Introduction
A total of ~85% of lung cancer cases are recognized
as non-small cell lung cancer (NSCLC), which is a leading cause of
human mortality worldwide with a critical 5-year survival (1) due to late diagnosis at advanced
stages (2). Despite adequate
treatment, which consists of thoracic radiotherapy (60-66 Gy in
30-33 daily fractions) with concomitant cisplatin-based
chemotherapy (3), 50% of medically
inoperable patients with advanced NSCLC experience local
recurrence; on the contrary, the avoidance of local recurrence in
the early stage is up to 85-90% (4). Adequate treatment depends on accurate
early diagnosis and effective treatment response, and peripheral
blood detection of NSCLC can reduce the detection cost and avoid
the potential damage caused by X-ray and pathological examination
(5). Carcinoembryonic antigen
(CEA) is a tumor marker, which represents the earliest tumor marker
for the diagnosis of lung cancer (6). Squamous cell carcinoma antigen
(SCCAg) is associated with the stage of lung cancer (7). Neuron specific enolase (NSE) is a
specific enzyme distributed in neuroendocrine cells and involved in
glycolysis (8). The clinical
diagnostic value of the three genes in NSCLC has been largely
discussed, but no single reliable index for the early diagnosis of
NSCLC has been discovered (9).
Molecular profiling of NSCLC in the past decade has revealed
numerous oncogenic driving events (such as epidermal growth factor
receptor mutation and anaplastic lymphoma kinase rearrangement)
(10) in NSCLC, leading to several
effective therapies, including tyrosine kinase inhibitors and
immunotherapy (11). The present
study aimed to unravel effective biomarkers in the peripheral blood
of patients with NSCLC to achieve early diagnosis and timely
intervention.
A number of studies have documented the involvement
of microRNAs (miRNAs/miRs) in gene expression and evolution and the
regulation of oncogene expression, which is closely associated with
cell proliferation and differentiation and the occurrence,
development and metastasis of malignant tumors (12,13).
In tumor tissues, changes in miRNA expression are tissue specific
with high levels of stability and detectability, and miRNAs can act
as oncogenes or tumor suppressors, which renders them novel targets
for cancer diagnosis and treatment (14,15).
Essentially, the abnormal expression of the miR-200 family may
promote the occurrence of NSCLC and may represent a potential
diagnostic biomarker of NSCLC (16,17).
miR-200a-3p is highly expressed in NSCLC tissues (18). Although it has been demonstrated
that miR-200a-3p is associated with NSCLC, the expression of
miR-200a-3p in the peripheral blood of patients with NSCLC and its
clinical diagnostic value have not been examined.
Transcription factor GATA-6 (GATA6) is a
transcriptional regulator of the GATA family. As a cancer
suppressor in several tumors, GATA6 is expressed at low level in
patients with NSCLC (19). GATA6
is downregulated in lung adenocarcinoma (LUAD) and associated with
LUAD metastasis and prognosis (20). In addition, the
GATA6-PCAT1-Fyn-related kinase axis has been indicated to be a
useful target for NSCLC (21).
Moreover, miR-141 of the miR-200 family has been reported to
regulate angiogenesis and affect cancer invasion and metastasis
through multiple targets, including GATA6(22). Therefore, we hypothesized that
miR-200a-3p may serve a regulatory role in the occurrence and
development of NSCLC by targeting GATA6. To the best of our
knowledge, there is no report at present on the expression and
clinical value of GATA6 in the peripheral blood of patients with
NSCLC, and whether miR-200a-3p can serve a regulatory role in the
occurrence and development of NSCLC by targeting GATA6. Therefore,
the present study aimed to examine the expression and clinical
significance of miR-200a-3p and GATA6 in the peripheral blood of
patients with NSCLC and the possible regulatory mechanism between
the two molecules, to provide a reference for the diagnosis,
treatment and prognosis of NSCLC.
Materials and methods
Study subjects and grouping
In the current study, 60 patients pathologically
diagnosed with NSCLC in Guizhou Provincial People's Hospital
(Guiyang, China) between February 2014 and January 2015 were
selected. The patients were free of mental illness, autoimmune
diseases and other cancers. There were 42 males and 18 females with
an age range of 39-80 years. Inclusion criteria were as follows: i)
According to the 2015 World Health Organization Lung Tumor
Classification (23), the patients
were initially diagnosed with NSCLC via histopathology; ii) none of
the patients received lung cancer-related surgery or chemotherapy
before enrollment; and iii) according to the TNM staging standard
of the International Anti-Cancer Alliance (24), the subjects were allocated into
stage I-II and III-IV. In addition, 60 individuals who underwent
physical examination in Guizhou Provincial People's Hospital during
the same period were included as the healthy control group,
including 42 males and 18 females with an age range of 38-71 years.
The clinicopathological data in the present study included age,
sex, smoking, tumor size, histological types, TNM staging, lymph
node metastasis (LNM) and distal metastasis. Written informed
consent was obtained from each subject. The current study was
approved by the Ethics Committee of Guizhou Provincial People's
Hospital (Guiyang, China).
Sample collection
A total of 3 ml venous blood was collected from
fasting patients without chemotherapy or radiotherapy before
operation and placed in a coagulation-promoting tube. After
standing for 30 min at room temperature, the blood sample was
centrifuged at 100 x g for 10 min at room temperature. Then, the
separated serum was stored at -80˚C until further use. The levels
of CEA, NSE and SCCAg in the serum were determined using a Roche
E601 electrochemiluminescence instrument [Roche Diagnostics
(Shanghai) Co., Ltd.] using matching original reagents in strict
accordance with the manufacturer's instructions. The normal
reference range was 0-5.2 ng/ml for CEA, 0-16.3 ng/ml for NSE and
0-2.5 ng/ml for SCCAg.
Patient follow-up and records
Patients with NSCLC were followed up until August
2020. All patients were followed up every 6 months through clinic
visits or telephone calls from the date of diagnosis. Each patient
was followed up for at least four times. If the patients were not
followed up, they were considered as lost cases and no longer
recorded.
