 cells induced by ox-LDL are altered following
HMGB1 overexpression
RT-qPCR was conducted to determine HMGB1 mRNA
expression levels in A7r5 cells induced by ox-LDL, and in A7r5
cells transfected with miR-129-5p mimics. Notably, the expression
level of HMGB1 in the con + miR-129-5p + pcDNA3.1 group was
significantly reduced compared with the control group (Fig. 5A). Furthermore, the expression level
of HMGB1 was significantly increased in the HMGB1 overexpression
group compared with the miR-129-5p mimics group (Fig. 5A). Additionally, the expression
level of HMGB1 was markedly increased in the ox-LDL40 + miR-129-5p
+ pcDNA3.1-HMGB1 group compared with the ox-LDL40 + miR-129-5p +
pcDNA3.1 group. A CCK-8 assay was conducted to analyze the
viability of A7r5 cells induced by 40 µg/ml ox-LDL. Compared with
the control group, the viability of A7r5 cells transfected with
miR-129-5p mimics was significantly reduced. Notably, the viability
of A7r5 cells induced by 40 µg/ml ox-LDL was significantly
increased following HMGB1 overexpression compared with the ox-LDL40
+ miR-129-5p + pcDNA3.1 group (Fig.
5B). In order to confirm transfection efficiency, HMGB1 mRNA
expression levels were detected using RT-qPCR analysis. The results
demonstrated that HMGB1 mRNA expression levels were significantly
increased in the con + pcDNA3.1-HMGB1 group compared with the con +
pcDNA3.1 group (Fig. 5C). Colony
formation and Transwell assays were subsequently performed to
determine the viability and migration rates of A7r5 cells induced
by ox-LDL. The results of the present study demonstrated that the
viability and migration of A7r5 cells induced by 40 µg/ml ox-LDL
were increased compared with control group. However, the viability
and migration rate of A7r5 cells induced by 40 µg/ml ox-LDL were
significantly increased following HMGB1 overexpression compared
with 40 µg/ml ox-LDL + pcDNA3.1-NC group (Fig. 5D and E).
 | Figure 5HMGB1 overexpression reverses the
miR-129-5p-mediated inhibition of ox-LDL-induced A7r5 cell
proliferation and migration. (A) RT-qPCR assays were used to
determine the mRNA expression levels of HMGB1 in A7r5 cells. The
following groups were established: Control group, con + miR129-5p +
pcDNA3.1, 40 µg/ml ox-LDL + miR129-5p + pcDNA3.1, con + miR-129-5p
+ pcDNA3.1-HMGB1 and 40 µg/ml ox-LDL + miR129-5p + pcDNA3.1-HMGB1.
(B) Cell Counting Kit-8 assays were used to determine the effects
of ox-LDL treatment on A7r5 cell viability. (C) RT-qPCR analysis
was used to detect the mRNA expression level of HMGB1. (D) Wound
closure of cells was detected using wound healing assays
(magnification, x100). (E) Migration of cells was determined using
Transwell assays (magnification, x100). Experiments were repeated
three times. *P<0.05, **P<0.01 vs.
control group; #P<0.05 and ##P<0.01 vs.
con + miR-129-5p + pcDNA3.1 group; &P<0.05,
&&P<0.01 and
&&&P<0.05 vs. ox-LDL40 + miR-129-5p +
pcDNA3.1 group. HMGB1, high-mobility group box 1 protein; miR,
microRNA; ox-LDL, oxidized low-density lipoprotein; con, control;
RT-qPCR, reverse transcription-quantitative PCR; NS,
non-significant. |
Effects of miR-129-5p on the
expression and phosphorylation of FAK and Akt
PI3k/Akt-related proteins were investigated in A7r5
cells following transfection with miR-129-5p mimics and treatment
with 40 µg/ml ox-LDL. As demonstrated in Fig. 6, the protein expression level of
PI3k, along with p-Akt and p-FAK levels were increased in A7r5
cells induced by 40 µg/ml ox-LDL compared with the control group.
Moreover, the expression levels of PI3k, and the levels of p-Akt
and p-FAK were markedly reduced in A7r5 cells following
transfection with the miR-129-5p mimic, compared with A7r5 cells
induced by 40 µg/ml ox-LDL (Fig.
6).
