Introduction
Sepsis is caused by a detrimental inflammatory
response to infection, and is associated with a high mortality rate
of >40% (1). In particular,
acute kidney injury (AKI) caused by sepsis accounts for ~50% of all
AKI cases (2). A previous study
revealed that various factors are related to sepsis-induced AKI,
including non-coding RNAs (3).
Whole-genome transcriptome sequencing analysis
showed that encoded protein genes account for ~2% of human
transcripts, while other transcripts are mostly non-protein-coding
genes (4,5). Previous studies have focused on the
important role of long non-coding RNAs (lncRNAs) in molecular
function (6), and >50,000 human
lncRNAs have been identified (7).
The transcript of the human gene lncRNA plasmacytoma
variant translocation gene 1 (lncRNA PVT1) is located in chromosome
8q24, which is widely recognized as a cancer-associated region
(8). PVT1 is involved in human
diseases, and plays a crucial role in the development and
progression of various cancer types, such as gallbladder cancer
(9), leukemia (10), hepatocellular cancer (11-13),
breast cancer (14) and ovarian
cancer (15). In addition, it has
been revealed that PVT1 plays a key role in a variety of
pathological conditions, including sepsis (16-18).
lncRNA PVT1 is upregulated in tissues of septic
models, and regulates sepsis-induced heart inflammation and cardiac
function by interacting with microRNA (miRNA/miR)-143 and the
mitogen-activated protein kinase (MAPK)/NF-κB pathway (17). Furthermore, PVT1 aggravated
lipopolysaccharide (LPS) induced myocardial injury via
miR-29a/HMGB1 axis (18). PVT1 also
promotes cell proliferation, invasion and epithelial-mesenchymal
transition by downregulating miR-16-5p in renal cell carcinoma
in vitro (19). Moreover, it
has been shown that PVT1 promotes the cell cycle and apoptosis of
cervical cancer cells by activating the NF-κB pathway (20). lncRNA PVT1 downregulation is also
involved in the increased rate of apoptosis and reduced
proliferation of acute lymphoblastic leukemia cells (21). Therefore, these studies suggest that
lncRNA PVT1 generally acts as an oncogene in a variety of cancer
types.
miRNAs are widely studied due to their essential
role in development, tissue-specific expression and close
relationship to human diseases (22). miR-27a is abnormally upregulated in
several types of cancer and has been identified as an oncogene
during tumorigenesis (23-26).
Furthermore, miR-27a-3p has been reported to participate in the
pathological processes of various diseases, including the immune
response and inflammatory response (27-29).
Among these disease types, miR-27a-3p is downregulated in sepsis
(30). It has also been shown that
miR-27a-3p is associated with inflammation and exerts an
anti-inflammatory effect in sepsis (30). However, the underlying mechanisms
remain unknown.
Therefore, the aims of the present study were to
investigate the regulation of lncRNA PVT1 and miR-27a-3p in sepsis
of AKI, and to identify the downstream mechanisms. Thus, the
present results may facilitate the development of novel treatments
for sepsis and related inflammation.
Materials and methods
Cell culture and transfection
The HK-2 cell line was purchased from the American
Type Culture Collection, and cultured in DMEM (Gibco; Thermo Fisher
Scientific, Inc.) with 10% FBS (Gibco; Thermo Fisher Scientific,
Inc.), 100 IU/ml penicillin and 100 mg/ml streptomycin (Beijing
Solarbio Science & Technology Co., Ltd.). Cells were grown and
maintained at 37˚C in a 5% CO2 humidified incubator. At
~60% confluence, HK-2 cells were treated with different
concentrations (0, 0.1, 2, 5 and 10 µg/ml) of LPS (Sigma-Aldrich;
Merck KGaA) at 37˚C for 8 h, and then 5 µg/ml was selected to
generate a sepsis cell model based on previously reported research
(16).
miR-27a-3p mimic (miR-27a-3p), oxidative stress
responsive kinase 1 (OXSR1) overexpression vector (OXSR1), PVT1
overexpression vector (pcDNA-PVT1) and their negative controls (NC
mimic or empty pcDNA3.1 vector) were purchased from Shanghai
GenePhama Co., Ltd. Small interfering (si)-PVT1, anti-miR-27a-3p
and their NCs (si-NC and anti-NC) were also purchased from Shanghai
GenePhama Co., Ltd. The sequences of siRNA, miR mimic, inhibitor
and NC were as follows: si-PVT1, 5'-GCUUGGAGGCUGAGG AGUUTT-3';
miR-27a-3p mimic, 5'-UUCACAGUGGCUAA GUUCCGC-3'; miR-27a-3p
inhibitor, 5'-GCGGAACTTAG CCACTGTGAA-3'; si-NC,
5'-UUCUCCGAACGUGUCA-3'; miRNA mimic NC, 5'-UUUGUACUACACAAAAGU
ACUG-3'; and miRNA inhibitor NC, 5'-CAGUACUUUUGUG UAGUACAAA-3'.
Cell were seeded in 6-well plates and cultured to 70% confluence.
siRNAs (50 nM), miR mimics (50 nM), miR inhibitors (100 nM) and
plasmids (5 µg) were transfected into HK-2 cells using
Lipofectamine® 3000 (Thermo Fisher Scientific, Inc.).
After transfection for 48 h, the cells were collected for further
experiments.
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA was extracted from HK-2 cells using
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc.). cDNAs of lncRNA and mRNA were generated using M-MLV reverse
transcriptase (Invitrogen; Thermo Fisher Scientific, Inc.) at 42˚C
for 1 h. A TaqMan miRNA RT kit (Thermo Fisher Scientific, Inc.) was
used for the RT-qPCR of miR-27a-3p (RT was performed at 16˚C for 30
min, 42˚C for 30 min and 85˚C for 5 min; qPCR was performed as
follows: initial denaturation at 95˚C for 10 min; hold at 55˚C for
2 min and 72˚C for 2 min; followed by 12 cycles of 95˚C for 15 sec
and 60˚C for 4 min). TaqMan Universal Master mix II (Thermo Fisher
Scientific, Inc.) was used for the qPCR of lncRNA and mRNA (initial
denaturation at 95˚C for 10 min followed by 40 cycles of 95˚C for
15 sec and 60˚C for 1 min). U6 and 18s rRNA were used for
normalization. All data were calculated with the 2-ΔΔCq
method (31). The following primer
sequences were used: PVT1 forward, 5'-TGA GAACTGTCCTTACGTGACC-3'
and reverse, 5'-AGAGCACC AAGACTGGCTCT-3'; miR-27a-3p forward,
5'-GCGCGTTCA CAGTGGCTAAG-3' and reverse, 5'-AGTGCAGGGTCCGAG
GTATT-3'; 18S rRNA forward, 5'-GGCCCTGTAATTGGAAT GAGTC-3' and
reverse, 5'-CCAAGATCCAACTACGA GCTT-3'; and U6 forward,
5'-CTCGCTTCGGCAGCACA-3' and reverse, 5'-AACGCTTCACGAATT
TGCGT-3'.
