Introduction
Regulation of gene expression is a complex process
that induces gene expression in the cells providing spatiotemporal
response to changes in environmental conditions, which serve as the
molecular basis of cellular differentiation, morphogenesis and
ontogeny in the organism (1). This
regulation can be carried out at multiple levels, including the
gene, transcriptional, post-transcriptional, translational and
post-translational levels (2-4).
Post-translational protein modification is important and occurs in
almost every protein during or after protein synthesis and can
change the stability, structure, localization and function of the
proteins (5). These modifications
include ubiquitination, phosphorylation and dephosphorylation,
glycosylation and deglycosylation, lipidation, methylation and
acetylation (6,7). RNA-binding proteins (RBPs) are
critical regulators of post-transcriptional gene regulation and a
number of RBPs are modified after the translation to influence
localization, stability and translation of the target mRNAs
(5,8). Dysregulated post-translational
modifications have been shown to influence pathological processes,
such as cancer, neurodegenerative disorders and cardiovascular
disease (9). RBPs exert
significant control over numerous cellular functions and thus
represent a popular area of investigations by scientists. Recent
developments in experimental identification of RBPs significantly
expanded the number of known RBPs. Although RBPs serve a crucial
role in post-transcriptional regulation of gene expression,
relatively few RBPs have been studied systematically (10,11).
Human antigen R (HuR; also known as HuA or ELAVL1)
is one of the most extensively studied RBPs and serves an important
role in the stabilization of mRNAs containing AU-rich elements
(AREs) and regulation of the expression of the target genes
(12). When cells are under normal
conditions, HuR is mainly located in the nucleus and is in an
inactivated state. When cells encounter certain pathological
environments, such as hypoxia, radiation damage and cytokine
stimuli, HuR translocates from the nucleus to the cytoplasm by
binding and interacting with various 3'-UTR AREs (8,12-14).
Thus, HuR can protect mRNAs from nuclease degradation in the
process of translocation from the nucleus to the cytoplasm and
increase the stability of mRNAs. HuR has been shown to serve
important roles in tumorigenesis and malignant transformation and
to regulate oncogenes, the cell cycle, apoptosis, inflammatory
factors, invasion, metastasis and related molecules by binding to
the target genes (15-18).
In addition, several studies have shown that HuR contributes to the
development of resistance to chemotherapy in multiple types of
cancer (14,19,20).
c-Myc, p53, cyclin D1, cyclin A, survivin, Bcl-2, cyclooxygenase
(COX)-2, VEGF and MMP-9 have been identified as the downstream
targets of HuR depending on specific cell type (21).
The skin is the largest outermost organ of the human
body. The primary role of the skin is to serve as a physical
barrier, protecting our bodies from potential assault by foreign
organisms or toxic substances. HuR is known to be modulated by
mitogenic and stress-causing agents, including UV radiation
(22). Stress-induced modulation
of HuR activity may be achieved by phosphorylation resulting in its
translocation to the cytoplasm. HuR binds to COX-2 mRNA in a
constitutive manner and forced overexpression of an HuR-GFP
construct stabilizes COX-2 mRNA in unstimulated HaCaT cells
(23). Given the importance of HuR
in gene regulation, the aim of the present study was to elucidate
further the role and regulatory mechanisms of HuR in proliferation,
senescence and radiosensitivity of the skin
Materials and methods
Vectors and viruses
In the present study, the third generation of
four-plasmid lentivirus vector system was used. The lentiviral
vector system was composed of four plasmids: The expression plasmid
and three packaging vectors, including 1.5 µg pMDLg-pRRE, 1.5 µg
pMD2.G and 1.5 µg pRSV-Rev Virus packaging helper plasmids.
Lentiviral vectors and packaging vectors were transfected into 293T
cells (Wuhan GeneCreate Biological Engineering Co., Ltd.). 293T
cells were seeded with DMEM (HyClone; Cytiva) supplemented with 10%
fetal bovine serum (FBS; cat. no. 04-001-1A; Biological Industries)
and cultured in a 37˚C incubator with a 6-well plate of 2 ml/well.
When the cell density reached 70-80%, it was used for transfection.