Reverse transcription quantitative
polymerase chain reaction (RT-qPCR)
Total RNA from the serum samples was extracted using
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc.). According to the manufacturer's instructions, PrimeScript RT
Reagent Kit (Takara Biotechnology Co., Ltd.) was employed to
reverse transcribe total RNA into cDNA. According to the
manufacturer's instructions, the cDNA products of miRNA were
obtained via RT using a SuperScript™ IV One-Step RT-PCR
System (Applied Biosystems; Thermo Fisher Scientific, Inc.). qPCR
was performed on ABI 7900HT fast real-time PCR system (Applied
Biosystems; Thermo Fisher Scientific, Inc.) using SYBR®
Premix Ex Taq™ II (Takara Biotechnology Co., Ltd.). The
PCR procedure was as follows: Pre-denaturation at 95˚C for 5 min,
followed by 40 cycles of denaturation at 95˚C for 15 sec, annealing
at 60˚C for 20 sec and extension at 72˚C for 35 sec. GAPDH was used
as the internal reference for GATA6 mRNA, and the expression of
miR-200a-3p was normalized using cel-miR-39 (25-27).
The data were analyzed based on the 2-ΔΔCq method
(28,29). The primer sequences of miR-200a-3p
were selected according to a previous study (28). Primer sequences were synthesized by
Sangon Biotech Co., Ltd., and are were presented in Table I.
 | Table IPrimer sequences used for reverse
transcription-quantitative PCR. |
Table I
Primer sequences used for reverse
transcription-quantitative PCR.
| Gene | Forward
(5'-3') | Reverse
(5'-3') |
|---|
| miR-200a-3p |
TAACACTGTCTGGTAACGATGT |
CATCTTACCGGACAGTGCTGGA |
| GATA6 |
GTGCCAACTGTCACACCACA |
GAGTCCACAAGCATTGCACAC |
| GAPDH |
CTCAGACACCATGGGGAAGGTGA |
ATGATCTTGAGGCTGTTGTCATA |
| cel-miR-39 |
CGTATGAGCGTCACCGGGTGTAAATCA |
CTCAAGTGTCGTGGAGTCGGCAA |
Bioinformatics analysis
The pan-cancer analysis platform [The Encyclopedia
of RNA Interactomes (ENCORI); http://starbase.sysu.edu.cn/panMirDiffExp.php] was
used to analyze and predict the expression of miR-200a-3p and GATA6
in NSCLC (GSE48568, GPL570, PRJNA185966) (30-32).
starBase v2.0 (http://starbase.sysu.edu.cn), miRDB (http://www.mirdb.org/) and TargetScan v7.1 (http://www.targetscan.org/) were employed to analyze
and predict whether there was a target binding relationship between
miR-200a-3p and GATA6.
Dual-luciferase reporter gene assay
and cell transfection
The binding sites of miR-200a-3p and GATA6 were
predicted via bioinformatics analysis as aforementioned. The
complementary wild-type (WT) binding sequence and the mutant (MUT)
sequence of the 3'-untranslated region of GATA6 were amplified and
cloned into pmiRGLO vector (Promega Corporation) to obtain
pmiRGLO-GATA6-WT and pmiRGLO-GATA6-MUT plasmids (Fig. S1). The WT sequence was randomly
scrambled and amplified to obtain the MUT sequence. The constructed
vectors were mixed with 60 nM mimic negative control (NC) or 60 nM
miR-200a-3p mimic (Shanghai GenePharma Co., Ltd.) and
co-transfected with Lipofectamine® 2000 (Invitrogen;
Thermo Fisher Scientific, Inc.) into 293T cells (Shanghai Institute
of Biochemistry and Cell Biology) cultured to 80% confluence at
37˚C. With Renilla luciferase as the internal reference, the
luciferase activities were detected using Dual-Luciferase Assay kit
(cat. no. ZY130595; Shanghai Zeye Biotechnology Co., Ltd.,
Shanghai, China) after 48 h according to the manufacturer's
instructions. 293T cells were cultured in high-glucose DMEM (cat.
no. SNM-002A; Sunncell Wuhan) containing 10% FBS (Gibco; Thermo
Fisher Scientific, Inc.) and 1% penicillin/streptomycin at 37˚C
containing 5% CO2.
In addition, the human NSCLC cell line NCI-H1437
(Shanghai Huiying biological technology Co., Ltd.) was transfected
with 60 nM mimic NC or 60 nM miR-200a-3p mimic and 120 nM inhibitor
NC or 120 nM miR-200a-3p inhibitor at 37˚C using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.). After 48 h, the expression of GATA6 in each
group was detected via RT-qPCR. NCI-H1437 cells were cultured in
RPMI-1640 medium (Procell Life Science & Technology Co., Ltd.)
with 10% FBS at 37˚C and sub-cultured at a 1:4 ratio.
miR-200a-3p mimic was the simulant of miR-200a-3p,
and its sequence was 5'-UAACACUGUCUGGUAACGAUGU-3', while the
sequence of mimic NC was 5'-CUGUUACACGCAGUUGAUGUAA-3'. miR-200a-3p
inhibitor was a chemically modified complementary single chain of
mature miR-200a-3p (cat. no. miR20000682-1-5), and inhibitor NC was
its negative control (cat. no. miR02102-1-1), which were designed
and synthesized by Guangzhou RiboBio Co., Ltd.
Statistical analysis
SPSS 21.0 (IBM Corp.) and GraphPad Prism 8.01
(GraphPad Software, Inc.) were used for statistical analysis and
plotting of the data. The normal distribution of the continuous
variables was verified using Kolmogorov-Smirnov test. Numerical
data are presented as the mean ± standard deviation and categorical
data as count and percentage. An unpaired Student's t-test was
employed for the analysis between two groups and one-way ANOVA was
used for the comparison among multiple groups (continuous
variables). Tukey's multiple comparisons test was utilized after
one-way ANOVA. The χ2 test was employed for the analysis
of categorical variables. Pearson correlation coefficient method
was applied to analyze correlations. The r value was the
correlation coefficient between variables in the sample. When r=0,
it indicates no correlation; when r≤0.3, it indicates poor
correlation; when 0.3<r≤ 0.8, it indicates medium correlation;
when r>0.8, it indicates high correlation. The cut-off point,
sensitivity, specificity and the area under the curve (AUC) of the
receiver operating characteristic (ROC) curves were obtained using
MedCalc Statistical Software version 14.8.1 (MedCalc Software
bvba). A multivariate logistic regression model was used to analyze
whether the expression of miR-200a-3p and GATA6 was independently
associated with the prognosis of NSCLC. The Cox proportional hazard
regression model and the Kaplan-Meier method were used to evaluate
the overall survival (OS), and the log-rank test was used to create
the survival curve. P<0.05 was considered to indicate a
statistically significant difference.