ELISA
The levels of TNF-α and IL-6 were measured by ELISA
using the cell culture medium. The cell culture medium was
centrifuged at 1,000 x g for 10 min at 4˚C, and the supernatant was
collected. Commercially available ELISA kits for TNF-α and IL-6
(cat. nos. 550610 and 550799, respectively; BD Biosciences) were
used according to the manufacturer's instructions. Results were
read at an optical density of 450 nm using a Spectra Max Plus plate
reader (Molecular Devices LLC). Measurements were performed in
triplicate, and P-values were computed using two-tailed Student's
t-tests.
Cell Counting Kit-8 (CCK-8) assay
HK-2 cells were seeded into 96-well plates at a
density of 10,000 cells/well. After 12 h of incubation at 37˚C,
cells were transfected for 24 h in the presence of 0-10 µg/ml LPS
and then washed three times with PBS. Then, 10 µl/well CCK-8
solution (Sigma-Aldrich; Merck KGaA) was pipetted into each well,
and the plate was incubated for 1.5 h at 37˚C. Absorbance was
measured at 450 nm using a microplate reader (Model 550; Bio-Rad
Laboratories, Inc.).
Cell apoptosis
HK-2 were seeded in 96-well plates at a density of
10,000 cells/well for 12 h. The cells were transfected as described
above. Then, the cells were prepared into single-cell suspension
and double-stained with 5 µl FITC-Annexin V and 5 µl propidium
iodide at room temperature for 15 min in the dark using the
FITC-Annexin V apoptosis detection kit (BD Biosciences). The
staining was followed by analysis using a flow cytometer (FACScan;
BD Biosciences) equipped with Cell Quest software (version 5.1; BD
Biosciences). The apoptosis rate was calculated as the percentage
of early + late apoptotic cells.
Western blotting
Total protein was extracted by RIPA buffer (Cell
Signaling Technology, Inc.). The concentration of protein was
measured with a bicinchoninic acid (BCA) protein assay kit (Bio-Rad
Laboratories, Inc.). Equal quantities of proteins (20 µg/lane) were
separated on 10% SDS-PAGE gels and transferred to PVDF membranes.
The membranes were blocked with 5% non-fat milk in 1X Tris-buffered
saline and 0.1% Tween-20 at room temperature for 1 h. Membranes
were then incubated overnight at 4˚C with primary antibodies for
Bcl-2 (1:1,000; cat. no. ab59348; Abcam), Bax (1:1,000; cat. no.
ab32503; Abcam), cleaved caspase-3 (C-caspase-3; 1:1,000; cat. no.
ab49822; Abcam), OXSR1 (1:1,000; cat. no. ab97694; Abcam),
phosphorylated (p)-inhibitor of κB (IκBα; 1:1,000; cat. no. 2859;
Cell Signaling Technology), total-IκBα (1:1,000; cat. no. 4812;
Cell Signaling Technology) p-p65 (1:1,000; cat. no. 3033; Cell
Signaling Technology), total-p65 (1:1,000; cat. no. 8242; Cell
Signaling Technology) and GAPDH (1:5,000; cat. no. ab9485; Abcam).
The membranes were then probed with appropriate secondary
antibodies at room temperature for 2 h [horseradish
peroxidase-conjugated goat anti-rabbit IgG H&L (1:5,000; cat.
no. ab205718; Abcam)]. The target protein levels were detected
using enhanced chemiluminescence reagents (Pierce; Thermo Fisher
Scientific, Inc.) and analyzed with Image J (version 1.8.0;
National Institutes of Health).
Dual-luciferase reporter assay
The putative binding sites between miR-27a-3p and
PVT1 or OXSR1 were predicted by StarBase 2.0 (http://starbase.sysu.edu.cn/starbase2/)
and TargetScanHuman 7.2 (http://www.targetscan.org/), respectively. The
fragments from the 3' untranslated regions (3'UTR) of PVT1 or
OXSR1, containing the predicted miR-27a-3p binding site, were
synthesized and cloned into the XhoI and NotI sites
of a psiCHECK-2 vector (Promega Corporation) to form the reporter
vectors PVT1-wild-type (wt) or OXSR1-wt. The corresponding mutants
(mut) were constructed by mutating the miR-27a-3p seed region
binding site, and were referred to as the reporter vectors PVT1-mut
or OXSR1-mut. Then, miR-27a-3p mimic or NC mimic (50 nM) were
co-transfected with the reporter vectors (5 µg) containing either
the targeting sequences or the corresponding mutants using
Nanofectin transfection reagent (Shanghai ExCell Biology, Inc.).
After transfection for 48 h, luciferase activity was determined
using a Dual-Luciferase Assay Kit (GeneCopoeia) and detected by
multimode detector (Beckman Coulter, Inc.). Firefly luciferase
activity was used for normalization.
RNA immunoprecipitation (RIP)
assay
RIP was performed using an EZ-Magna RIP RNA-Binding
protein immunoprecipitation kit (EMD Millipore). Cells stably
transfected with NC or miR-27a-3p were lysed by RIP lysis buffer,
and 100 µl cell lysates was incubated with RIP buffer containing
magnetic beads conjugated with human anti-argonaute 2 antibody
(1:500; cat. no. 04-642; EMD Millipore) or normal mouse IgG (1:500;
cat. no. 12-371; EMD Millipore), which was the negative control, at
4˚C overnight. Then, proteinase K buffer was used to digest the
protein from samples and the immunoprecipitated RNA was extracted
using TRIzol reagent (Invitrogen). Purified RNA was analyzed by
RT-qPCR to detect the expression levels of lncRNA PVT1 and OXSR1 in
the precipitates as aforementioned. The primer sequences for OXSR1
were 5'-AAAGACGTTTGTTGGCACCC-3' (forward) and 5'-GCC
CCTGTGGCTAGTTCAAT-3' (reverse).
