Introduction
The genus Rhus is widely distributed in
tropical, subtropical and temperate regions, and several
Rhus spp. are used for nutritional and medicinal purposes
(1). Of these, R. glabra is
used for bacterial infection (2),
whereas R. coriaria is used for wound healing (3). In addition, R. verniciflua has
been demonstrated to possess strong antioxidant properties,
attributed to its bioactive flavonoids, including quercetin,
butein, fustin and sulfuretin (4,5).
R. tripartita (Ucria) Grande, mainly distributed in North
Africa and the Arabian Peninsula, is traditionally used for
inflammatory, cardiovascular and gastrointestinal conditions
(6-8).
Further phytochemical and pharmacological studies of Rhus
spp., including R. tripartita, have identified a variety of
bioactive flavonoids, bioflavonoids and proanthocyanidins (1,9-11).
Recently, a novel catechin along with
epicatechin-3-O-rhamnoside from R. tripartita have
been isolated (12).
Hepatitis B virus (HBV)-induced chronic liver
diseases such as fulminant hepatitis, cirrhosis and carcinoma
account for substantial morbidity and mortality (13). While several efficacious
HBV-polymerase inhibitors (e.g., lamivudine, adefovir and
acyclovir) are available, their long-term use frequently produces
drug-resistant viral strains (14). To counter this issue, a range of
natural bioactive flavonoids, polyphenols, alkaloids, terpenes,
lignans and anthraquinones have been reported as potential anti-HBV
agents with no sign of resistance (15-21).
In line with these studies, R. coriaria has been
demonstrated to have marked anti-HBV activity via inhibition of HBV
surface or 's' antigen (HBsAg) secretion in cultured hepatoma cells
(22). Recently, robustaflavone
from R. succedanea has been reported as a potential
inhibitor of HBV replication in HepG2.2.15 cells (23). However, to the best of our
knowledge, the antiviral potential of Rhus tripartita or its
bioactive constituents has so far remained elusive. Therefore, the
present study assessed the anti-HBV efficacies of the R.
tripartita-derived new catechin and
epicatechin-3-O-rhamnoside, using in vitro as well as
in silico approaches.
Materials and methods
Plant collection, extraction and
isolation
R. tripartita, locally known as ‘Sumac’, was
collected from Hail, Saudi Arabia and authenticated (voucher
specimen no. SY 202/2013) by a plant taxonomist at College of
Pharmacy, King Saud University (Riyadh, Saudi Arabia). The
air-dried extract (80% ethanol) of the stem bark was further
subjected to fractionation with ethyl acetate and sub-fractionated
using column chromatography and thin-layer chromatography to yield
two yellow-amorphous, powdery compounds as described previously
(12). Subsequently,
high-resolution electrospray ionization mass spectrometry
(HRESIMS), ultraviolet (UV) and infrared (IR) spectroscopy,
1H and 13C DEPT-135 NMR spectroscopy, as well
as 2D 1H and 13C heteronuclear single quantum
correlation (HSQ)C analyses were performed to determine their
structures (12).
Cell culture and drugs
The human hepatoblastoma cell line HepG2 (ATCC
HB-065) and its HBV-reporter derivative HepG2.2.15 (SCC249, Merck)
were kind gift of Dr Shahid Jameel, Virology Group, International
Center for Genetic Engineering & Biotechnology. Cells were
cultured in RPMI-1640 medium (Gibco; Thermo Fisher Scientific Inc.)
containing 10% fetal calf serum (Gibco; Thermo Fisher Scientific,
Inc.) and 1X penicillin-streptomycin mix (HyClone; Cytiva) at 37˚C
with 5% CO2. HepG2.2.15 cells are HBV-infected HepG2
cells developed by stable transfection of HBV genomic DNA, which
expresses all viral genes and proteins [e.g. HBsAg and HBV pre-core
or ‘e’ antigen (HBeAg)], and are globally used to assess anti-HBV
agents (24). The cells were
further seeded (0.5x105 cells/100 µl/well) in a 96-well
plate (Corning, Inc.) and incubated overnight prior to an assay.