Results
miR-200a-3p is expressed at a high and
GATA6 at a low level in the peripheral blood of patients with
NSCLC
To examine the diagnostic value of miR-200a-3p and
GATA6 for NSCLC, the pan-cancer analysis platform ENCORI was used
to firstly predict the expression of miR-200a-3p and GATA6 in
NSCLC. Lung squamous cell carcinoma (LUSC) and LUAD account for
~90% of NSCLC (33), therefore
these groups were further explored. The results demonstrated that
compared with the normal group, miR-200a-3p was significantly
elevated in LUSC and LUAD (Fig.
1A), whereas GATA6 was significantly decreased in LUSC and LUAD
(Fig. 1B).
Therefore, the expression of miR-200a-3p and GATA6
in the peripheral blood of patients with NSCLC and healthy controls
was detected using RT-qPCR. The results revealed that miR-200a-3p
in the peripheral blood of patients with NSCLC was remarkably
higher than that of healthy controls (Fig. 1C), and GATA6 was lower than that of
healthy controls (Fig. 1D).
Subsequently, the ROC curve was used to analyze the
diagnostic value of miR-200a-3p and GATA6 in NSCLC. The AUC of
miR-200a-3p in the peripheral blood of patients with NSCLC was
0.721, with a cut-off value was 1.475, a sensitivity of 83.33% and
a specificity of 63.33% (Fig. 1E).
The AUC of GATA6 in the peripheral blood of patients with NSCLC was
0.774, with a cut-off value of 1.195, a sensitivity of 53.33% and a
specificity of 83.33% (Fig. 1F).
The results revealed that cut-off values of miR-200a-3p >1.475
and GATA6 <1.195 could assist in the early diagnosis of
NSCLC.
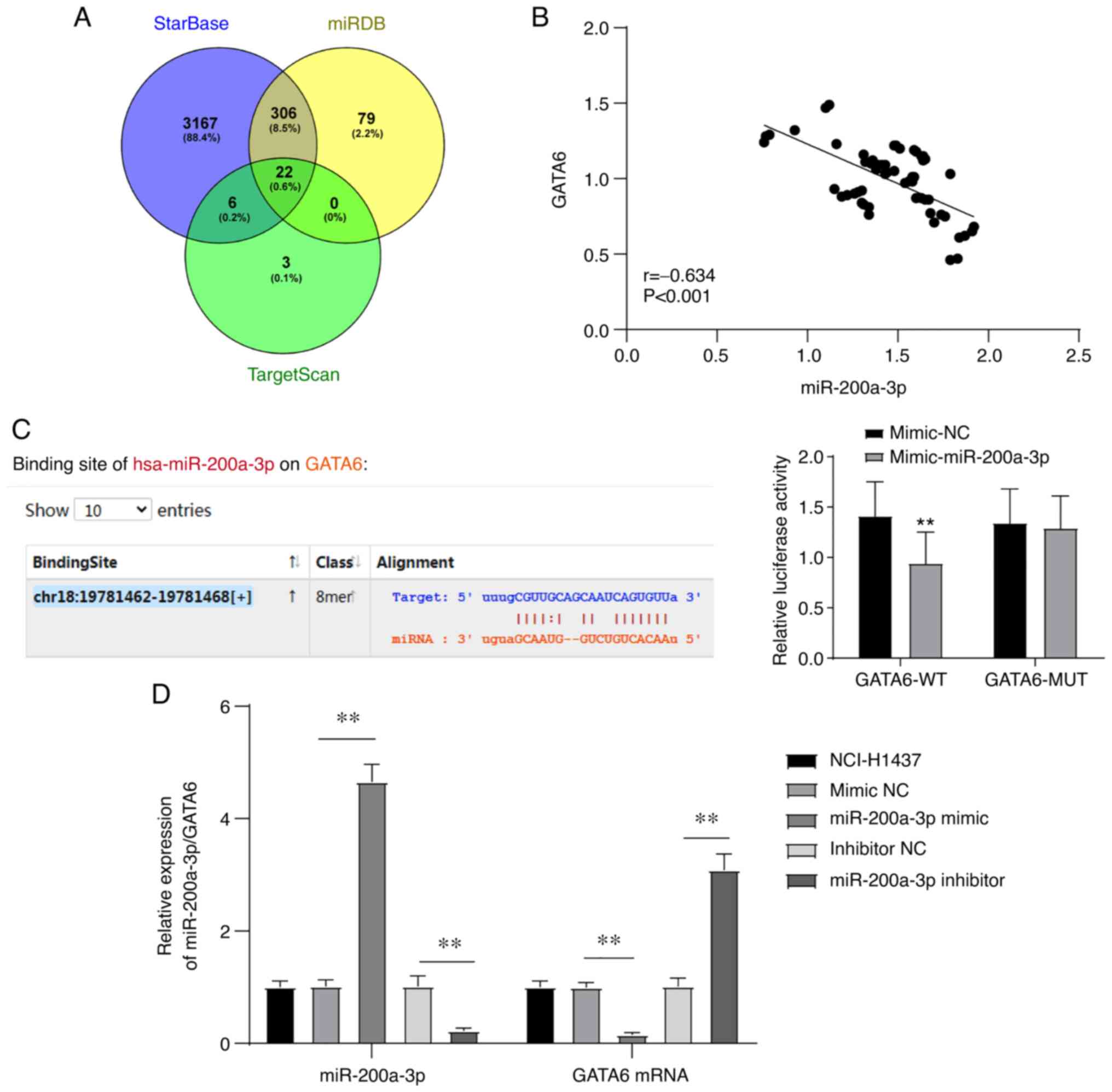
GATA6 is the target of
miR-200a-3p
Bioinformatics analysis was used to analyze and
predict whether there was a targeted binding relationship between
miR-200a-3p and GATA6. starBase, miRDB (score >70) and
TargetScan (preferentially conserved targeting >80) were used to
analyze and predict the downstream targets of miR-200a-3p, and 22
downstream target genes of miR-200a-3p were demonstrated to be
common in all three databases (Table
SI and Fig. 2A). Among these
genes, GATA6 was indicated to be downregulated in NSCLC tissues
compared with normal tissues, which may be used as a useful target
for the intervention of NSCLC. In addition, miR-141 of the miR-200
family can regulate angiogenesis via multiple targets, including
GATA6, thus affecting cancer invasion and metastasis (19,21,22).