RNA pull-down assay
Probe miR-27a-3p (50 nM) was biotinylated by a
biotinylation kit (Thermo Fisher Scientific, Inc.) and transfected
into HK-2 cells with Lipofectamine 3000. After transfection for 48
h, cells were lysed and incubated with Dynabeads M-280 Streptavidin
(10 mg/ml; Invitrogen; Thermo Fisher Scientific, Inc.) at 25˚C for
1 h. The complexes were isolated using streptavidin agarose beads
(Invitrogen; Thermo Fisher Scientific, Inc.). The bound RNAs were
detected using RT-qPCR as aforementioned.
Statistical analysis
All experiments were performed in triplicate. Data
are presented as the mean ± SD. Results were analyzed using
two-tailed Student's t-test for two groups, and one-way ANOVA for
multiple groups followed by Tukey's test. All statistical analyses
were performed using SPSS 19.0 (SPSS, Inc.). P<0.05 was
considered to indicate a statistically significant difference.
Results
lncRNA PVT1 expression is upregulated,
while miR-27a-3p is downregulated in LPS-induced HK-2 cells
To investigate the roles of lncRNA PVT1 and
miR-27a-3p in septic AKI progression in vitro, HK-2 cells
were selected for septic AKI model construction by LPS stimulation.
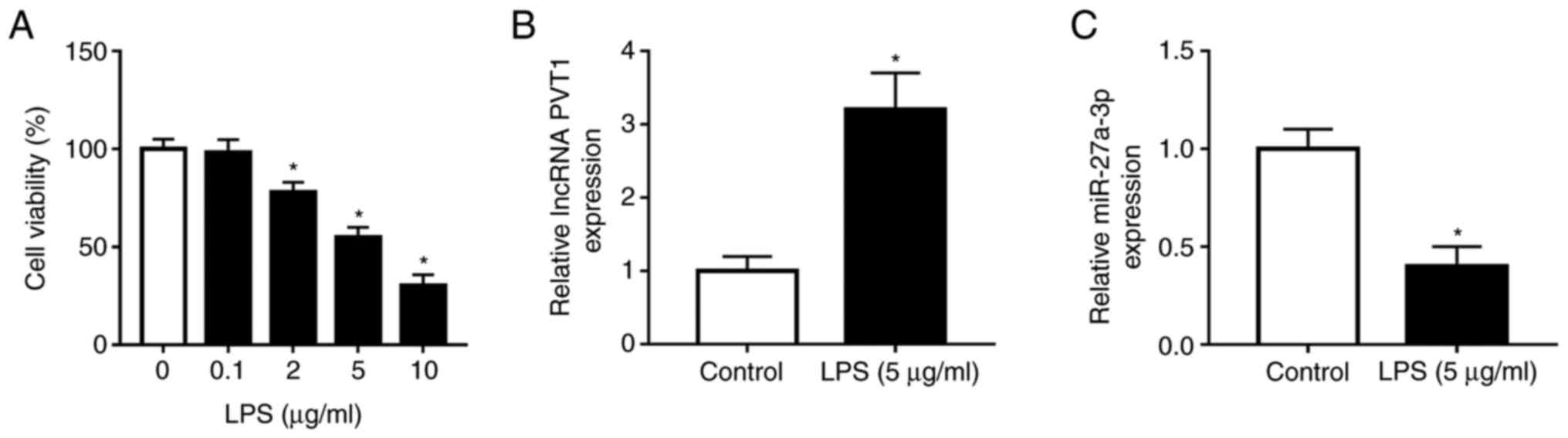
To detect cell viability after LPS treatment, cells were incubated
with LPS at various concentrations (0, 0.1, 2, 5 and 10 µg/ml) for
8 h, and it was found that low cell viability was negatively
associated with enhanced concentrations of LPS (Fig. 1A). Based on the significant
difference with the control group (0 µg/ml), 5 µg/ml LPS
administration was selected for further experiments. Moreover, the
mRNA expression levels of PVT1 and miR-27a-3p were examined by
RT-qPCR, and it was demonstrated that PVT1 was significantly
increased in LPS-treated cells, while the opposite was observed for
miR-27a-3p (Fig. 1B and C). Therefore, the present results
indicated that LPS-induced HK-2 cells exhibited enhanced expression
of PVT1 and downregulated expression of miR-27a-3p.
PVT1 knockdown decreases inflammatory
cytokines secretion, promotes cell survival and inhibits apoptosis
in LPS-treated HK-2 cells
To further investigate the effect of lncRNA PVT1 on
LPS-treated HK-2 cells, loss-of-function assays were performed. The
transfection efficiency of si-PVT1 was detected, and it was found
that this transfection significantly suppressed PVT1 expression
(Fig. S1A). Moreover, it was
identified that si-PVT1 significantly blocked the promotive effect
of LPS on PVT1 expression compared with the LPS+scramble group
(Fig. 2A). Subsequently, the levels
of the inflammatory cytokines TNF-α and IL-6 were detected. After
silencing PVT1 expression, the secretion levels of these immune
factors were significantly reduced compared with the LPS+scramble
group (Fig. 2B and C). Furthermore, it was found that cell
viability was significantly increased after PVT1 knockdown
(Fig. 2D). Then, flow cytometry was
used to assess the apoptotic rate, and it was identified that the
number of apoptotic cells decreased significantly after PVT1
silencing (Fig. 2E). Moreover, the
expression of the apoptosis inhibitor Bcl-2 was significantly
increased, while the protein expression levels of the apoptotic
factors Bax and C-caspase-3 were significantly decreased in the
LPS+si-PVT1 group (Fig. 2F).
Collectively, the present results suggested that knockdown of
lncRNA PVT1 in LPS-induced HK-2 cells decreased TNF-α and IL-6
secretion, promoted cell activity and inhibited apoptosis.
 | Figure 2PVT1 knockdown alleviates
inflammation, promotes cell survival and inhibits apoptosis. (A)
Relative expression of lncRNA PVT1 in control, LPS, LPS+scramble
and LPS+si-PVT1 groups were detected in HK-2 cells by reverse
transcription-quantitative PCR. Levels of the inflammatory
cytokines (B) TNF-α and (C) IL-6 were measured by ELISA. (D) Cell
viability and (E) apoptotic rates of control, LPS, LPS+scramble and
LPS+si-PVT1 groups were assessed with Cell Counting Kit-8 assay and
flow cytometry, respectively. (F) Protein expression levels of
Bcl-2, Bax and C-caspase 3 was measured in each group using western
blotting. *P<0.05. C-, cleaved; LPS,
lipopolysaccharide; PVT1, plasmacytoma variant translocation gene
1; lncRNA, long non-coding RNA; miR, microRNA; si, small
interfering RNA; TNF-α, tumor necrosis factor-α; IL, interleukin;
PI, propidium iodide |
miR-27a-3p overexpression decreases
inflammatory cytokines secretion, promotes cell survival and
inhibits apoptosis in HK-2 cells after LPS treatment
Next, the present study investigated the role of
miR-27a-3p in LPS-induced HK-2 cells. It was demonstrated that
miR-27a-3p overexpression was successful after transfection with
miR-27a-3p mimic (Fig. S1C). When
LPS-induced cells were overexpressed with miR-27a-3p, it was
identified that miR-27a-3p expression was significantly increased
(Fig. 3A). Moreover, the secretion
of TNF-α and IL-6 were significantly decreased after miR-27a-3p
mimic transfection (Fig. 3B and
C). It was also demonstrated that
miR-27a-3 overexpression increased cell viability (Fig. 3D), and the number of apoptotic cells
was significantly decreased (Fig.