The approved HBV polymerase-inhibitor, lamivudine triphosphate
(LAM; MilliporeSigma) and the anti-HBV flavonoid quercetin (QRC;
MilliporeSigma) were used as a standard in cell culture studies as
described elsewhere (20).
Cytotoxicity assay
The effect of R. tripartita-derived compounds
(catechin and epicatechin) on HepG2 cells viability or toxicity was
assessed using a TACS MTT Cell Proliferation Assay Kit (Bio-Techne)
and the optimal safe doses were estimated. In brief, each compound
was first dissolved in DMSO and then reconstituted in culture media
to obtain four test concentrations or doses (10, 20, 30, 60 and 120
µM). HepG2 cells grown in a 96-well plate were replenished with
fresh media containing the different drug doses or vehicle control
(0.1% DMSO) and then incubated at 37˚C for 72 h. All samples were
tested in triplicate and the experiment was repeated twice. The
optical density of the samples at 570 nm was recorded using a
microplate reader (ELx800; BioTek Instruments, Inc.) and non-linear
regression analysis was performed (Excel software 2010; Microsoft
Corp.) to determine the 50% inhibitory concentration.
HBsAg inhibition assay
First, dose-dependent inhibition of HBsAg expression
by the two test compounds (10, 20 and 30 µM each) was performed to
determine the optimal active concentration. HepG2.2.15 cells grown
in a 96-well plate were replenished with fresh media containing
three selected doses of the compounds as well as controls, and
incubated for 3 days. Following the determination of the optimal
dose, a time-course inhibition of HBsAg expression by the two
compounds was performed. Likewise, HepG2.2.15 cells were
replenished with fresh media containing the optimal dose (30 µM
each) of the compounds as well as controls, and incubated for up to
5 days. The culture supernatants were collected on days 1, 3 and 5
for analysis. The secretion of HBsAg into the culture supernatant
was quantitatively analyzed using the diagnostic ELISA kit (cat.
no. 72348; MonolisaHBsAg ULTRA; Bio-Rad Laboratories Inc.) as per
the manufacturer's protocol. The optical density of the samples at
450 nm was recorded using a microplate reader (ELx800; BioTek
Instruments, Inc.), and analyzed in relation to the untreated
control in Excel. All samples were tested in triplicate and the
experiment was repeated twice.
HBeAg inhibition assay
The test compounds (30 µM) were evaluated for their
time-course inhibitory effects on HBeAg synthesis. The
post-treatment HepG2.2.15 supernatants collected on days 1, 3 and 5
were quantitatively analyzed for HBeAg expression using the
diagnostic ELISA kit (cat. no. KAPG4BNE3; HBeAg/Anti-HBe Elisa Kit;
DIAsource ImmunoAssays® S.A.) according to the
manufacturer's protocol. The optical density of the samples at 450
nm was recorded using a microplate reader (ELx800; BioTek
Instruments, Inc.), and analyzed in relation to the untreated
control in Excel. All samples were tested in triplicate and the
experiment was repeated twice.
Molecular docking analysis
For molecular docking analysis, an in-house
constructed 3D structure of HBV polymerase (HBVpol) enzyme was
used, as described in a previous study by our group (20). The 3D structures of LAM and
co-crystalized entecavir triphosphate (ETV) were retrieved from the
PubChem database (https://pubchem.ncbi.nlm.nih.gov/) and used as ligand
controls. The 2D structures of the test compounds (ligands) were
drawn in ChemDraw Pro 8.0 (chemistrydocs.com/chemdraw-pro-8-0/), following
assignments of bond orders and bond angles. The structures of
target protein (NCBI GenBank: AGA95798.1) was prepared and
optimized using Maestro v12.3 LigPrep module (25), whereas the 2D and 3D visualizations
of the ligand-target interactions were generated using University
of California San Francisco ChimeraX (26). Prior to the docking of test
compounds, any water molecules or bound hetero atoms were removed
from the target. Further, the Gasteiger partial charges were
defined and energies were minimized for all ligands in the
Universal Force Field program (27). The docking analysis was performed
on the target's binding sites in AutoDock Vina 1.2 operated in
Linux OS (28). The docking
protocol was validated by re-docking the co-crystallized ligands
into the binding site and visual inspection and the Root Mean
Square Deviation (RMSD) was calculated.