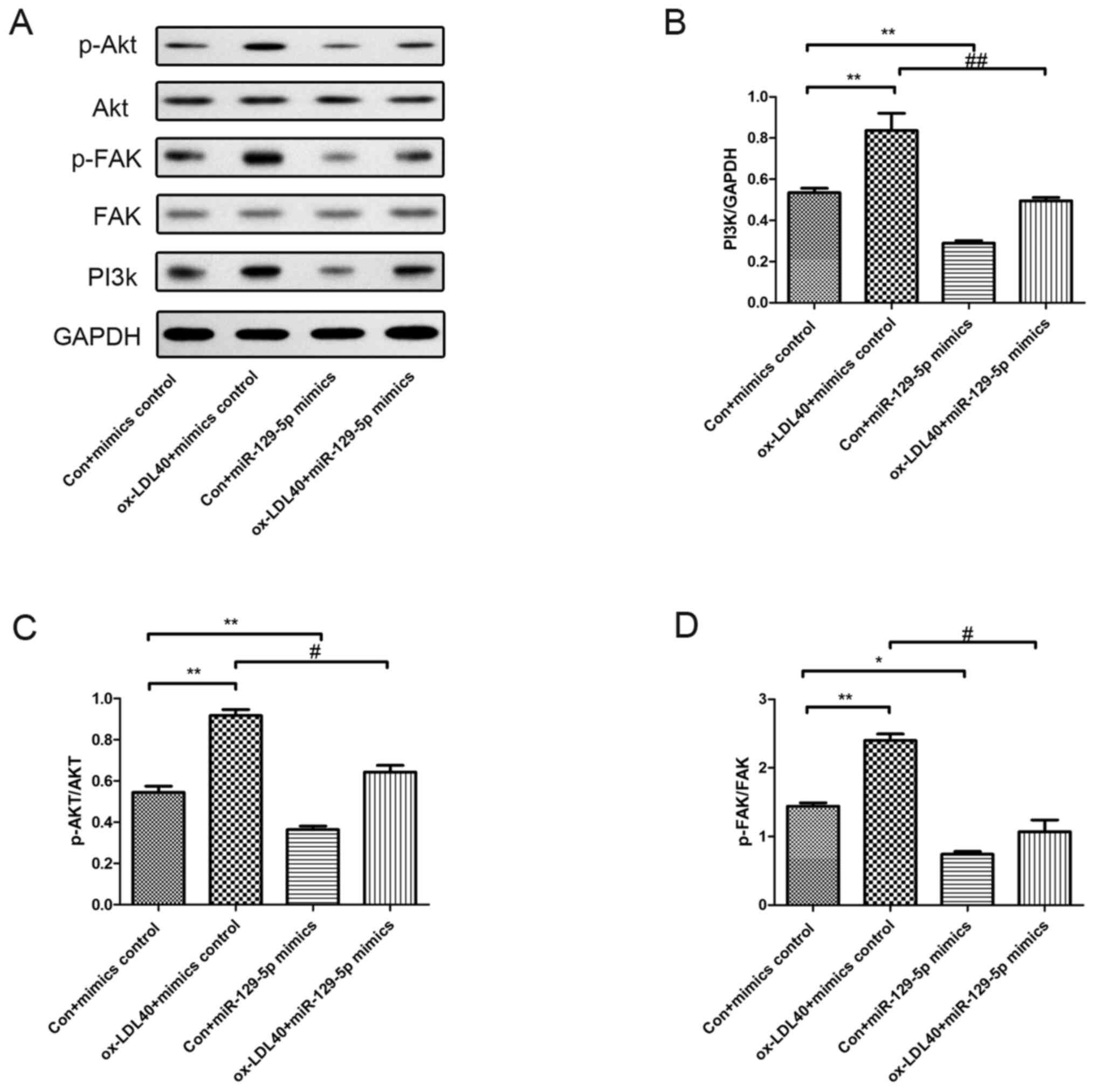 | Figure 6Effects of miR-129-5p on PI3k, p-FAK
and p-Akt protein expression levels. Western blot analyses were
conducted to determine the phosphorylation levels of FAK and Akt,
and the protein expression levels of PI3k. (A) Protein expression
levels in A7r5 cells transfected with mimics, 40 µg/ml ox-LDL +
mimics control group, miR-129-5p + mimics control group, and 40
µg/ml ox-LDL + miR-129-5p mimics group. Quantification of protein
expression levels of (B) PI3k, (C) p-Akt and (D) p-FAK. Experiments
were repeated three times. *P<0.05 and
**P<0.01 vs. con + mimics control group;
#P<0.05 and ##P<0.01 vs. ox-LDL40 +
mimics control group. miR, microRNA; p-, phosphorylated; FAK, focal
adhesion kinase; ox-LDL, oxidized low-density lipoprotein; con,
control. |
Discussion
AS is a main cause of CVDs and cerebrovascular
diseases that lead to the highest mortality and morbidity rates
worldwide (28). The American Heart
Association reported that 17.9 million people died of
cardiovascular disease in 2017, accounting for 31 percent of deaths
worldwide (29). The main risk
factors for AS include hyperglycemia, hyperlipidemia, smoking, poor
diet, obesity and diabetes mellitus (30). Epidemiological data have
demonstrated that the mortality rate of CVD is much higher than
that of other diseases worldwide (31). VSMC proliferation is one of the
major contributors to AS, and the associated risk factors remain
the focus of current research. For example, ox-LDL is associated
with AS, and is formed as a result of ox-LDL oxidation (32-34).