3E). Furthermore, after miR-27a-3p overexpression, the
expression of the apoptosis inhibitor Bcl-2 was promoted, while the
expression levels of the apoptotic pathway proteins Bax and
C-caspase-3 were significantly decreased (Fig. 3F). Thus, the present results
indicated that overexpression of miR-27a-3p in LPS-induced HK-2
cells reduced inflammation, promoted cell activity and inhibited
apoptosis, which was similar to PVT1 knockdown.
 | Figure 3Effect of miR-27a-3p overexpression
is similar to that of PVT1 knockdown in HK-2 cells. (A) Relative
expression of miR-27a-3p in control, LPS, LPS+NC and LPS+miR-27a-3p
groups was detected by reverse transcription-quantitative PCR.
Secretion levels of (B) TNF-α and (C) IL-6 were measured by ELISA.
(D) Cell viability and (E) apoptotic rates of control, LPS, LPS+NC
and LPS+miR-27a-3p groups were detected by Cell Counting Kit-8
analysis and flow cytometry, respectively. (F) Protein expression
levels of Bcl-2, Bax and C-caspase3 were measured in each group
using western blotting. *P<0.05. C-, cleaved; LPS,
lipopolysaccharide; PVT1, plasmacytoma variant translocation gene
1; lncRNA, long non-coding RNA; miR, microRNA; TNF-α, tumor
necrosis factor-α; IL, interleukin; NC, negative control. |
miR-27a-3p is a target miRNA of
PVT1
The relationship between PVT1 and miR-27a-3p was
predicted with StarBase, and it was identified that miR-27a-3p
contained complementary sequences with PVT1 (Fig. 4A). Then, dual-luciferase reporter
vectors were constructed, and miR-27a-3p mimic or miR-NC were
co-transfected with PVT1-wt or PVT1-mut in HK-2 cells. It was found
that miR-27a-3p, but not miR-NC, reduced the luciferase activity of
PVT1-wt, but did not affect that of PVT1-mut (Fig. 4B). Furthermore, RIP and pull-down
experiment results demonstrated that PVT1 directly targets
miR-27a-3p in HK-2 cells (Fig. 4C
and D). It was also identified that
transfection of pcDNA-PVT1 significantly upregulated the expression
of PVT1, thus exhibiting a successful overexpression efficiency
(Fig. S1B). The present results
suggested that miR-27a-3p may be a target miRNA of PVT1.
Furthermore, when PVT1 was overexpressed in HK-2 cells, the
expression of miR-27a-3p was significantly decreased; however, when
PVT1 was knocked down, the expression of miR-27a-3p was
significantly increased (Fig.
4E).
miR-27a-3p silencing reverses the
effect of PVT1 knockdown in LPS-treated HK-2 cells
To further examine the functional relationship
between PVT1 and miR-27a-3p, si-PVT1 and anti-miR-27a-3p were
co-transfected into HK-2 cells as the experimental group. The
transfection efficiency of anti-miR-27a-3p was determined, and it
was found that anti-miR-27a-3p significantly inhibited miR-27a-3p
expression (Fig. S1D). It was also
identified that the secretion of the inflammatory cytokines TNF-α
and IL-6 were elevated in LPS-induced HK-2 cells co-transfected
with si-PVT1 and anti-miR-27a-3p (Fig.
5A and B). Moreover, a
significant decrease in cell activity was detected using a CCK-8
assay (Fig. 5C), and flow cytometry
results demonstrated that the number of apoptotic cells was
significantly increased after miR-27a-3p silencing (Fig. 5D). Furthermore, the expression of
the apoptosis inhibitor Bcl-2 was significantly decreased, and the
expression levels of Bax and C-caspase-3 were significantly
elevated after miR-27a-3p silencing (Fig. 5E). Therefore, the results indicated
that miR-27a-3p knockdown in cells attenuated the effects of
si-PVT1 transfection, triggered inflammatory responses and
accelerated apoptosis.
 | Figure 5Decreasing the expression of
miR-27a-3p slows the effect of PVT1 knockdown. Levels of (A) TNF-α
and (B) IL-6 of LPS+scramble, LPS+si-PVT1, LPS+si-PVT1+anti-NC and
LPS+si-PVT1+anti-miR-27a-3p groups were detected in HK-2 cells by
ELISA. (C) Cell viability and (D) apoptotic rates of the groups
were detected by Cell Counting Kit-8 and flow cytometry,
respectively. (E) Protein expression levels of Bcl-2, Bax and
C-caspase 3 were measured in each group using western blotting.
*P<0.05. C-, cleaved; LPS, lipopolysaccharide; PVT1,
plasmacytoma variant translocation gene 1; miR, microRNA; si, small
interfering RNA; TNF-α, tumor necrosis factor-α; IL, interleukin;
NC, negative control. |
OXSR1 is the target gene of
miR-27a-3p
TargetScan identified that OXSR1 was a potential
target of miR-27a-3p (Fig. 6A). In
the present study, miR-27a-3p or miR-NC, along with OXSR1-wt or
OXSR1-mut were co-transfected into HK-2 cells. Luciferase reporter
assay results demonstrated that overexpression of miR-27a-3p
reduced the luciferase activity of cells with OXSR1-wt, but not
with OXSR1-mut (Fig. 6B).
Furthermore, RIP experiments indicated that miR-27a-3p directly
targeted OXSR1 in HK-2 cells (Fig.
6C). The present findings suggested that overexpression of
miR-27a-3p significantly decreased OXSR1 expression, while
anti-miR-27a-3p significantly increased OXSR1 expression in HK-2
cells (Fig. 6D).