Statistical analysis
Data analysis was performed using the SPSS
statistical package, version 17.0 (SPSS, Inc.). Data of all
triplicate samples, expressed as the mean ± standard error of the
mean were analyzed by one-way ANOVA, followed by Dunnett's-test.
P<0.05 was considered to indicate a statistically significant
difference.
Results and Discussion
Identification of isolated
compounds
Of the several known anti-HBV natural flavonoids,
including R. succedanea-derived robustaflavone, the flavonol
catechin and derivatives have been also reported for antiviral
activities against HBV (23,29).
In line with this, the HRESIMS, UV/IR and
1H-13C NMR spectroscopy, as well as 2D
1H-13C HSQC analyses of R.
tripartita-derived compounds led to their identification as
3,5,13,14-flavantetrol-(4β→8)-catechin (rhuspartin; RPT) and
epicatechin-3-O-rhamnoside (ECR) (Fig. 1, upper panel), as described
elsewhere (12).
Hepatocyte proliferative activities of
the catechin and epicatechin
Prior to their anti-HBV assessments, RPT and ECR
when tested on HepG2 cells and did not exhibit any cytotoxicity
even at the maximal dose (120 µM). Of note, while they had marginal
growth stimulatory activities at 30 µM, they exhibited significant
but comparable growth enhancement at 60 and 120 µM doses as
compared to the untreated cells (P<0.01; Fig. 1, lower panel) on day 3. Therefore,
further anti-HBV assays of RPT and ECR were conducted at 20 and 30
µM doses, and not continued for longer than 5 days due to cell
overgrowth and death.
Dose- and time-dependent inhibition of
HBV antigen synthesis by RPT and ECR
RPT and ECR exhibited a dose-dependent anti-HBV
activity, where the 20 and 30 µM doses led to maximal activities as
compared to the untreated cells (P<0.01; Fig. 2). As no significant differential
activity was observed between the two doses, 30 µM was selected as
the optimal dose for the further analyses. In the time-dependent
analysis of HBsAg, ECR and RPT suppressed its expression by 71.3%
(P<0.01) and 68.8% (P<0.01), respectively whereas QRC
inhibited it by 76.4% (P<0.001) as compared to the reference
drug LAM on day 5 (Fig. 3).
Furthermore, in the time-dependent analysis of HBeAg, ECR and RPT
maximally downregulated its synthesis by 71.2% (P<0.01) and
62.3% (P<0.01), respectively, whereas QRC suppressed it by 75.4%
(P<0.001) as compared to LAM on day 5 (Fig. 4). Thus, ECR had a marginally higher
anti-HBV activity than RPT. As RPT and ECR exerted growth
stimulatory effects on HepG2 cells at 60 µM or above on day 3
(Fig. 1, lower panel), a single
optimal dose of 30 µM was used in the antiviral assays performed
for up to 5 days. As compared to LAM, incubating HepG2.2.15 cells
with RPT and ECR at 30 µM for longer than 5 days also enhanced cell
growth (data not shown). In view of this, it was not possible to
compare the anti-HBV effects of RPT and ECR with those of LAM due
to differences in cell population and the amount of secreted HBV
antigens, and incubation was therefore terminated a day 5. In
previous studies, the antiviral activities of catechin derivatives
such as epicatechin-3-gallate and epigallocatechin-3-gallate have
been reported against herpes simplex virus, human immunodeficiency
virus, human T-cell leukemia virus, Epstein-Barr virus, influenza
virus, rotavirus and adenovirus as well as HBV and hepatitis C
virus (16,29). In addition, a previous study by our
group indicated moderate anti-HBV activities of Oncocalyx
glabratus-derived catechin, catechin-7-O-gallate,
catechin-7,4'-O-digallate and
catechin-7,3'-O-digallate in HepG2.2.15 cells (30).