Therefore, GATA6 was selected for further study.
Subsequently, the correlation between the expression
of miR-200a-3p and GATA6 in the peripheral blood of patients with
NSCLC was analyzed, and a negative correlation was observed between
them (r=-0.634; Fig. 2B). Next,
the targeted binding relationship between miR-200a-3p and GATA6 was
verified via dual-luciferase assay. Compared with the cells
co-transfected with pmiRGLO-GATA6-WT plasmid and mimic NC, the
luciferase enzyme activity of 293T cells co-transfected with
pmiRGLO-GATA6-WT and miR-200a-3p was significantly decreased, while
this difference was not observed between cells transfected with
pmiRGLO-GATA6-MUT and miR-200a-3p or mimic NC (Fig. 2C). In addition, miR-200a-3p was
overexpressed or knocked down by transfecting miR-200a-3p mimic or
miR-200a-3p inhibitor into the NSCLC cell line NCI-H1437. The mRNA
level of GATA6 was detected via RT-qPCR. Overexpression of
miR-200a-3p significantly decreased, while knockdown of miR-200a-3p
significantly increased the mRNA level of GATA6 (Fig. 2D). These results suggested that
GATA6 is a target gene of miR-200a-3p.
Association of miR-200a-3p and GATA6
with clinical indicators
To further analyze the clinical significance of
miR-200a-3p and GATA6 in the peripheral blood of patients with
NSCLC, the patients were assigned into miR-200a-3p high expression
and miR-200a-3p low expression group according to the median
expression of miR-200a-3p in the peripheral blood (median, 1.48;
range, 0.76-1.92). At the same time, the patients were classified
into GATA6 high expression and GATA6 low expression group according
to the median expression of GATA6 expression in the peripheral
blood (median, 1.02; range, 0.46-1.49). The clinicopathological
features of the two groups were analyzed (Table II). The results demonstrated that
miR-200a-3p and GATA6 in the peripheral blood were not associated
with age, sex, smoking history, histological type, tumor size and
distant metastasis, but were significantly associated with TNM
stage and LNM.
 | Table IIAssociation between the expression of
miR-200a-3p and GATA6 with clinical indicators. |
Table II
Association between the expression of
miR-200a-3p and GATA6 with clinical indicators.
| A, miR-200a-3p |
|---|
| | Expression | |
|---|
| Clinical data | Patient number
(n=60) | High, n (%) | Low, n (%) | P-value |
|---|
| Sex | | | | |
|
Male | 42 | 22 (52.38) | 20 (47.62) | |
|
Female | 18 | 8 (44.44) | 10 (52.38) | 0.573 |
| Age, years | | | | |
|
≤60 | 29 | 15 (51.72) | 14 (48.28) | |
|
>60 | 31 | 15 (48.39) | 16 (51.65) | 0.796 |
| Smoking
history | | | | |
|
Yes | 26 | 11 (42.31) | 15 (57.69) | |
|
No | 34 | 19 (55.88) | 15 (44.12) | 0.297 |
| Histological
types | | | | |
|
LUSC | 35 | 19 (54.29) | 16 (45.71) | |
|
LUAD | 25 | 11 (44.00) | 14 (56.00) | 0.432 |
| Tumor size, cm | | | | |
|
≤3 | 24 | 11 (45.83) | 13 (54.17) | |
|
>3 | 36 | 19 (52.78) | 17 (47.22) | 0.299 |
| TNM stage | | | | |
|
I-II | 42 | 16 (38.10) | 26 (61.90) | |
|
III-IV | 18 | 14 (77.78) | 4 (22.22) | 0.002 |
| LNM | | | | |
|
Yes | 22 | 15 (68.18) | 7 (31.82) | |
|
No | 38 | 15 (39.47) | 23 (60.53) | 0.016 |
| Distant
metastasis | | | | |
|
Yes | 32 | 20 (62.50) | 12 (37.50) | |
|
No | 28 | 10 (35.71) | 18 (64.29) | 0.019 |
| B, GATA6 |
| | Expression | |
| Clinical data | Patient number
(n=60) | High, n (%) | Low, n (%) | P-value |
| Sex | | | | |
|
Male | 42 | 19 (45.24) | 23 (54.76) | |
|
Female | 18 | 11 (61.11) | 7 (38.89) | 0.130 |
| Age, years | | | | |
|
≤60 | 29 | 16 (55.17) | 13 (44.83) | |
|
>60 | 31 | 14 (45.16) | 17 (54.84) | 0.219 |
| Smoking
history | | | | |
|
Yes | 26 | 14 (53.85) | 12 (46.15) | |
|
No | 34 | 16 (47.06) | 18 (52.94) | 0.301 |
| Histological
types | | | | |
|
LUSC | 35 | 17 (48.57) | 18 (51.43) | |
|
LUAD | 25 | 13 (52.00) | 12 (48.00) | 0.397 |
| Tumor size, cm | | | | |
|
≤3 | 24 | 11 (45.83) | 13 (54.17) | |
|
>3 | 36 | 19 (52.78) | 17 (47.22) | 0.299 |
| TNM stage | | | | |
|
I-II | 42 | 25 (59.52) | 17 (40.48) | |
|
III-IV | 18 | 5 (27.78) | 13 (72.22) | 0.012 |
| LNM | | | | |
|
Yes | 22 | 6 (27.27) | 16 (72.73) | |
|
No | 38 | 24 (63.16) | 14 (36.84) | 0.004 |
| Distant
metastasis | | | | |
|
Yes | 32 | 13 (40.63) | 19 (59.38) | |
|
No | 28 | 17 (56.67) | 11 (36.67) | 0.060 |
Subsequently, the expression of miR-200a-3p and
GATA6 in patients with NSCLC at different stages was further
explored. Compared with healthy controls, miR-200a-3p expression in
the peripheral blood of patients with NSCLC at stage I-II and
III-IV was significantly increased (Fig. 3A), and that of GATA6 was
significantly decreased (Fig. 3B).
In addition, compared with stage I-II, miR-200a-3p expression in
the peripheral blood of patients with NSCLC at stage III-IV was
significantly increased, while that of GATA6 was significantly
decreased. Therefore, it was speculated that the expression of
miR-200a-3p and GATA6 may be associated with the clinical
progression of NSCLC.