Replenishment of OXSR1 reverses the
effect of miR-27a-3p overexpression
The present study examined the expression of OXSR1
in HK-2 cells by western blotting, and found that the protein
expression of OXSR1 was significantly increased after LPS treatment
(Fig. 7A). Furthermore, western
blot analysis suggested that the protein expression of OXSR1 was
significantly increased in HK-2 cells transfected with OXSR1
(Fig. S1E). To assess the
functional relationship between miR-27a-3p and OXSR1, miR-27a-3p,
miR-27a-3p+vector or miR-27a-3p+OXSR1 were transfected into HK-2
cells. When miR-27a-3p was co-transfected with OXSR1, it was found
that the secretion of TNF-α and IL-6 significantly increased
(Fig. 7B and C). Moreover, a significant decrease in
cell viability was detected (Fig.
7D), and the number of apoptotic cells was significantly
increased in the miR-27a-3p+OXSR1 group (Fig. 7E). In addition, the expression of
Bcl-2 was significantly decreased, and the expression levels of Bax
and C-caspase-3 were significantly increased (Fig. 7F). Thus, the present results
indicated that overexpression of OXSR1 reversed the effect of
miR-27a-3p overexpression in LPS-treated HK-2 cells.
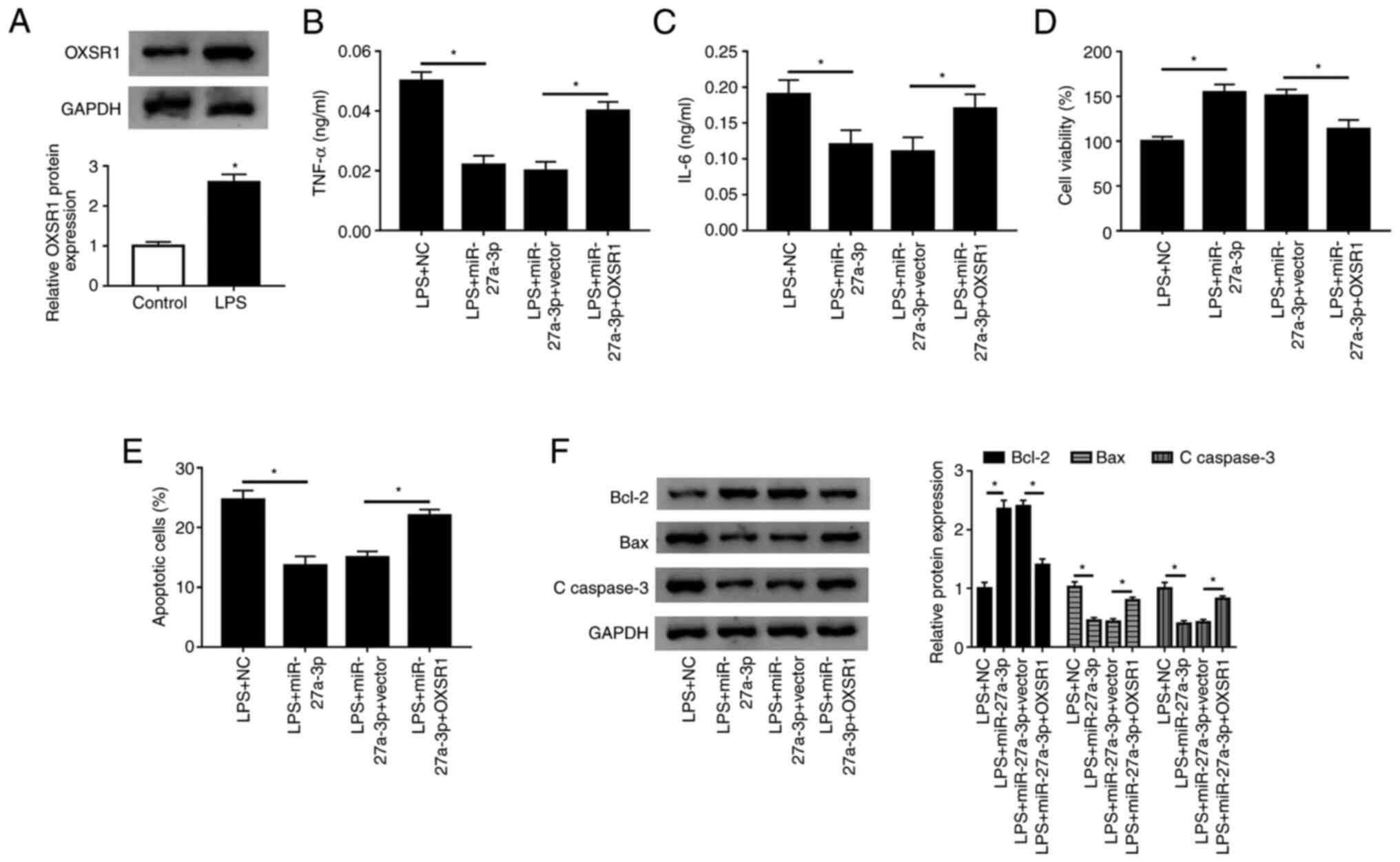 | Figure 7Overexpression of OXSR1 reverses the
effect of miR-27a-3p overexpression. (A) Protein expression of
OXSR1 was measured in HK-2 cells using western blotting after LPS
treatment. Secretion levels of (B) TNF-α and (C) IL-6 in LPS+NC,
LPS+miR-27a-3p, LPS+miR-27a-3p+vector and LPS+miR-27a-3p+OXSR1
groups were detected in HK-2 cells by ELISA. (D) Cell viability and
(E) apoptotic rates of groups were detected by Cell Counting Kit-8
and flow cytometry, respectively. (F) Protein expression levels of
Bcl-2, Bax and C-caspase 3 were measured in each group using
western blotting. *P<0.05 vs. control or as
indicated. C-, cleaved; LPS, lipopolysaccharide; PVT1, plasmacytoma
variant translocation gene 1; miR, microRNA; TNF-α, tumor necrosis
factor-α; IL, interleukin; NC, negative control; OXSR1, oxidative
stress responsive kinase 1. |
PVT1 regulates OXSR1 expression and
NF-κB pathway activation via miR-27a-3p
To investigate the interactions between PVT1,
miR-27a-3p and OXSR1, si-PVT1 and anti-miR-27a-3p were transfected
into LPS-treated HK-2 cells. It was identified that the expression
level of OXSR1 was significantly increased when miR-27a-3p and PVT1
were knocked down (Fig. 8A).
Moreover, when the expression levels of miR-27a-3p and PVT1 were
decreased, the expression levels of p-IκBα and p-p65 were
significantly increased (Fig. 8B).