 | Figure 2Anti-HBV assay indicating
dose-dependent (10, 20 and 30 µM) inhibition of HBsAg by R.
tripartita-derived catechin RPT and epicatechin ECR in
HepG2.2.15 cells. LAM (2 µM) served as a positive control, while
DMSO (0.1%) was used as a negative control. Values are expressed as
the mean ± standard error of the mean (n=3). **P<0.01
vs. LAM. HBsAg, HBV surface antigen; ECR,
epicatechin-3-O-rhamnoside; RPT, rhuspartin,
3,5,13,14-flavantetrol-catechin; LAM, lamivudine triphosphate. |
 | Figure 3Anti-HBV assay indicating
time-dependent inhibition of HBsAg by R. tripartita-derived
catechin RPT (30 µM) and epicatechin ECR (30 µM) in HepG2.2.15
cells. LAM (2 µM) and QRC (27 µM) served as positive controls,
while DMSO (0.1%) was used as an NC. Values are expressed as the
mean ± standard error of the mean (n=3). ●●P<0.01,
●●●P<0.001 vs. NC; *P<0.05,
**P<0.01, ***P<0.001 vs. LAM. QRC,
quercetin; NC, negative control; HBsAg, HBV surface antigen; ECR,
epicatechin-3-O-rhamnoside; RPT, rhuspartin,
3,5,13,14-flavantetrol-catechin; LAM, lamivudine triphosphate. |
 | Figure 4Anti-HBV assay demonstrating
time-dependent inhibition of HBeAg by R. tripartita-derived
catechin RPT (30 µM) and epicatechin ECR (30 µM) in HepG2.2.15
cells. LAM (2 µM) and QRC (27 µM) served as positive controls,
while DMSO (0.1%) acted as NC. Values are expressed as the mean ±
standard error of the mean (n=3). ●●P<0.01,
●●●P<0.001 vs. BC. *P<0.05,
**P<0.01, ***P<0.001 vs. LAM. HBeAg,
HBV pre-core or ‘e’ antigen; QRC, quercetin; NC, negative control;
ECR, epicatechin-3-O-rhamnoside; RPT, rhuspartin,
3,5,13,14-flavantetrol-catechin; LAM, lamivudine triphosphate. |
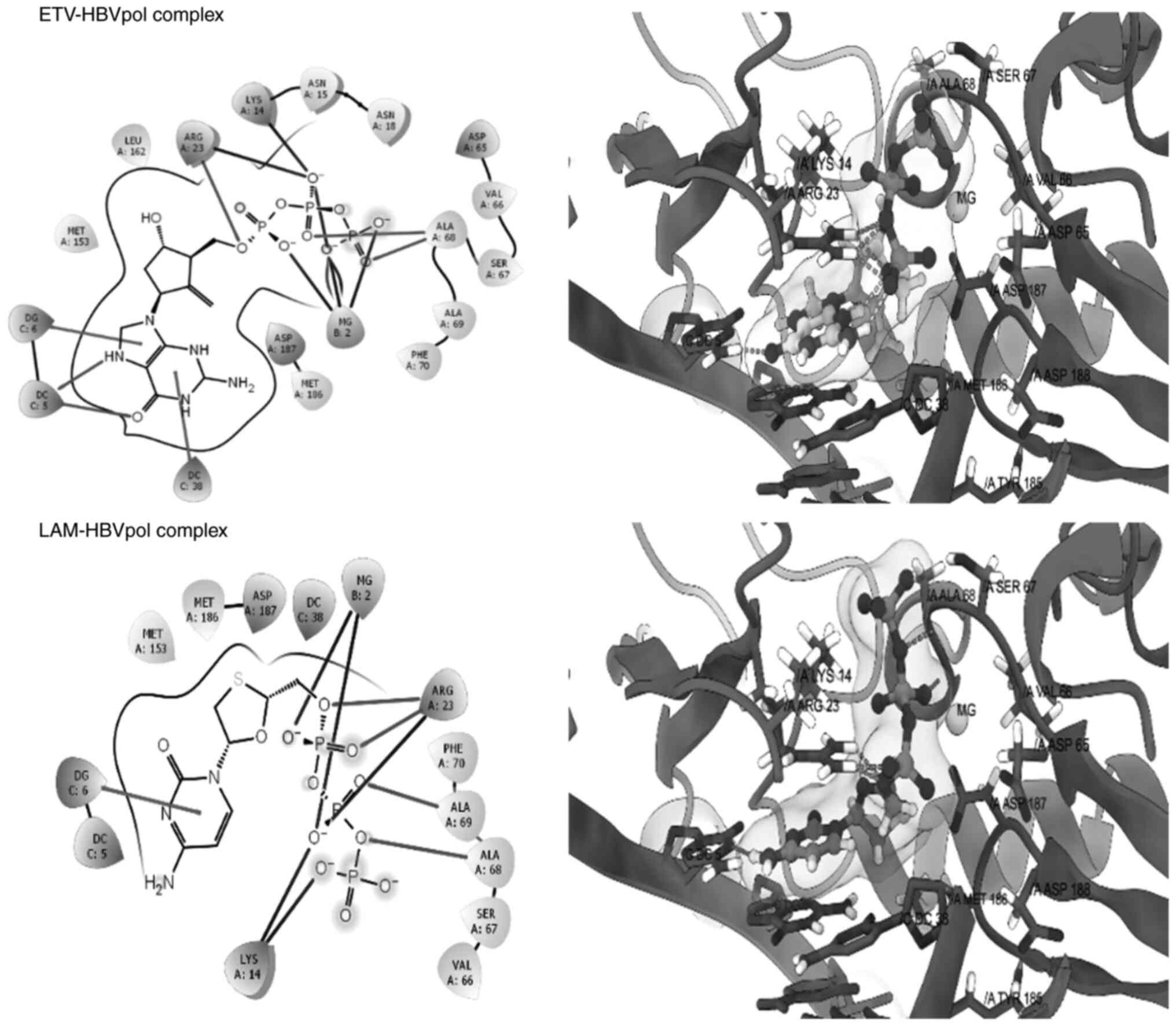
Structure-based virtual interaction of
RPT and EPR with HBVpol
Molecular docking is a widely used in silico
technique in drug discovery projects to predict the best