Correlation of miR-200a-3p and GATA6
with serum tumor markers
In addition, the serum tumor marker CEA, NSE and
SCCAg levels in the peripheral blood of patients with NSCLC were
detected [CEA (21.0±6.9 ng/ml), NSE (23.6±9.7 ng/ml), SCCAg
(4.6±2.8 ng/ml)], and their correlation with the levels of
miR-200a-3p and GATA6 was analyzed (Fig. 4A-F). The results demonstrated that
miR-200a-3p and GATA6 were positively correlated with CEA and
SCCAg, but not with NSE.
High expression of miR-200a-3p and low
expression of GATA6 predicts poor prognosis in patients with
NSCLC
Finally, the prognostic significance of miR-200a-3p
and GATA6 expression in the peripheral blood of patients with NSCLC
was analyzed. The patients were followed up regularly every 6
months. Among them, 31 patients died or were lost during the
follow-up, including 11 cases in miR-200a-3p low expression group,
20 cases in miR-200a-3p high expression group, 19 cases in GATA6
low expression group and 12 cases in GATA6 high expression group.
Furthermore, the cumulative survival rates of the two groups of
patients with NSCLC were compared and analyzed. The results
demonstrated that the cumulative survival rate of the miR-200a-3p
high expression group was markedly lower than that of the
miR-200a-3p low expression group (Fig.
5A). The cumulative survival rate of the GATA6 low expression
group was notably lower than that of the GATA6 high expression
group (Fig. 5B). In addition, the
expressions of miR-200a-3p and GATA6 in the peripheral blood were
further compared and analyzed using the ROC curve to predict the
adverse prognosis of NSCLC. The AUC of miR-200a-3p for NSCLC
adverse prognosis was 0.653, with a sensitivity of 64.52% and a
specificity of 68.97%. The AUC of GATA6 for NSCLC adverse prognosis
was 0.616, with a sensitivity of 48.39% and a specificity of
75.86%. There were no significant differences between them
(Fig. 5C). It was suggested that
high expression of miR-200a-3p and low expression of GATA6
predicted poor prognosis.
Furthermore, to further analyze whether there was an
independent association between the expression of miR-200a-3p and
GATA6 and the prognosis of NSCLC, the indexes with P<0.1 from
the clinicopathological associations presented in Table II and the serum tumor markers CEA
and SCCAg that were significantly correlated with miR-200a-3p and
GATA6 were included in a multivariate logistic regression model.
After adjusting for TNM stage, lymph node metastasis, distant
metastasis, GATA6, CEA, NSE and SCCAg in the logistic regression
model, it was indicated that high expression of miR-200a-3p
significantly increased the risk of death in patients with NSCLC
(OR, 17.917; 95% CI, 1.822-176.157; Table III), indicating that the
expression of miR-200a-3p was independently associated with poor
prognosis in NSCLC.
 | Table IIIMultivariate logistic regression
analysis of prognostic indicators for non-small cell lung
cancer. |
Table III
Multivariate logistic regression
analysis of prognostic indicators for non-small cell lung
cancer.
| Variable | P-value | OR | 95% CI |
|---|
| TNM stage | 0.115 | 0.220 | 0.034-1.447 |
| Lymph node
metastasis | 0.119 | 4.583 | 0.676-31.071 |
| Distant
metastasis | 0.377 | 0.495 | 0.104-2.354 |
| miR-200a-3p | 0.013 | 17.917 | 1.822-176.157 |
| GATA6 | 0.230 | 0.039 | 0.000-7.801 |
| CEA | 0.111 | 0.888 | 0.768-1.028 |
| SCCAg | 0.139 | 0.754 | 0.518-1.096 |
| NSE | 0.261 | 0.960 | 0.894-1.031 |
Discussion
In general, miRNAs are stably expressed in the blood
and determining their expression levels is relatively simple
(34). A previous study explored
the association between the levels of certain peripheral blood
markers, such as absolute neutrophil count, lymphocyte count,
monocyte count and eosinophil count, as well as serum C-reactive
protein and lactate dehydrogenase levels, and immune-related
adverse events, OS and progression-free survival in NSCLC (35,36).
The present study explored the expression and clinical significance
of miR-200a-3p and GATA6, and demonstrated that the abnormal
expression of miR-200a-3p/GATA6 in the peripheral blood of patients
with NSCLC has a high clinical diagnostic efficiency and prognostic
value.
miRNAs represent molecular biomarkers and potential
targets for therapeutic interventions in NSCLC, especially the
miR-200 family members (16,37).
The pan-cancer analysis platform ENCORI used in the current study
demonstrated that miR-200a-3p was elevated, whereas GATA6 was
significantly decreased in LUSC and LUAD, which was also confirmed
in the peripheral blood of clinical patients with NSCLC. The
crucial and complex roles of the miR-200 family have been discussed
in lung cancer (16). miR-200a-3p
is upregulated in lung cancer and the subtype LUSC (38-40).
GATA6 is a potent and clinically relevant tumor suppressor gene in
LUAD (41). Disease-related miRNAs
have been extensively studied, and several blood-based miRNA tests
have been developed for lung cancer diagnosis, with reasonable
sensitivity and specificity (42-44).
Subsequently, the ROC curve method was used to analyze the
diagnostic value of miR-200a-3p and GATA6. The AUC of miR-200a-3p
in the peripheral blood of patients with NSCLC was 0.721, and the
cut-off value was 1.475 with a sensitivity of 83.33% and a
specificity of 63.33%. The AUC of GATA6 in the peripheral blood of
patients with NSCLC was 0.774, and the cut-off value was 1.195 with
a sensitivity of 53.33% and a specificity of 83.33%. To sum up,
miR-200a-3p >1.475 and GATA6 <1.195 may assist in the early
diagnosis of NSCLC.
Through database prediction and dual-luciferase
assay, it was validated that GATA6 is a target gene of miR-200a-3p.
miR-200a-3p upregulation has been indicated to independently lower
TNF-α-induced expression of transcription factor GATA6(45). Next, the clinical significance of
miR-200a-3p and GATA6 were analyzed. The patients were allocated
into miR-200a-3p and GATA6 high/low expression groups according to
the median expression of miR-200a-3p and GATA6. The analysis of
clinicopathological features demonstrated that the levels of
miR-200a-3p and GATA6 in the peripheral blood were not associated
with age, sex, smoking history, histological type and tumor size,
but were significantly associated with TNM stage, LNM and distal
metastasis (not for GATA6). Consistently, another study has
revealed that miR-200c overexpression is significantly associated
with LNM, advanced TNM and poor survival rates in NSCLC (46). Higher level of GATA6 in LUAD was
significantly associated with longer survival of patients in all
stages (41). Furthermore,
compared with stage I-II, the expression of miR-200a-3p in the
peripheral blood of patients with NSCLC at stage III-IV was
increased while that of GATA6 was significantly decreased.