Collectively, these findings suggested that PVT1 regulated OXSR1
expression and activation of the NF-κB pathway via miR-27a-3p.
 | Figure 8Long non-coding PVT1 regulates OXSR1
expression and NF-κB pathway activation via miR-27a-3p. HK-2 cells
were subjected to LPS+scramble, LPS+si-PVT1, LPS+si-PVT1+anti-NC,
and LPS+si-PVT1+anti-miR-27a-3p. (A) Western blot analysis was used
to measure the protein expression of OXSR1 in each group. (B)
Protein expression levels of p-IκBα and p-p65 of HK-2 cells were
measured using western blotting. *P<0.05. NC,
negative control; OXSR1, oxidative stress responsive kinase 1; p,
phosphorylated; LPS, lipopolysaccharide; PVT1, plasmacytoma variant
translocation gene 1; miR, microRNA; si, small interfering RNA;
IκBα, inhibitor of κB. |
Discussion
Previous studies have shown that AKI is a common
symptom in patients with sepsis, with 26-50% of patients in
developed nations with sepsis suffering from AKI, compared with
7-10% of patients with primary kidney disease-associated AKI
(32-36).
However, the exact mechanisms of AKI are not fully understood.
Recently, clinical studies have shown that AKI is associated with
increased mortality in patients with sepsis (37,38).
lncRNA PVT1 is abundant in several solid tumors, and
its abnormal expression is associated with tumor metastasis and
recurrence (39,40). A previous study has revealed that
transient expression of PVT1 promotes cell proliferation and tumor
formation in nude mice, whereas deletion of PVT1 in tumor cells
reduces tumorigenicity (7). In
small cell lung cancer (SCLC), PVT1 has been evaluated as a
candidate biomarker for diagnosis and prognosis, as high expression
of PVT1 is positively associated with the status of clinical stage,
overall survival time, lymph node metastasis and distant metastasis
in patients with SCLC (41).
Moreover, PVT1 expression in paraffin-embedded tissues also
indicates poor prognosis in nasopharyngeal carcinoma, and PVT1
knockdown may induce apoptosis (42). Although PVT1 has been shown to
facilitate cell proliferation and inhibit apoptosis in various
cancer types, the function of PVT1 in healthy tissues is unknown. A
previous study reported that PVT1 was induced by LPS and PVT1
knockdown alleviated LPS induced myocardial injury s (18). In line with these studies, the
present results suggested that the expression of lncRNA PVT1 was
increased in HK-2 cells after LPS administration. Furthermore, it
was found that knockdown of PVT1 could inhibit the secretion of
inflammatory cytokines, promote cell survival and inhibit
apoptosis. Moreover, luciferase reporter assay, RIP assay and
pull-down assay results indicated that miR-27a-3p may be a target
miRNA of lncRNA PVT1. It was also demonstrated that after PVT1
knockdown, the expression of miR-27a-3p was significantly
increased.
miRNAs are evolutionarily conserved, small
non-coding RNAs 21-25 nucleotides in length that interact with
short motifs in the 3'UTR of target mRNAs, leading to their
transcript destabilization, translational inhibition or both
(43). miRNAs are critical
post-transcriptional regulators of the majority of human genes that
can respond to physiological and pathological stress, with a
primary function to maintain the systemic homeostatic balance
(44). The present results
suggested that miR-27a-3p overexpression inhibited TNF-α and IL-6
secretion, rescued cell proliferation and inhibited apoptosis. To
further understand the regulatory mechanism of miR-27a-3p, OXSR1
was predicted to be a potential target gene of miR-27a-3p.
Moreover, dual-luciferase reporter assay and RIP assay results
identified that OXSR1 was a downstream target gene of miR-27a-3p.
It was also found that OXSR1 expression was altered following
transfection with the miR-27a-3p mimic or anti-miR-27a-3p.
OXSR1, a member of Ser/Thr kinase family, regulates
downstream kinases in response to environmental stress, which is
critical for its phosphatase activity and tumor suppressor function
(45,46). It has been reported that OXSR1 and
the inflammation marker IL-6 are significantly increased in serum
specimens from patients with diabetes undergoing hemodialysis
compared with healthy participants (47). Qin et al (48) showed that overexpression of OXSR1
increases the expression of Bcl-2, but suppresses Bax and
C-caspase-3 expression levels. Moreover, it was hypothesized that
this effect may be involved in the regulation of p38/MAPK/NF-κB
signaling pathway, and the upregulated protein expression levels of
p-p38 and p-p65(48).
NF-κB is a critical nuclear transcription factor
from different Rel family proteins, including p50, p65, p52 and
RelB (49). p65 is one of the most
common subtypes localized in the cytoplasm, and can be translocated
into the nucleus by binding to the promoter region of the target
gene, regulating the expression of various genes encoding pain and
inflammatory mediators (50). IκBα
combines with NF-κB proteins to form a complex, which cannot enter
the nucleus (50). After IκBα is
phosphorylated, NF-κB proteins are released (50). In the present study, it was found
that PVT1 knockdown significantly decreased the ratios of
p-p65/total p65 and p-IκBα/total IκBα, and silencing miR-27a-3p
reversed these effects. Therefore, the present results indicated
that PVT1 knockdown inhibited the NF-κB signaling pathway by
regulating miR-27a-3p.
In conclusion, the present results suggested that
lncRNA PVT1 regulated OXSR1 expression and activation of the NF-κB
pathway via miR-27a-3p to respond to AKI inflammation.
Supplementary Material
Transfection efficiency of vectors and
RNAs. Relative expression of target RNAs or proteins in HK-2 cells
following transfection with (A) si-PVT1, (B) pcDNA-PVT1, (C)
miR-27a-3p, (D) anti-miR-27a-3p and (E) OXSR1 as detected by
reverse transcription-quantitative PCR or western blotting.
*P<0.05 vs. scramble, pcDNA-NC, NC, anti-NC or
vector. miR, microRNA; si, small interfering RNA; OXSR1, oxidative
stress responsive kinase 1; PVT1, plasmacytoma variant
translocation gene 1; NC, negative control.
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
QY, QS and PJ conceived and designed the present
study. QS and PJ collected the data and performed the experiments.
QS performed the data analysis and interpretation. QY and QS were
involved in the preparation of manuscript. QY and QS confirm the
authenticity of all the raw data. All authors read and approved the
final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Napolitano LM: Sepsis 2018: Definitions
and Guideline Changes. Surg Infect (Larchmt). 19:117–125.
2018.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Bellomo R, Kellum JA, Ronco C, Wald R,
Martensson J, Maiden M, Bagshaw SM, Glassford NJ, Lankadeva Y,
Vaara ST, et al: Acute kidney injury in sepsis. Intensive Care Med.