conformation for a molecule and its potential affinity to a
specific molecular target. As HBVpol enzyme is essential for HBV
DNA replication, this enzyme remains an important drug target.
Therefore, a molecular docking technique was employed to support
the in vitro data towards delineating the possible
mechanisms of anti-HBV activity of the isolated catechin and
epicatechin. The good re-alignment of the co-crystallized ligand
ETV prior to and after docking inside the binding cavity of HBVpol
along with the low RMSD values between the two structures indicated
a valid docking protocol (Fig. 5,
upper panel). Furthermore, the docked complex of LAM-HBVpol was
observed to adopt a conformation quite similar to that of the
ETV-HBVpol complex (Fig. 5, lower
panel), confirming robustness and reliability of the protocol. The
estimated binding free energies for the ETV-HBVpol and LAM-HBVpol
complex were -8.21 and -9.24 kcal/mol, respectively (Table I). Furthermore, RPT and ECR
exhibited affinity for the HBVpol in proximity of its
‘Tyr-Met-Asp-Asp (YMDD)’ motif, similar to LAM (Fig. 6). In addition to the ‘YMDD’ motif,
Lys14, Ser67 and Ala68 also surrounded the ligand compounds, which
may potentially contribute to ligand-target complex stabilities. In
addition, both compounds adopted relatively similar orientations
and aligned inside the active site of HBVpol as compared to the
control ligands. ECR engaged in interactions with the key residues
of HBVpol, including a hydrogen bonding with a charged Arg23
residue, a face-to-face π-stacking with the nucleotides of the DNA
and a metal coordination with an Mg+2 ion embedded
inside the pocket (Fig. 6). It is
thought that coordination with Mg+2 ions has an integral
part in stabilizing a ligand-target complex (31). By contrast, the only key
interaction of RPT was its metal coordination by Mg+2.