In addition, further research has focused on improving and
preventing the development of AS. The role of ox-LDL in
hyperlipidemia has also been demonstrated as a key risk factor in
the development of AS (35).
Moreover, the pharmacological inhibition of miRNAs is being used as
treatment for a number of human diseases, and ongoing research has
focused on the development of RNA-based therapeutics for clinical
applications, such as for the treatment of CVD (36). As one of the most important cells in
the arterial mesangium, VSMCs serve a key role in the formation of
AS through excessive proliferation (37,38). A
number of studies have used VSMCs as models to elucidate the
pathological mechanism underlying AS. Furthermore, miRNAs have
become an integral part in determining the mechanisms associated
with AS, and have the potential to act as biomarkers (39,40).
Ruan et al (41)
demonstrated that miR-155 overexpression inhibited apoptosis in
different cells including vascular endothelial cells and vascular
smooth muscle cells by suppressing the expression of p85α to block
Akt activation, which supports a potential therapeutic role in AS.
Thus, it was hypothesized that miRNA may regulate the
proliferation, migration and cell viability of A7r5 cells induced
by ox-LDL.
To understand the functional mechanisms underlying
miRNAs, the identification of potential targets involved in their
regulation is required. Luo et al (42) reported that miR-129-5p attenuates
irradiation-induced autophagy and decreases the radio-resistance of
breast cancer cells by targeting HMGB1 (42,43).
In the present study, the role of miR-129-5p on AS, and the
potential underlying molecular mechanisms were investigated. The
effects of miR-129-5p on the viability and migration of A7r5 cells
induced by ox-LDL were also investigated. HMGB1 is a
well-established target of the of PI3k/Akt signaling pathway.
Moreover, the results of the present study demonstrated that
miR-129-5p regulated the PI3k/Akt signaling pathway by targeting
HMGB1. Thus, the results of the present study identified HMGB1 as a
direct functional target of miR-129-5p in A7r5 cells, and also an
important regulator of the PI3k/Akt signaling pathway in VSMCs. For
example, the results of the present study demonstrated that the
3'UTR of HMGB1 contained a binding site that matched the miR-129-5p
seed sequence. Furthermore, compared with control group,
overexpression of miR-129-5p decreased the luciferase activity
upstream of the WT 3'UTR of HMGB1, whereas a site mutation in
miR-129-5p abolished miR-129-5p regulation. Inhibition of
miR-129-5p led to an increase in luciferase activity of WT HMGB1
3'UTR in A7r5 cells. Finally, transfection of the miR-129-5p
inhibitor into A7r5 cells suppressed HMGB1 expression levels. Thus,
the results of the present study demonstrated that miR-129-5p
regulated HMGB1 expression by directly binding to its 3'UTR.
Previous studies (44,45)
have indicated the potential roles of HMGB1 in VSMCs. HMGB1 serves
a role in the regulation of the PI3k/Akt signaling pathway
(44), which is crucial for the
development or progression of AS in a number of ways, such as cell
growth, migration and apoptosis of A7r5 cells induced by ox-LDL.
The effective inhibition of VSMC proliferation at the early stage
of AS formation is an important method for the prevention and
treatment of vascular hyperplastic lesions and restenosis after
angioplasty (46).
In conclusion, miR-129-5p mediated HMGB1 to regulate
the PI3k/Akt signaling pathway and further inhibited the
proliferation and migration of A7r5 cells induced by ox-LDL. Thus,
the present study may provide new insights into the treatment of AS
through the regulation of VSMC proliferation and migration.