43:816–828. 2017.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Ren GL, Zhu J, Li J and Meng XM: Noncoding
RNAs in acute kidney injury. J Cell Physiol. 234:2266–2276.
2019.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Gibbs RA, Weinstock GM, Metzker ML, Muzny
DM, Sodergren EJ, Scherer S, Scott G, Steffen D, Worley KC, Burch
PE, et al: Rat Genome Sequencing Project Consortium: Genome
sequence of the Brown Norway rat yields insights into mammalian
evolution. Nature. 428:493–521. 2004.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Yin KJ, Hamblin M and Chen YE: Non-coding
RNAs in cerebral endothelial pathophysiology: Emerging roles in
stroke. Neurochem Int. 77:9–16. 2014.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Mercer TR, Dinger ME and Mattick JS: Long
non-coding RNAs: Insights into functions. Nat Rev Genet.
10:155–159. 2009.PubMed/NCBI View
Article : Google Scholar
|
|
7
|
Iyer MK, Niknafs YS, Malik R, Singhal U,
Sahu A, Hosono Y, Barrette TR, Prensner JR, Evans JR, Zhao S, et
al: The landscape of long noncoding RNAs in the human
transcriptome. Nat Genet. 47:199–208. 2015.PubMed/NCBI View
Article : Google Scholar
|
|
8
|
Barsotti AM, Beckerman R, Laptenko O,
Huppi K, Caplen NJ and Prives C: p53-Dependent induction of PVT1
and miR-1204. J Biol Chem. 287:2509–2519. 2012.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Chen J, Yu Y, Li H, Hu Q, Chen X, He Y,
Xue C, Ren F, Ren Z, Li J, et al: Long non-coding RNA PVT1 promotes
tumor progression by regulating the miR-143/HK2 axis in gallbladder
cancer. Mol Cancer. 18(33)2019.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Hu J, Han Q, Gu Y, Ma J, McGrath M, Qiao
F, Chen B, Song C and Ge Z: Circular RNA PVT1 expression and its
roles in acute lymphoblastic leukemia. Epigenomics. 10:723–732.
2018.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Guo J, Hao C, Wang C and Li L: Long
noncoding RNA PVT1 modulates hepatocellular carcinoma cell
proliferation and apoptosis by recruiting EZH2. Cancer Cell Int.
18(98)2018.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Yang L, Peng X, Jin H and Liu J: Long
non-coding RNA PVT1 promotes autophagy as ceRNA to target ATG3 by
sponging microRNA-365 in hepatocellular carcinoma. Gene.
697:94–102. 2019.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Xu Y, Luo X, He W, Chen G, Li Y, Li W,
Wang X, Lai Y and Ye Y: Long Non-Coding RNA PVT1/miR-150/ HIG2 Axis
Regulates the Proliferation, Invasion and the Balance of Iron
Metabolism of Hepatocellular Carcinoma. Cell Physiol Biochem.
49:1403–1419. 2018.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Guan Y, Kuo WL, Stilwell JL, Takano H,
Lapuk AV, Fridlyand J, Mao JH, Yu M, Miller MA, Santos JL, et al:
Amplification of PVT1 contributes to the pathophysiology of ovarian
and breast cancer. Clin Cancer Res. 13:5745–5755. 2007.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Yang Q, Yu Y, Sun Z and Pan Y: Long
non-coding RNA PVT1 promotes cell proliferation and invasion
through regulating miR-133a in ovarian cancer. Biomed Pharmacother.
106:61–67. 2018.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Ghafouri-Fard S, Omrani MD and Taheri M:
Long noncoding RNA PVT1: A highly dysregulated gene in malignancy.
J Cell Physiol. 235:818–835. 2020.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Feng F, Qi Y, Dong C and Yang C: PVT1
regulates inflammation and cardiac function via the MAPK/NF-κB
pathway in a sepsis model. Exp Ther Med. 16:4471–4478.
2018.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Luo YY, Yang ZQ, Lin XF, Zhao FL, Tu HT,
Wang LJ, Wen MY and Xian SX: Knockdown of lncRNA PVT1 attenuated
macrophage M1 polarization and relieved sepsis induced myocardial
injury via miR-29a/HMGB1 axis. Cytokine. 143(155509)2021.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Ren Y, Huang W, Weng G, Cui P, Liang H and
Li Y: LncRNA PVT1 promotes proliferation, invasion and
epithelial-mesenchymal transition of renal cell carcinoma cells
through downregulation of miR-16-5p [Corrigendum]. OncoTargets
Ther. 12:5649–5650. 2019.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Wang C, Zou H, Yang H, Wang L, Chu H, Jiao
J, Wang Y and Chen A: Long non coding RNA plasmacytoma variant
translocation 1 gene promotes the development of cervical cancer
via the NF-κB pathway. Mol Med Rep. 20:2433–2440. 2019.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Yazdi N, Houshmand M, Atashi A, Kazemi A,
Najmedini AA and Zarif MN: Long noncoding RNA PVT1: Potential
oncogene in the development of acute lymphoblastic leukemia. Turk J
Biol. 42:405–413. 2018.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Barres BA: The mystery and magic of glia:
A perspective on their roles in health and disease. Neuron.
60:430–440. 2008.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Mertens-Talcott SU, Chintharlapalli S, Li
X and Safe S: The oncogenic microRNA-27a targets genes that
regulate specificity protein transcription factors and the G2-M
checkpoint in MDA-MB-231 breast cancer cells. Cancer Res.
67:11001–11011. 2007.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Guttilla IK and White BA: Coordinate
regulation of FOXO1 by miR-27a, miR-96, and miR-182 in breast
cancer cells. J Biol Chem. 284:23204–23216. 2009.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Liu T, Tang H, Lang Y, Liu M and Li X:
MicroRNA-27a functions as an oncogene in gastric adenocarcinoma by
targeting prohibitin. Cancer Lett. 273:233–242. 2009.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Chintharlapalli S, Papineni S, Abdelrahim
M, Abudayyeh A, Jutooru I, Chadalapaka G, Wu F, Mertens-Talcott S,
Vanderlaag K, Cho SD, et al: Oncogenic microRNA-27a is a target for
anticancer agent methyl
2-cyano-3,11-dioxo-18beta-olean-1,12-dien-30-oate in colon cancer
cells. Int J Cancer. 125:1965–1974. 2009.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Wang Y, Cao L, Wang Q, Huang J and Xu S:
LncRNA FOXD2-AS1 induces chondrocyte proliferation through sponging
miR-27a-3p in osteoarthritis. Artif Cells Nanomed Biotechnol.