However, ECR exhibited a better binding affinity as compared to
RPT, which may be attributed to the hydrophobic contact between the
ligand and the target (Fig. 5).
Though slightly lower than that for LAM, the calculated binding
free energies for ECR and RPT were -8.483 and -7.949 kcal/mol,
respectively (Table I).
 | Table IEstimated docking energies of R.
tripartita-derived compounds and standard drugs against HBV
polymerase. |
Table I
Estimated docking energies of R.
tripartita-derived compounds and standard drugs against HBV
polymerase.
| Compound | Docking energy,
kcal/mol |
|---|
|
Rhuspartina | -7.949 |
|
Epicatechin-3-O-rhamnosidea | -8.483 |
| Lamivudine
triphosphateb | -9.245 |
| Entecavir
triphosphateb | -8.212 |
Flavonoids are plant phenols, which, according to
their variations in their heterocyclic carbon
(C6-C3-C6) ring, have been
classified as flavones, flavonols, flavanones, isoflavones,
anthocyanidins and catechins (32). Of note, the compounds examined in
the present study belong to the subclass of ‘catechins’, where ECR
is a monomeric (epi)catechin conjugated with a sugar moiety i.e.,
glycone, and RPT is a dimeric catechin, i.e., aglycon (Fig. 1, upper panel). In view of the
structure-activity relationship of the compounds, the observed
marginal difference in the bioactivity of ECR and RPT may therefore
be attributed to the differences in their chemical structures.
Furthermore, in the metabolic process, a glycone is generally
poorly absorbed as compared to its aglycone counterpart (32). However, hydrolysis of the glycone
furnishes a free aglycone, which easily gets absorbed and performs
its activity. By contrast, the breakdown of a dimeric aglycone is
relatively difficult and therefore, it has poor absorption and low
activity. Taken together, this information strongly supports the
differential structure-activity of the studied catechin and
epicatechin.
In conclusion, the present data, for the first time,
demonstrated the promising anti-HBV therapeutic potential of the
R. tripartita-derived compounds ECR and RPT in an
HBV-reporter cell culture model, supported by molecular docking
analysis. This warrants their further molecular and pharmacological
study towards developing novel and efficacious natural therapeutics
against HBV infection.
Acknowledgements
Not applicable.
Funding
Funding: The authors gratefully acknowledge the Researchers
Supporting Project (grant no. RSP-2021/379), King Saud University
(Riyadh, Saudi Arabia) for supporting this work.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
MKP and MSAD conceptualized and designed the study,
performed in vitro assays, collected and analyzed data and
wrote the manuscript. MASA performed the in silico analysis
and contributed to manuscript writing. ASA and ARA prepared the
compounds and performed statistical analysis. MKP and MASA confirm
the authenticity of all the raw data. All authors have read and
approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Opiyo SA, Njoroge PW, Ndirangu EG and
Kuria KM: A review of biological activities and phytochemistry of
Rhus Species. Am J Chem. 11:28–36. 2021.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Erichsen-Brown C: Medicinal and Other uses
of North American Plants: A Historical Survey with Special
Reference to the Eastern Indian Tribes. Dover Publications,
Mineola, NY, 1989.
|
|
3
|
Sezik E, Tabata M, Yesilada E, Honda G,
Goto K and Ikeshiro Y: Traditional medicine in Turkey. Folk
medicine in Northeast Anatolia. J Ethnopharmacol. 35:191–196.