The cells were cultured in serum-free medium before transfection. 2
µg expression plasmid, 1.5 µg PMDLG-PRRE, 1.5 µg PMD2.g and 1.5 µg
PRSV-Rev virus packing assistant plasmid were diluted in 500 µl
serum-free medium. Lipofectamine® 2000 (15 µl;
Invitrogen; Thermo Fisher Scientific, Inc.) was diluted with 500 µl
serum-free medium. After standing for 5 min, The DNA solution was
mixed with Lipofectamine® 2000 solution and stood for 20
min at room temperature. Serum-free medium (1 ml) was taken from
the 6-well plate and 1 ml plasmid and Lipofectamine®
2000 were dropped into the 6-well plate at 37˚C and 5%
CO2 for 8 h. Following transfection, the culture medium
was exchanged with DMEM (HyClone; Cytiva) supplemented with 10%
fetal bovine serum (FBS; cat. no. 04-001-1A; Biological
Industries). After 48 h, the supernatant containing the retroviral
particles was collected and then concentrated by centrifugation at
4,000 x g for 10 min at 4˚C. The cell supernatant was filtered by a
0.45 µm filter into a 50 ml ultrafast centrifuge tube and 5X
PEG8000 was added to precipitate at 4˚C overnight. After
centrifugation at 7,000 x g, the supernatant was discarded and the
precipitation was redissolved with 10 ml PBS. The viral
supernatants were added to the upper layer of 20% sucrose solution,
centrifuged at 20,000 x g for 2 h and 4˚C. The precipitate was
suspended with 1 ml PBS and filtered by a 0.22 µm filter for
sterilization. The virus suspension was separated into 50 µl and
stored at -80˚C. A total of 1x105 cells/well were
transduced with viral supernatants. The cells were collected for
subsequent experiments after being cultured in fresh medium for 48
h at 37˚C.
HuR overexpression and short hairpin
(sh)RNA lentiviruses
The human HuR coding region was amplified by
polymerase chain reaction (PCR) using a primer pair specific to
HuR. The fragments were inserted into the lentiviral expression
vectors (LV-HuR) and then packaged into viral particles. The
negative control lentiviral (LV-NC) was constructed by not
inserting any sequences. Lentivirus silencing HuR through shRNAs
were obtained from Hanbio Biotechnology (containing RFP). The
targeting sequences of shRNA control (sh-NC) and four shRNA
targeting HuR (sh-HuR-1, sh-HuR-2, sh-HuR-3 and sh-HuR-4) are
listed in Table SI.
Cell culture and transfection
Human keratinocyte HaCaT (cat. no. iCell-h006, iCell
Bioscience Inc.) and human skin fibroblast WS1 (cat. no. CRL-1502T;
ATCC) cells were maintained in DMEM supplemented with 10% FBS and
100 U/ml penicillin-streptomycin at 37˚C and a 5% CO2
atmosphere. For transfection, cells were transfected by
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) with viral particles. Cells were exposed to a
single dose (20 Gy) of X-rays using the linear accelerator
(RadSource) at a dose rate of 1.15 Gy/min. The HaCaT cell line was
obtained from iCell Bioscience Inc. DNA was extracted with Axygen
genome extraction kit (cat. no. AP-GX-250; Axygen®
AxyPrep DNA Gel Extraction Kit; Corning, Inc.) and amplified with
21-str amplification scheme. STR loci and sex gene Amelogenin were
detected on an ABI 3730XL genetic analyzer (Applied Biosytems;
Thermo Fisher Scientific, Inc.). The results showed that the cell
line was completely matched by DNA typing in cell line retrieval
and the cell name was HACAT and the cell number was 771 according
to DSMZ database. No multiple alleles were found in this cell
line.
Reverse transcription-quantitative
(RT-q) PCR
After cells reached 90% confluence, total RNA was
extracted using TRIzol® reagent (cat. no. 15596018;
Thermo Fisher Scientific, Inc.) in accordance with the
manufacturer's instructions. RNA was reverse transcribed using a
reverse transcription kit (cat. no. K1691; RevertAid RT Reverse
Transcription kit; Thermo Fisher Scientific, Inc.) under 42˚C for
60 min and 70˚C for 5 min. The mRNA level of HuR gene in
each sample was measured by RT-PCR. The reaction was performed at
37˚C for 15 min followed by 85˚C for 5 sec. cDNA was amplified via
qPCR with TB Green dye (cat. no. RR420S; Takara Bio, Inc.) on a
Prism 7500 RT-PCR machine (Applied Biosystems; Thermo-Fisher
Scientific, Inc.). The thermocycling conditions included an initial
denaturation step of 95˚C for 60 sec, followed by 40 cycles of
amplification at 95˚C for 15 sec and then annealing at 60˚C for 30
sec. GAPDH was used as an endogenous standard. The relative
amount of target genes was carried out according to the
2-ΔΔCq algorithm (24).
The experiments were repeated three times (HUR: Forward Primer
5'-GGGTGACATCGGGAGAACG-3', Reverse Primer
5'-CTGAACAGGCTTCGTAACTCAT-3'; GAPDH: Forward Primer
5'-GGAGCGAGATCCCTCCAAAAT-3', Reverse Primer
5'-GGCTGTTGTCATACTTCTCATGG-3').
Western blotting analysis
The cells were lysed in lysis buffer (Promega
Corporation) and centrifuged at 4˚C, 12,000 x g for 10 min. The
supernatant was collected and subjected to western blotting.