Similarly, miR-200a has been associated with cancer metastasis
(47). Therefore, we hypothesized
that miR-200a-3p and GATA6 expression may be associated with the
clinical progression of NSCLC.
In the treatment process of NSCLC, blood-based
biomarkers can help guide treatment decisions by monitoring tumor
load, minimal residual or recurrent disease and genetic alterations
(48). CEA, SCCAg and NSE are
typical serum tumor markers (6,8,49)
and exhibit high clinical reliability in NSCLC. Therefore, we
evaluated their expression and correlation with miR-200a-3p and
GATA6. The results demonstrated that miR-200a-3p and GATA6 were
correlated with CEA and SCCAg, but not with NSE. There is no report
on the correlation of the three biomarkers with miR-200a-3p and
GATA6 in the peripheral blood of patients with NSCLC, to the best
of our knowledge, therefore the present study demonstrated this
analysis for the first time. Finally, the prognostic significance
of miR-200a-3p and GATA6 was analyzed. The cumulative survival rate
of the miR-200a-3p high expression group was markedly lower than
that of the miR-200a-3p low expression group, and the cumulative
survival rate of the GATA6 low expression group was notably lower
than that of GATA6 high expression group. In addition, miR-200a-3p
predicted NSCLC adverse prognosis with an AUC of 0.653, a
sensitivity of 64.52% and a specificity of 68.97%. GATA6 predicted
NSCLC adverse prognosis with an AUC of 0.616, a sensitivity of
48.39% and a specificity of 75.86%, indicating that high expression
of miR-200a-3p and low expression of GATA6 predicted poor
prognosis. Furthermore, after adjusting for TNM stage, lymph node
metastasis, distant metastasis, GATA6, CEA, NSE and SCCAg in the
logistic regression model, high miR-200a-3p expression was
indicated to increase the risk of death in patients with NSCLC,
indicating that miR-200a-3p expression was independently associated
with the poor prognosis of NSCLC. miR-200a/b/c are potential
prognostic indicators of LUSC (40). GATA6 is decreased in high-grade
LUAD (50,51), and its reduction can enhance the
metastatic potency in LUAD (20).
In summary, high expression of miR-200a-3p and low expression of
GATA6 predicted poor prognosis in patients with NSCLC.
In conclusion, high expression of miR-200a-3p and
low expression of GATA6 in the peripheral blood were associated
with clinical features and serum markers, and predicted poor
prognosis in patients with NSCLC. Restoration of GATA6 expression
or inhibition of miR-200a-3p may therefore represent a possibility
for NSCLC early treatment. This was a case-control study, which
revealed the clinical diagnostic efficacy of miR-200a-3p and GATA6
in NSCLC. However, there are still certain limitations in the
present study: i) The sample size was small and the diagnostic and
prognostic value of miR-200a-3p and GATA6 still requires further
verification in a cohort with an appropriate sample size; ii) serum
and plasma miRNAs mainly come from exosomes: Because of their
increased number when diseases invade the body, stable existence in
body fluids, easy access and resistance to enzymatic hydrolysis,
they are considered to be ideal biomarkers for early cancer
diagnosis. Serum exosomal miRNAs not only can be used for the early
diagnosis of lung cancer, but also have the potential to
distinguish NSCLC and SCLC (52).
Whether the level of miR-200a-3p in the peripheral blood of
patients with NSCLC included in the present study is associated
with the level of serum exosomal miR-200a-3p, and whether the level
of serum exosomal miR-200a-3p has the value of early diagnostic and
prognostic evaluation of NSCLC still needs to be further studied;
iii) the expression of miR-200a-3p and GATA6 in tumor samples were
not detected; iv) the downstream targets of miR-200a-3p and GATA6
are not fully understood, and the regulatory mechanism of NSCLC
still requires to be further elucidated. In the future, the
regulatory upstream or downstream mechanism of miR-200a-3p and
GATA6 in NSCLC will be further investigated, and in-depth
functional research on miR-200a-3p and GATA6 will be conducted.
Furthermore, the present study demonstrated that the expression of
miR-200a-3p and GATA6 may be used for the early diagnosis and
treatment of NSCLC, indicating that it may be associated with the
intensity of tumor load in patients with NSCLC. Whether the
postoperative expression of miR-200a-3p and GATA6 is also
associated with the tumor load intensity remains to be explored.
Detection of their expression following patient operation will help
to evaluate the effect of radical operation and provide reference
for further treatment after the operation.
Supplementary Material
Construction of dual-luciferase assay
vector. UTR, untranslated region; WT, wild-type; MUT, mutant;
miRNA, microRNA; Luc, luciferase; GATA6, transcription factor
GATA-6; RISC, RNA-induced silencing complex; IRES, internal
ribosome entry site.
A total of 22 common genes in
starBase, miRDB and TargetScan.
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
JY, XYH and YBX contributed to the study concept and
study design. CJF, HXW and LH contributed to the literature search.
CJF and HXW contributed to manuscript preparation and YBX
contributed to manuscript editing and review. JY, XYH, YBX and LH
contributed to performing the experiments and data acquisition.
CJF, HXW and LH contributed to data analysis and statistical
analysis. JY and XYH confirm the authenticity of all the raw data.
All authors have read and approved the final manuscript.
Ethics approval and consent to
participate
The current study was approved by the Ethics
Committee of Guizhou Provincial People's Hospital (approval no.
LLS-2019123012; Guiyang, China). Written informed consent was
obtained from each subject.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Mukherjee TK, Malik P and Hoidal JR: The
emerging role of estrogen related receptorα in complications of
non-small cell lung cancers. Oncol Lett. 21(258)2021.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Min HY and Lee HY: Mechanisms of
resistance to chemotherapy in non-small cell lung cancer. Arch
Pharm Res. 44:146–164. 2021.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Postmus PE, Kerr KM, Oudkerk M, Senan S,
Waller DA, Vansteenkiste J, Escriu C and Peters S: ESMO Guidelines
Committee. Early and locally advanced non-small-cell lung cancer
(NSCLC): ESMO clinical practice guidelines for diagnosis, treatment
and follow-up. Ann Oncol. 28 (Suppl 4):iv1–iv21. 2017.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Alcibar OL, Nadal E, Romero Palomar I and
Navarro-Martin A: Systematic review of stereotactic body
radiotherapy in stage III non-small cell lung cancer. Transl Lung
Cancer Res. 10:529–538. 2021.PubMed/NCBI View Article : Google Scholar
|
|
5
|
D'Aniello C, Berretta M, Cavaliere C,
Rossetti S, Facchini BA, Iovane G, Mollo G, Capasso M, Pepa CD,
Pesce L, et al: Biomarkers of prognosis and efficacy of
anti-angiogenic therapy in metastatic clear cell renal cancer.