47:1241–1247. 2019.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Zaccaria V, Curti V, Di Lorenzo A, Baldi
A, Maccario C, Sommatis S, Mocchi R and Daglia M: Effect of Green
and Brown Propolis Extracts on the Expression Levels of microRNAs,
mRNAs and Proteins, Related to Oxidative Stress and Inflammation.
Nutrients. 9(9)2017.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Ndzi EN, Indu Viswanath AN, Adzemye NG,
Tamgue O, Nsongka MV, Nair AS and Nkenfou CN: Upregulated bovine
tuberculosis microRNAs Trigger oncogenic pathways: An In silico
perception. Int J Mycobacteriol. 8:70–74. 2019.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Jiang X, Li D, Shen W, Shen X and Liu Y:
LncRNA NEAT1 promotes hypoxia-induced renal tubular epithelial
apoptosis through downregulating miR-27a-3p. J Cell Biochem.
120:16273–16282. 2019.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) Method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Bagshaw SM, George C, Bellomo R and
Committee ADM: ANZICS Database Management Committee. Early acute
kidney injury and sepsis: A multicentre evaluation. Crit Care.
12(R47)2008.PubMed/NCBI View
Article : Google Scholar
|
|
33
|
Bagshaw SM, Uchino S, Bellomo R, Morimatsu
H, Morgera S, Schetz M, Tan I, Bouman C, Macedo E, Gibney N, et al:
Beginning and Ending Supportive Therapy for the Kidney (BEST
Kidney) Investigators: Septic acute kidney injury in critically ill
patients: Clinical characteristics and outcomes. Clin J Am Soc
Nephrol. 2:431–439. 2007.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Vincent JL, Sakr Y, Sprung CL, Ranieri VM,
Reinhart K, Gerlach H, Moreno R, Carlet J, Le Gall JR and Payen D:
Sepsis Occurrence in Acutely Ill Patients Investigators. Sepsis in
European intensive care units: Results of the SOAP study. Crit Care
Med. 34:344–353. 2006.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Cruz DN, Bolgan I, Perazella MA, Bonello
M, de Cal M, Corradi V, Polanco N, Ocampo C, Nalesso F, Piccinni P,
et al: North East Italian Prospective Hospital Renal Outcome Survey
on Acute Kidney Injury (NEiPHROS-AKI) Investigators: North East
Italian Prospective Hospital Renal Outcome Survey on Acute Kidney
Injury (NEiPHROS-AKI): Targeting the problem with the RIFLE
Criteria. Clin J Am Soc Nephrol. 2:418–425. 2007.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Kolhe NV, Stevens PE, Crowe AV, Lipkin GW
and Harrison DA: Case mix, outcome and activity for patients with
severe acute kidney injury during the first 24 hours after
admission to an adult, general critical care unit: Application of
predictive models from a secondary analysis of the ICNARC Case Mix
Programme database. Crit Care. 12 (Suppl 1)(S2)2008.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Peerapornratana S, Manrique-Caballero CL,
Gómez H and Kellum JA: Acute kidney injury from sepsis: Current
concepts, epidemiology, pathophysiology, prevention and treatment.
Kidney Int. 96:1083–1099. 2019.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Poston JT and Koyner JL: Sepsis associated
acute kidney injury. BMJ. 364(k4891)2019.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Zeng X, Liu Y, Zhu H, Chen D and Hu W:
Downregulation of miR-216a-5p by long noncoding RNA PVT1 suppresses
colorectal cancer progression via modulation of YBX1 expression.
Cancer Manag Res. 11:6981–6993. 2019.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Ding C, Yang Z, Lv Z, Du C, Xiao H, Peng
C, Cheng S, Xie H, Zhou L, Wu J, et al: Long non-coding RNA PVT1 is
associated with tumor progression and predicts recurrence in
hepatocellular carcinoma patients. Oncol Lett. 9:955–963.
2015.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Huang C, Liu S, Wang H, Zhang Z, Yang Q
and Gao F: LncRNA PVT1 overexpression is a poor prognostic
biomarker and regulates migration and invasion in small cell lung
cancer. Am J Transl Res. 8:5025–5034. 2016.PubMed/NCBI
|
|
42
|
He Y, Jing Y, Wei F, Tang Y, Yang L, Luo
J, Yang P, Ni Q, Pang J, Liao Q, et al: Long non-coding RNA PVT1
predicts poor prognosis and induces radioresistance by regulating
DNA repair and cell apoptosis in nasopharyngeal carcinoma. Cell
Death Dis. 9(235)2018.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Goedeke L, Rotllan N, Canfrán-Duque A,
Aranda JF, Ramírez CM, Araldi E, Lin CS, Anderson NN, Wagschal A,
de Cabo R, et al: MicroRNA-148a regulates LDL receptor and ABCA1
expression to control circulating lipoprotein levels. Nat Med.
21:1280–1289. 2015.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297.
2004.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Mercier-Zuber A and O'Shaughnessy KM: Role
of SPAK and OSR1 signalling in the regulation of NaCl
cotransporters. Curr Opin Nephrol Hypertens. 20:534–540.
2011.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Both J, Krijgsman O, Bras J, Schaap GR,
Baas F, Ylstra B and Hulsebos TJ: Focal chromosomal copy number
aberrations identify CMTM8 and GPR177 as new candidate driver genes
in osteosarcoma. PLoS One. 9(e115835)2014.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Totan A, Balcangiu-Stroescu A-E, Imre MM,
Miricescu D, Balan D, Stanescu I-I, Ionescu D, Timofte D, Tanasescu
MD and Greabu M: XOR-Possible Correlations with Oxidative Stress
and Inflammation Markers in the Context of Diabetic Kidney Disease.
Rev de Chim. 70:1396–1398. 2019.
|
|
48
|
Qin Y, Wang G and Peng Z: MicroRNA-191-5p
diminished sepsis-induced acute kidney injury through targeting
oxidative stress responsive 1 in rat models. Biosci Rep.
39(39)2019.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Jimi E and Fukushima H: [NF-κB signaling
pathways and the future perspectives of bone disease therapy using
selective inhibitors of NF-κB]. Clin Calcium. 26:298–304.
2016.PubMed/NCBI
|
|
50
|
Niederberger E and Geisslinger G: The
IKK-NF-kappaB pathway: A source for novel molecular drug targets in
pain therapy? FASEB J. 22:3432–3442. 2008.PubMed/NCBI View Article : Google Scholar
|