Protein concentration was subsequently measured using a BCA Protein
Assay kit (cat. no. P0012; Beyotime Institute of Biotechnology).
Protein (50 µg) from each lysate was fractionated by 10% SDS-PAGE.
The samples were electrophoresed for 2 h and transferred onto
polyvinylidene difluoride membranes (MilliporeSigma). After being
blocked with 5% BSA in TBS-0.1% Tween-20 (TBST) for 1 h at room
temperature, the membranes were blotted with HuR (Abcam; cat. no.
ab220342) or α-Tubulin (Beyotime Institute of Biotechnology; cat.
no. AF0001) primary antibodies at 1:1,000 dilutions. The membranes
were then incubated with the appropriate horseradish
peroxidase-coupled secondary antibody (Beyotime Institute of
Biotechnology; cat. no. A0277) at a 1:2,000 dilution for 1 h at
room temperature. After the membranes were washed with TBST, the
blots were incubated with enhanced chemiluminescence (ECL) stable
peroxide solution (Beyotime Institute of Biotechnology). All blots
were visualized using a FluoroChem MI imaging system (Alpha
Innotech Corporation) at room temperature. Densitometry was
performed using ImageJ v1.8.0-172 software (National Institutes of
Health).
Cell viability assay
Cell viability was evaluated using the Cell Counting
Kit-8 (CCK-8; Dojindo Laboratories, Inc.) assay. HaCaT and WS1
cells were plated in 96-well plates and cultured for 12 h at 37˚C
in a CO2 incubator. The cells were transfected with
LV-NC, LV-HuR, sh-NC, sh-HuR-1 and sh-HuR-2 viral particles. After
treatment for 24 and 48 h at 37˚C in a CO2 incubator,
the cells were then incubated with 10 µl CCK-8 for 4 h. Then the
optical density (OD) at 450 nm was measured using a Microplate
Reader (Bio-Rad Laboratories, Inc.). The viability index was
calculated as experimental OD value/control OD value. Three
independent experiments were performed in quadruplicate.
EdU assay
The proliferation of HaCaT and WS1 cells transfected
with different viral particles was determined by
5-Ethynyl-2'-deoxyuridine (EdU) assay kit. Cells were pre-infected
with LV-NC, LV-HuR, sh-NC, sh-HuR-1 and sh-HuR-2 viral particles
for 24 h at 37˚C in a CO2 incubator before receiving
sham or 20 Gy X-ray irradiation, after irradiation for 48 h at 37˚C
in a CO2 incubator, cells were labeled with 50 µM EdU
(Guangzhou RiboBio Co., Ltd.) for 4 h at 37˚C in a CO2
incubator. Then, the cells were fixed with 4% formaldehyde for 15
min at room temperature and treated with 0.5% Triton X-100 for 20
min at room temperature. The cells were washed with PBS for three
times and treated with 100 µl of 1X ApolloR (EdU; Guangzhou RiboBio
Co., Ltd.) reaction cocktail in the dark at room temperature for 30
min. Subsequently, the DNA of each well of cells were stained with
4',6-diamidino-2-phenylindole dihydrochloride (DAPI;
MilliporeSigma) for 30 min at room temperature and observed under a
fluorescence microscope (Olympus Corporation).
Clonogenic assay
For standard clonogenic assays, cells were
re-suspended and seeded into six-well plates at 200 cells/well,
cells were pre-infected with LV-NC, LV-HuR, sh-NC, sh-HuR-1 and
sh-HuR-2 viral particles 24 h at 37˚C in a CO2 incubator
before receiving sham or 20 Gy X-ray irradiation. The cells were
grown from 7-10 days to allow for colony formation and were
subsequently fixed and stained using crystal violet. Colonies
consisting of >50 cells were counted as a clone.
Apoptosis analysis
Apoptosis was measured using propidium iodide
(PI)/Annexin-V double staining following manufacturer's
instructions (BD Biosciences). Cells were pre-infected with LV-NC,
LV-HuR, sh-NC, sh-HuR-1 and sh-HuR-2 viral particles 24 h at 37˚C
in a CO2 incubator before receiving sham or 20 Gy X-ray
irradiation. After irradiation for 48 h, HaCaT and WS1 cells were
harvested and apoptotic fractions were measured using flow
cytometry (Beckman Coulter, Inc.). The Annexin-V+/PI- cells are
early in the apoptotic process, the Annexin-V+/PI+ cells indicating
late apoptosis. The percentage of both types of cells was counted.
To compute the percentage of apoptotic cells, a flow cytometer
(FACSCalibur; BD Biosciences) with ModFit's LT v.3.0 software (BD
Diagnostics) was used for data analysis.