1991.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Jang JY, Shin H, Lim JW, Ahn JH, Jo YH,
Lee KY, Hwang BY, Jung SJ, Kang SY and Lee MK: Comparison of
antibacterial activity and phenolic constituents of bark, lignum,
leaves and fruit of Rhus verniciflua. PLoS One.
13(e0200257)2018.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Kang SY, Kang JY and Oh MJ: Antiviral
activities of flavonoids isolated from the bark of Rhus
verniciflua stokes against fish pathogenic viruses in vitro. J
Microbiol. 50:293–300. 2012.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Itidel C, Chokri M, Mohamed B and Yosr Z:
Antioxidant activity, total phenolic and flavonoid content
variation among Tunisian natural populations of Rhus
tripartita (Ucria) Grande and Rhus pentaphylla Desf. Ind
Crops Prod. 51:171–177. 2013.
|
|
7
|
El-Mokasabi F: The state of the art of
traditional herbal medicine in the Eastern mediterranean coastal
region of Libya. Middle East J Sci Res. 21:575–582. 2014.
|
|
8
|
Shahat AA, Alsaid MS, Rafatullah S,
Al-Sohaibani MO, Parvez MK, Al-Dosari MS, Exarchou V and Pieters L:
Treatment with Rhus tripartita extract curtails
isoproterenol-elicited cardiotoxicity and oxidative stress in rats.
BMC Complement Altern Med. 16(351)2016.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Mahjoub MA, Ammar S and Mighri Z: A new
biflavonoid and an isobiflavonoid from Rhus tripartitum. Nat
Prod Res. 19:723–729. 2005.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Alimi H, Mbarki S, Barka ZB, Feriani A,
Bouoni Z, Hfaeidh N, Sakly M, Tebourbi O and Rhouma KB:
Phytochemical, antioxidant and protective effect of Rhus
tripartitum root bark extract against ethanol-induced ulcer in
rats. Gen Physiol Biophys. 32:115–127. 2013.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Mohammed AE-SI: Phytoconstituents and the
study of antioxidant, antimalarial and antimicrobial activities of
Rhus tripartita growing in Egypt. J Pharmacogn Phytochem.
4:276–281. 2015.
|
|
12
|
Alqahtani AS, Abdel-Mageed WM, Shahat AA,
Parvez MK, Al-Dosari MS, Malik A, Abdel-Kader MS and Alsaid MS:
Proanthocyanidins from the stem bark of Rhus tripartita
ameliorate methylgloxal-induced endothelial cell apoptosis. J Food
Drug Anal. 27:758–765. 2019.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Tang LSY, Covert E, Wilson E and Kottilil
S: Chronic hepatitis B infection: A review. JAMA. 319:1802–1813.
2018.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Devi U and Locarnini S: Hepatitis B
antivirals and resistance. Curr Opin Virol. 3:495–500.
2013.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Wang G, Zhang L and Bonkovsky HL: Chinese
medicine for treatment of chronic hepatitis B. Chin J Integr Med.
18:253–255. 2012.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Parvez MK, Arab AH, Al-Dosari MS and
Al-Rehaily AJ: Antiviral natural products against chronic hepatitis
B: Recent developments. Curr Pharm Des. 3:286–293. 2016.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Parvez MK, Tabish Rehman M, Alam P,
Al-Dosari MS, Alqasoumi SI and Alajmi MF: Plant-derived antiviral
drugs as novel hepatitis B virus inhibitors: Cell culture and
molecular docking study. Saudi Pharm J. 27:389–400. 2019.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Parvez MK, Al-Dosari MS, Alam P, Rehman M,
Alajmi MF and Alqahtani AS: The anti-hepatitis B virus therapeutic
potential of anthraquinones derived from Aloe vera. Phytother Res.