However, the present study was limited in that it
primarily focused on the viability, migration and apoptosis of
ox-LDL-induced A7r5 cells. For example, despite there being
discussions surrounding the pathological mechanisms underlying the
atherosclerotic cell model, these were not verified using any in
vivo experimental data. In addition, agonists or inhibitors
were not used in the present study to explore the signaling
pathway. Thus, Akt inhibitors, such as MK2206, should be used in
future investigations to further explore the role of the PI3k/Akt
signaling pathway regulated by miR-129-5p in CVD.
Acknowledgements
Not applicable.
Funding
Funding: The present study was supported by the funded project
of National Natural Science Foundation of China (grant. no.
8166020210).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
HJ wrote the manuscript and analyzed the data. RG
and YW participated in experimental design, modification, data
analysis and submission. RG participated in experiments and
literature review. HJ and YW confirm the authenticity of all the
raw data. All authors have read and approved the final
manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Jackson J, Patterson AJ, MacDonald-Wicks L
and McEvoy M: The role of inorganic nitrate and nitrite in CVD.
Nutr Res Rev. 30:247–264. 2017.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Veerbeek JH, Brouwers L, Koster MP, Koenen
SV, van Vliet EO, Nikkels PG, Franx A and van Rijn BB: Spiral
artery remodeling and maternal cardiovascular risk: The spiral
artery remodeling (SPAR) study. J Hypertens. 34:1570–1577.
2016.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Sivasinprasasn S, Wikan N, Tocharus J,
Chaichompoo W, Suksamrarn A and Tocharus C: Pelargonic acid
vanillylamide and rosuvastatin protect against oxidized low-density
lipoprotein-induced endothelial dysfunction by inhibiting the
NF-κB/NLRP3 pathway and improving cell-cell junctions. Chem Biol
Interact. 345(109572)2021.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Groenen AG, Halmos B, Tall AR and
Westerterp M: Cholesterol efflux pathways, inflammation, and
atherosclerosis. Crit Rev Biochem Mol Biol. 56:426–439.
2021.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Valanti EK, Dalakoura-Karagkouni K, Siasos
G, Kardassis D, Eliopoulos AG and Sanoudou D: Advances in
biological therapies for dyslipidemias and atherosclerosis.
Metabolism. 116(154461)2021.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Zhou Z, Subramanian P, Sevilmis G, Globke
B, Soehnlein O, Karshovska E, Megens R, Heyll K, Chun J,
Saulnier-Blache JS, et al: Lipoprotein-derived lysophosphatidic
acid promotes atherosclerosis by releasing CXCL1 from the
endothelium. Cell Metab. 13:592–600. 2011.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Pirillo A, Norata GD and Catapano AL:
LOX-1, OxLDL, and atherosclerosis. Mediators Inflamm.
2013(152786)2013.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Yu Q, Chen S, Tang H, Zhang X, Tao R, Yan
Z, Shi J, Guo W and Zhang S: Veratric acid alleviates liver
ischemia/reperfusion injury by activating the Nrf2 signaling
pathway. Int Immunopharmacol. 101(108294)2021.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Gu Y, Xiao ZH, Wu J, Guo M, Lv P and Dou
N: Anti-atherosclerotic effect of afrocyclamin A against vascular
smooth muscle cells is mediated via p38 MAPK signaling pathway.
Cell J. 23:191–198. 2021.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Yang Y, Yang L, Liang X and Zhu G:
MicroRNA-155 promotes atherosclerosis inflammation via targeting
SOCS1. Cell Physiol Biochem. 36:1371–1381. 2015.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Toba H, Cortez D, Lindsey ML and Chilton
RJ: Applications of miRNA technology for atherosclerosis. Curr
Atheroscler Rep. 16(386)2014.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Zhang Z, Pan X, Yang S, Ma A, Wang K, Wang
Y, Li T and Liu S: miR-155 promotes ox-LDL-induced autophagy in
human umbilical vein endothelial cells. Mediators Inflamm.
2017(9174801)2017.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Arai K, Jin G, Navaratna D and Lo EH:
Brain angiogenesis in developmental and pathological processes:
Neurovascular injury and angiogenic recovery after stroke. FEBS J.
276:4644–4652. 2009.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Geng Z, Xu F and Zhang Y:
MiR-129-5p-mediated Beclin-1 suppression inhibits endothelial cell
autophagy in atherosclerosis. Am J Transl Res. 8:1886–1894.