Cell senescence staining
The cell senescence of HaCaT and WS1 cells
transfected with different viral particles was determined by
β-galactosidase staining. After the cells were transfected with
LV-NC, LV-HuR, sh-NC, sh-HuR-1 and sh-HuR-2 viral particles for 24
h at 37˚C in a CO2 incubator. The cell senescence
staining was performed according to the β-galactosidase staining
kit (Beyotime Institute of Biotechnology).
Cell senescence assay
HaCaT and WS1 cells were fixed in 2%
formaldehyde/0.2% glutaraldehyde for 5 min at room temperature.
β-Galactosidase staining solution containing X-gal (cat. no. C0602;
Beyotime Institute of Biotechnology) was added after rinsing with
PBS. The cells were then incubated for 6-10 h in a 37˚C incubator
without CO2. Senescent cells (stained blue) were
observed and images were captured using light microscopy (Olympus
Corporation; magnification, x10) and positive staining areas were
calculated by determining the percentage of SA-β-gal+
cells in five random fields in each of the three wells.
Reactive oxygen species (ROS)
generation assay
ROS levels after irradiation were determined using
the ROS assay kit (Beyotime Institute of Biotechnology). HaCaT and
WS1 cells were pre-infected with sh-NC, sh-HuR-1 and sh-HuR-2 viral
particles 24 h at 37˚C in a CO2 incubator before
receiving 20 Gy X-ray irradiation. After irradiation for 24 h,
cells were labeled with 10 µM 2,7-dichlorofluorescein diacetate
(DCFH-DA) at 37˚C for 30 min. Then, the cells were fixed with 4%
formaldehyde for 15 min at room temperature and washed with PBS for
three times, after then treated with 5 µg/ml Hoechst 33258
(Beyotime Institute of Biotechnology) for 30 min and observed under
a fluorescence microscope (PerkinElmer, Inc.).
Sample preparation for RNA-seq
Total RNA was extracted from HaCaT cells infected
with LV-NC, LV-HuR, sh-NC or sh-HuR-1 lentiviruses (n=3) using
TRIzol® reagent (Thermo Fisher Scientific, Inc.). For
RNA high-throughput sequencing, RNA libraries were created from
each group using the NEBNext Ultra Directional RNA Library
preparation kit from Illumina, Inc. The main steps in the workflow
involved the removal of ribosomal RNA, the fragmentation of total
RNA, reverse transcription and second-strand complementary DNA
(cDNA) synthesis, end repair, dA tailing and adaptor ligation. The
products of these reactions were purified and enriched by
polymerase chain reaction to create the final cDNA library. The
libraries were then sequenced using an Illumina HiSeq2500
(paired-end sequencing; Illumina, Inc.).
RNA-seq data acquisition and quality
control
The RNA sequence row data was obtained by RNA-seq on
the Illumina platform. Then, the reads were clipped and trimmed to
avoid low-quality data using Trim Galore (http://www.bioinformatics.babraham.ac.uk/projects/trim_galore/).
Trimmed sequence files underwent quality control analysis using
FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/).
The present study utilized several parameters to evaluate the read
quality, including the number of the reads, guanine-cytosine (GC)
contents and average length of the reads.
Differential expression analysis of
genes
For the analysis of differentially expressed genes,
the clean data for each sample were aligned to the rat reference
genome
(ftp://ftp.ensembl.org/pub/release-83/fasta/rattus_norvegicus/dna/)
using TopHat (version 2.0.10) software (http://ccb.jhu.edu/software/tophat/index.shtml).
The alignment files were assembled using Cufflinks (http://cole-trapnell-lab.github.io/cufflinks/) and the
StringTie software (http://ccb.jhu.edu/software/stringtie/) based on the
location of the known transcript and the transcripts of all samples
were assembled again using Cuffmerge (http://cole-trapnell-lab.github.io/cufflinks/cuffmerge/index.html).
The differential gene expressed values of each sample were
normalized using the fragments per kilobase of transcript per
million fragments mapped. The differential expression levels were
calculated and the statistical significance of detected genes
evaluated. Genes were considered as significantly differentially
expressed with false discovery rates (FDRs) ≤0.05 and fold-changes
≥1.0.
Functional enrichment analysis
The database for annotation, visualization and
integrated discovery (DAVID; http://david.abcc.ncifcrf.gov/) was used, which
leveraged the Gene Ontology (GO) to determine the most functional
annotation and classification of significant differently expressed
genes and DEL targets. To demonstrate GO or molecular pathway
enrichment, DAVID calculates a modified Fisher exact P-value.
P-value <0.05 was considered to be strongly enriched in the
annotation category. In addition, the present study used the Kyoto
Encyclopedia of Genes and Genomes (KEGG) database (http://www.genome.ad.jp/kegg/) to analyze the roles of
differently expressed genes and DEL targets in the pathways.