33:2960–2970. 2019.PubMed/NCBI View
Article : Google Scholar
|
|
19
|
Parvez MK, Al-Dosari MS, Arbab AH,
Al-Rehaily AJ and Abdelwahid MAS: Bioassay-guided isolation of
anti-hepatitis B virus flavonoid myricetin-3-O-rhamnoside
along with quercetin from Guiera senegalensis leaves. Saudi
Pharm J. 28:550–559. 2020.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Parvez MK, Ahmed S, Al-Dosari MS,
Abdelwahid MAS, Arbab AH, Al-Rehaily AJ and Al-Oqail MM: Novel
Anti-Hepatitis B virus activity of Euphorbia schimperi and
its quercetin and kaempferol derivatives. ACS Omega. 6:29100–29110.
2021.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Parvez MK, Al-Dosari MS, Rehman MT,
Al-Rehaily AJ, Alqahtani AS and Alajmi MF: The anti-hepatitis B
virus and anti-hepatotoxic efficacies of solanopubamine, a rare
alkaloid from Solanum schimperianum. Saudi Pharm J: Feb 7,
2022 (Epub ahead of print). doi: https://doi.org/10.1016/j.jsps.2022.02.001.
|
|
22
|
Amin FG, Farzaneh S, Mohsen K, Mohammad K,
Hessam M, Dawood MNS and Ali AN: Effects of Rhus Coriaria L.
(Sumac) extract on hepatitis B virus replication and HBs Ag
secretion. J Rep Pharm Sci. 7:100–107. 2018.
|
|
23
|
Zembower DE, Lin YM, Flavin MT, Chen FC
and Korba BE: Robustaflavone, a potential non-nucleoside
anti-hepatitis B agent. Antiviral Res. 39:81–88. 1998.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Sells MA, Chen ML and Acs G: Production of
hepatitis B virus particles in Hep G2 cells transfected with cloned
hepatitis B virus DNA. Proc Natl Acad Sci USA. 84:1005–1009.
1987.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Schrödinger Release 2022-1: Maestro,
Schrödinger, LLC, New York, NY, 2021.
|
|
26
|
Goddard TD, Huang CC, Meng EC, Pettersen
EF, Couch GS, Morris JH and Ferrin TE: UCSF ChimeraX: Meeting
modern challenges in visualization and analysis. Protein Sci.
27:14–25. 2018.PubMed/NCBI View
Article : Google Scholar
|
|
27
|
Eberhardt J, Santos-Martins ED, Tillack AF
and Forli F: AutoDock Vina 1.2.0: New docking methods, expanded
force field, and python bindings. J Chem Inf Model. 61:3891–3898.
2021.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Trott O and Olson AJ: AutoDock Vina:
Improving the speed and accuracy of docking with a new scoring
function, efficient optimization, and multithreading. J Comput
Chem. 31:455–461. 2010.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Badshah SL, Faisal S, Muhammad A, Poulson
BG, Emwas AH and Jaremko M: Antiviral activities of flavonoids.
Biomed Pharmacother. 40(111596)2021.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Ahmed S, Al-Rehaily AJ, Ahmad MS, Yousaf
M, Alam MN, Parvez MK, Al-Dosari MS, Noman OM, Khan SI and Khan IA:
Chemical constituents from Oncocalyx glabratus and their
bioactivities. Phytochemistry Lett. 20:128–132. 2017.
|
|
31
|
Bertoletti N, Chan AH, Schinazi RF, Yin YW
and Anderson KS: Structural insights into the recognition of
nucleoside reverse transcriptase inhibitors by HIV-1 reverse
transcriptase: First crystal structures with reverse transcriptase
and the active triphosphate forms of lamivudine and entecavir.
Protein Sci. 28:1664–1675. 2019.PubMed/NCBI View
Article : Google Scholar
|
|
32
|
Hollman PCH: Absorption, bioavailability
and metabolism of flavonoids. Pharm Biol. 42:74–83. 2004.
|