2016.PubMed/NCBI
|
|
15
|
Sun X, Icli B, Wara AK, Belkin N, He S,
Kobzik L, Hunninghake GM and Vera MP: MICU Registry. Blackwell TS,
et al: MicroRNA-181b regulates NF-κB-mediated vascular
inflammation. J Clin Invest. 122:1973–1990. 2012.PubMed/NCBI View
Article : Google Scholar
|
|
16
|
Liu X, Cheng Y, Yang J, Xu L and Zhang C:
Cell-specific effects of miR-221/222 in vessels: Molecular
mechanism and therapeutic application. J Mol Cell Cardiol.
52:245–255. 2012.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Xin M, Small EM, Sutherland LB, Qi X,
McAnally J, Plato CF, Richardson JA, Bassel-Duby R and Olson EN:
MicroRNAs miR-143 and miR-145 modulate cytoskeletal dynamics and
responsiveness of smooth muscle cells to injury. Genes Dev.
23:2166–2178. 2009.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Liu YR, Chen JJ and Dai M: Paeonol
protects rat vascular endothelial cells from ox-LDL-induced injury
via downregulating microRNA-21 expression and TNF-α release. Acta
Pharmacol Sin. 35:483–488. 2014.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Xu C, Shao Y, Xia T, Yang Y, Dai J, Luo L,
Zhang X, Sun W, Song H, Xiao B and Guo J: lncRNA-AC130710 targeting
by miR-129-5p is upregulated in gastric cancer and associates with
poor prognosis. Tumor Biol. 35:9701–9706. 2014.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Long XH, Zhou YF, Peng AF, Zhang ZH, Chen
XY, Chen WZ, Liu JM, Huang SH and Liu ZL: Demethylation-mediated
miR-129-5p up-regulation inhibits malignant phenotype of osteogenic
osteosarcoma by targeting Homo sapiens valosin-containing protein
(VCP). Tumor Biol. 36:3799–3806. 2015.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Feng J, Guo J, Wang JP and Chai BF:
MiR-129-5p inhibits proliferation of gastric cancer cells through
targeted inhibition on HMGB1 expression. Eur Rev Med Pharmacol Sci.
24:3665–3673. 2020.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Pang X, Zhang Y and Zhang S: High-mobility
group box 1 is overexpressed in cervical carcinoma and promotes
cell invasion and migration. Oncol Rep. 37:831–840. 2017.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Wu CY, Zhou ZF, Wang B, Ke ZP, Ge ZC and
Zhang XJ: MicroRNA-328 ameliorates oxidized low-density
lipoprotein-induced endothelial cells injury through targeting
HMGB1 in atherosclerosis. J Cell Biochem, Oct 15, 2018 (Epub ahead
of print). doi: 10.1002/jcb.27469.
|
|
24
|
Moreno JA, Sastre C, Madrigal-Matute J,
Muñoz-García B, Ortega L, Burkly LC, Egido J, Martín-Ventura JL and
Blanco-Colio LM: HMGB1 expression and secretion are increased via
TWEAK-Fn14 interaction in atherosclerotic plaques and cultured
monocytes. Arterioscler Thromb Vasc Biol. 33:612–620.
2013.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Chen Y, Jiang J, Miao H, Chen X, Sun X and
Li Y: Hydrogen-rich saline attenuates vascular smooth muscle cell
proliferation and neointimal hyperplasia by inhibiting reactive
oxygen species production and inactivating the Ras-ERK1/2-MEK1/2
and Akt pathways. Int J Mol Med. 31:597–606. 2013.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Xiao S, Zhang D, Liu Z, Jin W, Huang G,
Wei Z, Wang D and Deng C: Diabetes-induced glucolipotoxicity
impairs wound healing ability of adipose-derived stem cells-through
the miR-1248/CITED2/HIF-1α pathway. Aging (Albany NY).
12:6947–6965. 2020.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Soares AC and Fonseca DA: Cardiovascular
diseases: A therapeutic perspective around the clock. Drug Discov
Today. 25:1086–1098. 2020.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Benjamin EJ, Virani SS, Callaway CW,
Chamberlain AM, Chang AR, Cheng S, Chiuve SE, Cushman M, Delling
FN, Deo R, et al: Heart disease and stroke statistics-2018 update:
A report from the american heart association. Circulation.