RNA immunoprecipitation (RIP) and
sequencing
RIP was performed as described previously (21). This radiation dose (5 Gy X-ray) was
selected because WS1 cells are more sensitive to radiation. The
doses in cell studies were the same as previous publications of
others and our own (25,26). In short, WS1 cells were washed with
PBS and harvested by adding 150 ml of immunoprecipitation buffer
(10 mM Tris-HCl, pH 7.4, 50 mM NaCl, 0.5 mM EDTA, 1 mM
phenylmethanesulfonylfluoride and 1% Triton X-100). After the cells
were sonicated (25 KHz, 5 sec/time) for 2 min on ice, insoluble
material was removed by centrifugation at 2,500 x g, 15 min, 4˚C.
Supernatants were collected and pre-cleared by agarose-coupled
protein A. The agarose beads were removed by centrifugation at
2,500 x g, 5 min, 4˚C. HuR (CST; cat. no. MA1-167) antibody and
GAPDH (Abcam; cat. no. ab181602;) antibody were added to the
supernatants (1:100 ratio) and the reaction was incubated at 4˚C
overnight with gentle rotation. Agarose-coupled protein A (50 µl)
were added to capture the antibody-protein complexes by rotating
for 2 h at 4˚C. The supernatants were then removed by centrifuging
at 2,000 x g and 4˚C for 5 min. RNA was extracted using
TRIzol® following the manufacturer's instructions
(Thermo Fisher Scientific, Inc.). rRNAs were removed from the
immunoprecipitated RNA and input RNA samples by using Ribo-Zero
rRNA Removal kit (Illumina, Inc.). RNA libraries were constructed
by using rRNA-depleted RNAs with TruSeq Stranded Total RNA Library
Prep kit (Illumina, Inc.) according to the manufacturer's
instructions. Libraries were controlled for quality and quantified
using the BioAnalyzer 2100 system (Agilent Technologies, Inc.).
Libraries (10 pM) were denatured as single-stranded DNA molecules,
captured on Illumina flow cells, amplified in situ as
clusters and finally sequenced for 150 cycles on an Illumina HiSeq
Sequencer according to the manufacturer's instructions by
CloudSeq.
Statistical analysis
The data were evaluated using either unpaired
two-sided Student's t-tests or one-way analysis of variance to
determine statistical significance after confirming that the data
met appropriate assumptions. For all experiments, three biological
replicates were analyzed for each condition and presented as the
mean ± standard error of the mean. Differences between two groups
were determined using a paired Student's t-test and differences
among >2 groups were analyzed by one-way analysis of variance
and Tukey post hoc tests. Statistical analysis was performed using
Prism 7 software (GraphPad Software, Inc.). Data are expressed as
mean ± standard error of the mean. P<0.05 was considered to
indicate a statistically significant difference.
Results
Overexpression and silencing of HuR in
skin cells
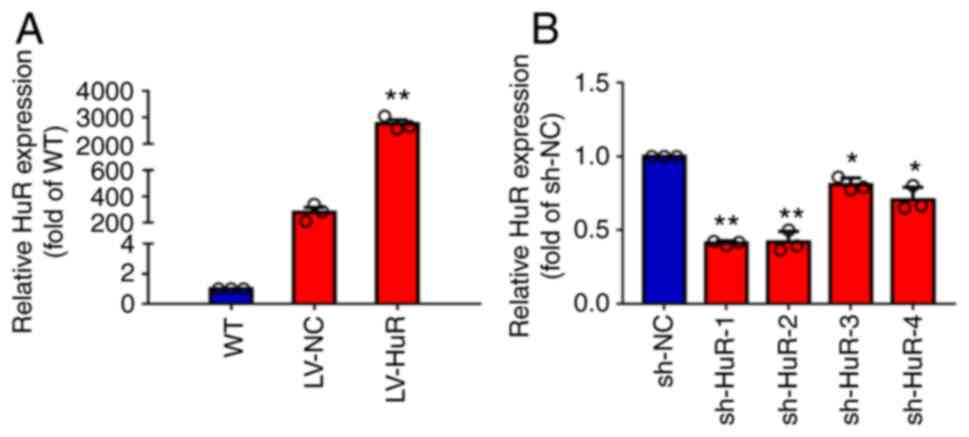
To test the overexpression of LV-HuR, the present
study measured HuR expression by RT-qPCR and western blotting. The
results showed that the expression of HuR mRNA in LV-HuR cells was
significantly higher compared with that in LV-NC cells (Fig. 1A). In addition, the silencing
effect of sh-NC and four shRNAs targeting HuR (designated sh-HuR-1,
sh-HuR-2, sh-HuR-3 and sh-HuR-4) was also tested. The results
showed that four sh-HuRs significantly reduced the expression of
HuR and sh-HuR-1 and sh-HuR-2 had the strongest silencing effect
(Fig. 1A); subsequent experiments
were performed using these two sh-HuR vectors.