Front Oncol. 9(1400)2019.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Dall'Olio FG, Abbati F, Facchinetti F,
Massucci M, Melotti B, Squadrilli A, Buti S, Formica F, Tiseo M and
Ardizzoni A: CEA and CYFRA 21-1 as prognostic biomarker and as a
tool for treatment monitoring in advanced NSCLC treated with immune
checkpoint inhibitors. Ther Adv Med Oncol.
12(1758835920952994)2020.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Hatzakis KD, Froudarakis ME, Bouros D,
Tzanakis N, Karkavitsas N and Siafakas NM: Prognostic value of
serum tumor markers in patients with lung cancer. Respiration.
69:25–29. 2002.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Wu H, Wang Q, Liu Q, Zhang Q, Huang Q and
Yu Z: The serum tumor markers in combination for clinical diagnosis
of lung cancer. Clin Lab. 66:2020.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Popper HH, Ryska A, Tímár J and Olszewski
W: Molecular testing in lung cancer in the era of precision
medicine. Transl Lung Cancer Res. 3:291–300. 2014.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Guo Y, Cao R, Zhang X, Huang L, Sun L,
Zhao J, Ma J and Han C: Recent progress in rare oncogenic drivers
and targeted therapy for non-small cell lung cancer. Onco Targets
Ther. 12:10343–10360. 2019.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Alzofon N and Jimeno A: Capmatinib for
non-small cell lung cancer. Drugs Today (Barc). 57:17–25.
2021.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Li LJ, Chang WM and Hsiao M: Aberrant
expression of microrna clusters in head and neck cancer development
and progression: Current and future translational impacts.
Pharmaceuticals (Basel). 14(194)2021.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Papanota AM, Karousi P, Kontos CK,
Ntanasis-Stathopoulos I, Scorilas A and Terpos E: Multiple myeloma
bone disease: Implication of MicroRNAs in its molecular background.
Int J Mol Sci. 22(2375)2021.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Hammouz RY, Kołat D, Kałuzińska Ż,
Płuciennik E and Bednarek AK: MicroRNAs: Their role in metastasis,
angiogenesis, and the potential for biomarker utility in bladder
carcinomas. Cancers (Basel). 13(891)2021.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Raue R, Frank AC, Syed SN and Brüne B:
Therapeutic targeting of MicroRNAs in the tumor microenvironment.
Int J Mol Sci. 22(2210)2021.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Liu C, Hu W, Li LL, Wang YX, Zhou Q, Zhang
F, Song-Yang YY, Zhu W, Sun CC and Li DJ: Roles of miR-200 family
members in lung cancer: More than tumor suppressors. Future Oncol.
14:2875–2886. 2018.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Xie K, Wang C, Qin N, Yang J, Zhu M, Dai
J, Jin G, Shen H, Ma H and Hu Z: Genetic variants in regulatory
regions of microRNAs are associated with lung cancer risk.
Oncotarget. 7:47966–47974. 2016.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Wei S, Wang K, Huang X and Zhao Z and Zhao
Z: LncRNA MALAT1 contributes to non-small cell lung cancer
progression via modulating miR-200a-3p/programmed death-ligand 1
axis. Int J Immunopathol Pharmacol.
33(2058738419859699)2019.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Li H, Feng C and Shi S: miR-196b promotes
lung cancer cell migration and invasion through the targeting of
GATA6. Oncol Lett. 16:247–252. 2018.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Cheung WK, Zhao M, Liu Z, Stevens LE, Cao
PD, Fang JE, Westbrook TF and Nguyen DX: Control of alveolar
differentiation by the lineage transcription factors GATA6 and HOPX
inhibits lung adenocarcinoma metastasis. Cancer Cell. 23:725–738.
2013.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Zang Q, Xu L, Li J and Jia H: GATA6
activated long non-coding RNA PCAT1 maintains stemness of non-small
cell lung cancer by mediating FRK. J BUON. 25:2371–2381.
2020.PubMed/NCBI
|
|
22
|
Dong H, Weng C, Bai R, Sheng J, Gao X, Li
L and Xu Z: The regulatory network of miR-141 in the inhibition of
angiogenesis. Angiogenesis. 22:251–262. 2019.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Travis WD, Brambilla E, Nicholson AG,
Yatabe Y, Austin JHM, Beasley MB, Chirieac LR, Dacic S, Duhig E,
Flieder DB, et al: The 2015 World Health Organization
classification of lung tumors: Impact of genetic, clinical and
radiologic advances since the 2004 classification. J Thorac Oncol.
10:1243–1260. 2015.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Ren Y, Qiu H, Yuan Y, Ye J, Tian Y, Wen B,
Zhang W and Li Q: Evaluation of 7th edition of AJCC staging system
for nasopharyngeal carcinoma. J Cancer. 8:1665–1672.
2017.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Cheng Y, Qu X, Dong Z, Zeng Q, Ma X, Jia
Y, Li R, Jiang X, Williams C, Wang T and Xia W: Comparison of serum
exosome isolation methods on co-precipitated free microRNAs. PeerJ.
8(e9434)2020.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Madadi S and Soleimani M: Comparison of
miR-16 and cel-miR-39 as reference controls for serum miRNA
normalization in colorectal cancer. J Cell Biochem. 120:4802–4803.
2019.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Mitchell PS, Parkin RK, Kroh EM, Fritz BR,
Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O'Briant
KC, Allen A, et al: Circulating microRNAs as stable blood-based
markers for cancer detection. Proc Natl Acad Sci USA.