137:e67–e492. 2018.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Pastore I, Bolla AM, Montefusco L, Lunati
ME, Rossi A, Assi E, Zuccotti GV and Fiorina P: The impact of
diabetes mellitus on cardiovascular risk onset in children and
adolescents. Int J Mol Sci. 21(4928)2020.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Ezzatvar Y, Izquierdo M, Núñez J,
Calatayud J, Ramírez-Vélez R and García-Hermoso A:
Cardiorespiratory fitness measured with cardiopulmonary exercise
testing and mortality in patients with cardiovascular disease: A
systematic review and meta-analysis. J Sport Health Sci.
10:609–619. 2021.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Mehta D and Malik AB: Signaling mechanisms
regulating endothelial permeability. Physiol Rev. 86:279–367.
2006.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Koshman YE, Patel N, Chu M, Iyengar R, Kim
T, Ersahin C, Lewis W, Heroux A and Samarel AM: Regulation of
connective tissue growth factor gene expression and fibrosis in
human heart failure. J Card Fail. 19:283–294. 2013.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Soltani A, Argani H, Rahimipour H,
Soleimani F, Rahimi F and Kazerouni F: Oxidized LDL: As a risk
factor for cardiovascular disease in renal transplantation. J Bras
Nefrol. 38:147–152. 2016.PubMed/NCBI View Article : Google Scholar : (In English,
Portuguese).
|
|
35
|
Maiolino G, Rossitto G, Caielli P, Bisogni
V, Rossi GP and Calò LA: The role of oxidized low-density
lipoproteins in atherosclerosis: The myths and the facts. Mediators
Inflamm. 2013(714653)2013.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Chang TY, Tsai WC, Huang TS, Su SH, Chang
CY, Ma HY, Wu CH, Yang CY, Lin CH, Huang PH, et al: Dysregulation
of endothelial colony-forming cell function by a negative feedback
loop of circulating miR-146a and -146b in cardiovascular disease
patients. PLoS One. 12(e0181562)2017.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Li W, Zhi W, Liu F, He Z, Wang X and Niu
X: Atractylenolide I restores HO-1 expression and inhibits
Ox-LDL-induced VSMCs proliferation, migration and inflammatory
responses. Exp Cell Res. 353:26–34. 2017.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Zhang L, Cheng H, Yue Y, Li S, Zhang D and
He R: H19 knockdown suppresses proliferation and induces apoptosis
by regulating miR-148b/WNT/β-catenin in ox-LDL-stimulated vascular
smooth muscle cells. J Biomed Sci. 25(11)2018.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Madrigal-Matute J, Rotllan N, Aranda JF
and Fernández-Hernando C: MicroRNAs and atherosclerosis. Curr
Atheroscler Rep. 15(322)2013.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Florijn BW, Bijkerk R, van der Veer EP and
van Zonneveld AJ: Gender and cardiovascular disease: Are sex-biased
microRNA networks a driving force behind heart failure with
preserved ejection fraction in women? Cardiovasc Res. 114:210–225.
2018.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Ruan Z, Chu T, Wu L, Zhang M, Zheng M,
Zhang Q, Zhou M and Zhu G: miR-155 inhibits oxidized low-density
lipoprotein-induced apoptosis in different cell models by targeting
the p85α/AKT pathway. J Physiol Biochem. 76:329–343.
2020.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Luo J, Chen J and He L: mir-129-5p
attenuates irradiation-induced autophagy and decreases
radioresistance of breast cancer cells by targeting HMGB1. Med Sci
Monit. 21:4122–4129. 2015.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Han JS, Kim K and Lee M: A high mobility
group B-1 box A peptide combined with an artery wall binding
peptide targets delivery of nucleic acids to smooth muscle cells. J
Cell Biochem. 107:163–170. 2009.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Lamb FS, Choi H, Miller MR and Stark RJ:
TNFα and reactive oxygen signaling in vascular smooth muscle cells
in hypertension and atherosclerosis. Am J Hypertens. 33:902–913.
2020.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Zhu XS, Zhou HY, Yang F, Zhang HS and Ma
KZ: miR-381-3p inhibits high glucose-induced vascular smooth muscle
cell proliferation and migration by targeting HMGB1. J Gene Med.
23(e3274)2021.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Chen Z, Pan X, Sheng Z, Yan G, Chen L and
Ma G: Baicalin suppresses the proliferation and migration of
Ox-LDL-VSMCs in atherosclerosis through upregulating miR-126-5p.
Biol Pharm Bull. 42:1517–1523. 2019.PubMed/NCBI View Article : Google Scholar
|




