The effects of HuR on proliferation
and apoptosis of skin cells
Initially, the present study investigated the effect
of HuR on the proliferation of skin cells by a CCK-8 assay. The
results showed that overexpression of HuR promoted the growth of
HaCaT and WS1 cells (Fig. 2A) and
silencing HuR inhibited the growth of these cells (Fig. 2B). Then, EdU staining was used to
confirm the role of HuR in the proliferation of skin cells. The
results also showed a positive association of HuR with the
percentage of EdU-positive HaCaT and WS1 cells (Fig. 2C and D). The results of a colony formation
assay showed that clonogenicity of HaCaT and WS1 cells was
significantly increased or reduced when HuR was overexpressed or
silenced, respectively (Fig. 3A
and B). The results of a flow
cytometry-based apoptosis assay showed that overexpression of HuR
reduced apoptosis of HaCaT and WS1 cells (Fig. 3C). Conversely, downregulation of
HuR increased apoptosis in the two skin cell lines (Fig. 3D).
The effects of HuR on skin cell
senescence
Temporal and environmental aging influences skin
structure and functions, including the skin barrier and elastic and
mechanical properties of cutaneous tissue associated with
alterations of biomechanical properties of skin cells (27). Thus, the present study evaluated
the effect of HuR expression levels on HaCaT and WS1 cell
senescence. Cellular senescence was evaluated by β-galactosidase
staining (28). The results of
senescence staining in Fig. 4A
showed that senescence of HuR-overexpressing WS1 and HaCaT cells
was reduced by 50%. The results of Fig. 4B indicated that senescence was
increased by 60% in the cells with silenced HuR. These results
showed that overexpression of HuR reduced senescence of HaCaT and
WS1 cells and that HuR silencing increased senescence.
HuR modulated the expression of mRNAs
in skin cells
Since HuR influences the stability of mRNAs
(7,8), the present study sought to explore
the landscape of HuR-influenced mRNAs in skin cells. RNA-Seq was
performed in WS1 cells with HuR overexpression or downregulation.
The RNA-Seq data are accessible from the GEO database (accession
number GSE161811). The results showed that comparison with the
control group (LV-NC) detected 2947 differentially expressed genes
in the overexpression group (LV-HuR; Fig. 5A and B), including 1,449 upregulated genes and
1,498 downregulated mRNAs. A total of 134 differentially expressed
genes were detected in the silenced group (sh-HuR vs. sh-NC),
including 99 upregulated and 35 downregulated genes (Fig. 5C and D). Combined sequencing data indicated
that HuR positively regulated 52 mRNAs and negatively regulated 25
mRNAs (Fig. S1). These mRNAs
included CALB2, INHBA, ACSS2, OLFM4 and TMPRSS11E.
The effect of HuR on skin cell
radiosensitivity
Radiation-induced skin injury is a common
complication after radiation accidents, tumor radiation therapy and
bone marrow transplantation pretreatment (29,30).
ROS produced by ionizing radiation in the skin are an important
cause of skin injury (31,32). The present study explored the
effect of HuR on intracellular ROS levels after radiation and the
results showed that the level of ROS in the sh-HuR group was
significantly increased after 20 Gy X-ray irradiation (Fig. 6A). In addition, we investigated the
effects of HuR on the proliferation and apoptosis of HaCaT and WS1
cells after irradiation. The results of EdU staining showed that
overexpression of HuR promoted cell proliferation after 20 Gy X-ray
irradiation (Fig. 6B) and
downregulation of HuR enhanced inhibition of cell proliferation
after irradiation (Fig. 6C). The
results of a colony formation assay showed that overexpression of
HuR significantly increased cell survival after X-ray irradiation
at 20 Gy (Fig. 6D); however,
downregulation of HuR significantly reduced cell survival after
irradiation (Fig. 6E). The results
of a cellular apoptosis assay showed that overexpression of HuR
reduced apoptosis of HaCaT and WS1 cells after X-ray irradiation at
20 Gy (Fig. 6F) and HuR silencing
increased apoptosis after irradiation (Fig. 6G). Overall, these results indicated
that HuR modulated radiosensitivity of skin cells in
vitro.