105:10513–10518. 2008.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Lee H, Kim C, Kang H, Tak H, Ahn S, Yoon
SK, Kuh HJ, Kim W and Lee EK: microRNA-200a-3p increases
5-fluorouracil resistance by regulating dual specificity
phosphatase 6 expression. Exp Mol Med. 49(e327)2017.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Campbell JD, Alexandrov A, Kim J, Wala J,
Berger AH, Pedamallu CS, Shukla SA, Guo G, Brooks AN, Murray BA, et
al: Distinct patterns of somatic genome alterations in lung
adenocarcinomas and squamous cell carcinomas. Nat Genet.
48:607–616. 2016.PubMed/NCBI View
Article : Google Scholar
|
|
31
|
Cancer Genome Atlas Research Network.
Comprehensive genomic characterization of squamous cell lung
cancers. Nature. 489:519–525. 2012.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Cancer Genome Atlas Research Network.
Comprehensive molecular profiling of lung adenocarcinoma. Nature.
511:543–550. 2014.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Zhang J, Zhu N and Chen X: A novel long
noncoding RNA LINC01133 is upregulated in lung squamous cell cancer
and predicts survival. Tumour Biol. 36:7465–7471. 2015.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Zhong S, Golpon H, Zardo P and Borlak J:
miRNAs in lung cancer. A systematic review identifies predictive
and prognostic miRNA candidates for precision medicine in lung
cancer. Transl Res. 230:164–196. 2021.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Peng L, Wang Y, Liu F, Qiu X, Zhang X,
Fang C, Qian X and Li Y: Peripheral blood markers predictive of
outcome and immune-related adverse events in advanced non-small
cell lung cancer treated with PD-1 inhibitors. Cancer Immunol
Immunother. 69:1813–1822. 2020.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Tanizaki J, Haratani K, Hayashi H, Chiba
Y, Nakamura Y, Yonesaka K, Kudo K, Kaneda H, Hasegawa Y, Tanaka K,
et al: Peripheral blood biomarkers associated with clinical outcome
in non-small cell lung cancer patients treated with nivolumab. J
Thorac Oncol. 13:97–105. 2018.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Weidle UH, Birzele F and Nopora A:
MicroRNAs as potential targets for therapeutic intervention with
metastasis of non-small cell lung cancer. Cancer Genomics
Proteomics. 16:99–119. 2019.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Chen Y, Peng W, Lu Y, Chen J, Zhu YY and
Xi T: MiR-200a enhances the migrations of A549 and SK-MES-1 cells
by regulating the expression of TSPAN1. J Biosci. 38:523–532.
2013.PubMed/NCBI View Article : Google Scholar
|
|
39
|
He Q, Fang Y, Lu F, Pan J, Wang L, Gong W,
Fei F, Cui J, Zhong J, Hu R, et al: Analysis of differential
expression profile of miRNA in peripheral blood of patients with
lung cancer. J Clin Lab Anal. 33(e23003)2019.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Shen Y, Pan X and Yang J: Gene regulation
and prognostic indicators of lung squamous cell carcinoma:
TCGA-derived miRNA/mRNA sequencing and DNA methylation data. J Cell
Physiol. 234:22896–22910. 2019.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Chen W, Chen Z, Zhang M, Tian Y, Liu L,
Lan R, Zeng G, Fu X, Ru G, Liu W, et al: GATA6 exerts potent lung
cancer suppressive function by inducing cell senescence. Front
Oncol. 10(824)2020.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Fumagalli C, Bianchi F, Raviele PR,
Vacirca D, Bertalot G, Rampinelli C, Lazzeroni M, Bonanni B,
Veronesi G, Fusco N, et al: Circulating and tissue biomarkers in
early-stage non-small cell lung cancer. Ecancermedicalscience.
11(717)2017.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Halvorsen AR, Bjaanæs M, LeBlanc M, Holm
AM, Bolstad N, Rubio L, Peñalver JC, Cervera J, Mojarrieta JC,
López-Guerrero JA, et al: A unique set of 6 circulating microRNAs
for early detection of non-small cell lung cancer. Oncotarget.
7:37250–37259. 2016.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Wang C, Ding M, Xia M, Chen S, Van Le A,
Soto-Gil R, Shen Y, Wang N, Wang J, Gu W, et al: A Five-miRNA panel
identified from a multicentric case-control study serves as a novel
diagnostic tool for ethnically diverse non-small-cell lung cancer
patients. EBioMedicine. 2:1377–1385. 2015.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Fan X, Chen X, Feng Q, Peng K, Wu Q,
Passerini AG, Simon SI and Sun C: Downregulation of GATA6 in
mTOR-inhibited human aortic endothelial cells: Effects on
TNF-α-induced VCAM-1 expression and monocytic cell adhesion. Am J
Physiol Heart Circ Physiol. 316:H408–H420. 2019.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Si L, Tian H, Yue W, Li L, Li S, Gao C and
Qi L: Potential use of microRNA-200c as a prognostic marker in
non-small cell lung cancer. Oncol Lett. 14:4325–4330.
2017.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Mongroo PS and Rustgi AK: The role of the
miR-200 family in epithelial-mesenchymal transition. Cancer Biol
Ther. 10:219–222. 2010.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Xu-Welliver M and Carbone DP: Blood-based
biomarkers in lung cancer: Prognosis and treatment decisions.
Transl Lung Cancer Res. 6:708–712. 2017.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Sun X, Wang M, Xu R, Zhang D, Liu A, Wang
Y, Lu T, Xin Y, Zhao Y, Xuan Y, et al: Prognostic model based on
circular RNA circPDK1 for resected lung squamous cell carcinoma.
Transl Lung Cancer Res. 8:907–919. 2019.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Hu H, Sun Z, Li Y, Zhang Y, Li H, Zhang Y,
Pan Y, Shen L, Wang R, Sun Y and Chen H: The histologic
classifications of lung adenocarcinomas are discriminable by unique
lineage backgrounds. J Thorac Oncol. 11:2161–2172. 2016.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Nakajima N, Yoshizawa A, Nakajima T,
Hirata M, Furuhata A, Sumiyoshi S, Rokutan-Kurata M, Sonobe M,
Menju T, Miyamoto E, et al: GATA6-positive lung adenocarcinomas are
associated with invasive mucinous adenocarcinoma morphology,
hepatocyte nuclear factor 4α expression, and KRAS mutations.
Histopathology. 73:38–48. 2018.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Masaoutis C, Mihailidou C, Tsourouflis G
and Theocharis S: Exosomes in lung cancer diagnosis and treatment.
From the translating research into future clinical practice.
Biochimie. 151:27–36. 2018.PubMed/NCBI View Article : Google Scholar
|