Characterization of HuR-interacting
mRNAs after irradiation
The present study used RIP combined with RNA-Seq to
detect changes in the HuR-binding sequence and expression profile
of WS1 cells after 0 or 5 Gy irradiation (Fig. 7A). The RNA-Seq data are accessible
from the GEO database (accession number GSE111823). The results
indicated the lack of significant changes in the binding locations
(Fig. 7C). Hundreds of HuR-binding
sites were changed after 5 Gy irradiation (Fig. 7B). Combined data of RIP-Seq and
RNA-Seq indicated that 14 mRNAs were associated with changes in HuR
binding and expression after irradiation (Fig. 7D), indicating that the abundance of
these mRNAs may be modulated by HuR binding (Figs. S2 and S3). For example, the levels of NLRP10,
SERPINE1 and CLCA2 mRNAs were increased in WS1 cells after 5 Gy
irradiation. NLRP10 is expressed at high levels in the skin and
contributes to cell-autonomous responses against invasive bacteria.
HUR exerted its influence through wide spectrum targets, which was
consistent with its behavior in other types of cells. These genes
were explored by RIP-Seq and RNA Seq in the present study. However,
which one could be the exact target for HUR that leads to cell
proliferation, senescence and radiosensitivity needs to be further
studied.
Discussion
HuR is an RNA-binding protein that recognizes
U/AU-rich elements in diverse RNAs and post-transcriptionally
regulates the fate of target RNAs. HuR regulates cellular responses
to differentiation, senescence, inflammatory factors and immune
stimuli by tightly controlling the post-transcriptional fate of
specific mRNAs (33,34). HuR is expressed at high levels in a
number of types of cancers and is believed to promote tumorigenesis
by interacting with mRNAs encoding proteins implicated in cell
proliferation and survival, angiogenesis, invasion and metastasis
(23). For example, HuR serves
pro-proliferative and anti-apoptotic roles in colorectal cancer
cells (5). In the present study,
the role of HuR in modulating proliferation and senescence of skin
cells was explored using HuR overexpression and silencing.
Consistent with previous reports in other cell lines, HuR
facilitated the proliferation of skin cells and inhibited their
senescence, suggesting a new therapeutic strategy for cosmetic
treatments and to combat skin injury.
HuR has been shown to associate with numerous
transcripts, including coding and noncoding transcripts, and
controls their splicing, localization, stability and translation
(16). HuR is predominantly
nuclear in unstimulated cells and shuttles to the cytoplasm in
response to various stimuli, including stress signals and mitogens
(5). HuR stabilizes a large subset
of target mRNAs, including mRNAs implicated in various pathologies,
particularly cancer and inflammation (16). Known HuR-regulated mRNAs include
cyclin family proteins (A2, B1, E1 and D1), cyclin-dependent kinase
inhibitor p21, inducible nitric oxide synthase,
granulocyte-macrophage colony-stimulating factor, murine double
minute 2, VEGF, TGF-β, TNF-α, Bcl-2, COX-2, p53, Toll-like receptor
4, MMP-9 and MAPK (35). In the
present study, 77 mRNAs were positively or negatively associated
with HuR expression levels. However, the majority of these mRNAs
were not reported to be regulated by HuR in other types of cells,
indicating the presence of a cell type- or organ-specific
regulatory network. Calcium retinal protein 2 (calretinin, calb2),
a calcium binding protein, is mainly expressed in the nervous
system, ovary, adrenal glands and testis (36). This protein functions as a sensor
and buffer of intracellular Ca2+ to prevent
Ca2+ overloading. Increasing evidence indicates that
calretinin is involved in multiple physiological processes by
regulating Ca2+ (37).
Ca2+ has been identified as a messenger that coordinates
endoplasmic reticulum (ER)-mitochondrial interactions that regulate
apoptosis (38). Induction of ER
stress has been reported to enhance chemotherapy sensitization
(39). Therefore, the regulation
of Ca2+ release is tightly controlled and a number of
Ca2+-binding proteins, such as calretinin, may function
downstream of ER Ca2+ release to modulate apoptosis or
other cell functions. NLRP10 is involved in the innate immune
response by contributing to proinflammatory cytokine release in
response to invasive bacterial infection (40) and contributes to T-cell-mediated
inflammatory responses in the skin. SERPINE1 is required for
stimulation of keratinocyte migration during cutaneous injury
repair (41) and is involved in
cellular and replicative senescence (42). These mRNAs may be novel and direct
targets of HuR for cosmetic treatments and to combat skin
injury.
Skin aging is a slow and complex process subjected
to intrinsic alterations at the cellular, molecular and genetic
levels and by exposure to extrinsic factors (43). Aging affects all layers of the
skin; in particular, the underlying epidermis and dermis undergo
numerous cellular and molecular changes due to aging (44). The skin, as an external organ, is
inevitably exposed to radiation that induces various skin
reactions. Ionizing radiation is widely used in military,
industrial, agricultural, medical and pollution treatments.
Ionizing radiation causes both direct and indirect damage to the
cells (45). DNA is the major
cellular target of ionizing radiation and can be damaged directly
by ionizing radiation, resulting in DNA double-strand breaks, or
indirectly through the generation of ROS (46). HuR facilitates radiation resistance
in triple-negative breast cancer (TNBC) by protecting against
radiation-induced DNA damage (47). Silencing HuR results in
radiosensitization of TNBC cells due to enhanced ROS accumulation
along with inhibition of the thioredoxin reductase system (47).
The results of RIP-Seq analyses identified 14 mRNAs
that preferentially interacted with HuR after ionizing radiation. A
number of these genes are known to participate in certain key
cellular processes, including proliferation, apoptosis, immune
response and metastasis. For example, ENDOCAN is a novel human
endothelial cell-specific molecule mainly expressed in endothelial
cells in various tissues (48) and
is expressed and secreted by vascular endothelial cells, including
the skin (49). ENDOCAN is known
to be involved in the development of vascular tissue in health and
disease (50). IL-33 has been
reported to be constitutively expressed in the nuclei of
endothelial and epithelial cells and is released into the
extracellular space as an alarmin after tissue damage to alert the
immune system (51). The level of
alarmin IL-33 is quickly elevated in the bone marrow serum after
ionizing radiation and confers radioprotection (52). Studies have shown that PID1
inhibits the phosphorylation of Akt and Erk, suggesting that PID1
may modulate the signaling pathways involved in cell proliferation
and survival (53-55).
Previous studies have shown that CLCA2 is a stress-inducible gene
and is strongly upregulated by p53 in response to cell detachment,
DNA damage and other stressors (56,57).
CLCA2 is a novel UVB target gene that may serve a role in epidermal
differentiation and UV-dependent skin malignancies (58). These findings suggest that these 14
HuR-interacting mRNAs may contribute to enhanced regeneration and
damage repair potential following irradiation. Therefore, HuR is
likely to modulate skin cell radiosensitivity through complex
mechanisms.
In order for HuR to exert its effects, it must
dimerize prior to binding its targets. Thus, targeting of HuR may
offer the ability to modulate a broad range of HuR-mediated
effects, by interfering with the actions of a single target. HUR
served a positive role in modulating proliferation, senescence and
radiosensitivity of skin cells and modulated downstream mRNAs
implicated in multiple pathways in skin cells, providing a new
therapeutic strategy for cosmetic treatments and to combat skin
injury.
Supplementary Material
77 common dysregulated genes in LV-HuR
vs. LV-NC and sh-HuR vs. sh-NC groups. HuR, Human antigen R; ROS,
reactive oxygen species; sh, short hairpin; NC, negative control;
LV, lentiviral.
KEGG pathway enrichment analysis of
mRNA sequencing profiles. (A) KEGG pathway distribution diagram of
upregulated and downregulated mRNA in LV-HuR vs. LV-NC and sh-HuR
vs. sh-NC groups. (B) KEGG pathway enrichment of the dysregulated
mRNAs from sh-HuR vs. sh-NC group. (C) KEGG pathway enrichment of
the dysregulated mRNAs from LV-HuR vs. LV-NC group. KEGG, Kyoto
Encyclopedia of Genes and Genomes; HuR, Human antigen R; ROS,
reactive oxygen species; sh, short hairpin; NC, negative control;
LV, lentiviral.
The list of upregulated genes of HuR
RIP and corresponding mRNA after 5 Gy X-ray irradiation. HuR, Human
antigen R; RIP, RNA immunoprecipitation.
Targeting sequences of the four shRNAs
against HuR used in this study.
Acknowledgements
Not applicable.
Funding
Funding: The present study supported by the National Natural
Science Foundation of China (grant nos. 32071238, 82073477 and
31770909), Natural Science Foundation of Sichuan Province (grant
no. 2020YJ0194), Natural Science Project of Chengdu Medical College
(grant nos. CYZ19-31 and CYZZD20-01) and Young Talent Program of
China National Nuclear Corporation.
Availability of data and materials
The datasets generated and/or analyzed during the
current study are available in the Gene Expression Omnibus
repository, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?Acc=GSE161811
and GSE111823.
Authors' contributions
DY and SZ confirm the authenticity of all the raw
data. DY and SZ conceived and designed the study. DY, KF and ZJ
carried out the molecular biology studies. DY, YF and TY drafted
the manuscript and figures, prepared the samples for RNA-seq and
carried out the senescence studies. SZ, YS and JZ performed the
statistical analysis. DY and SZ edited the manuscript. All authors
read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Pope SD and Medzhitov R: Emerging
principles of gene expression programs and their regulation. Mol
Cell. 71:389–397. 2018.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Zerdes I, Matikas A, Bergh J, Rassidakis G
and Foukakis : Genetic, transcriptional and
post-translational regulation of the programmed death protein
ligand 1 in cancer: Biology and clinical correlations. Oncogene.
37:4639–4661. 2018.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Zhang Q and Cao X: Epigenetic regulation
of the innate immune response to infection. Nat Rev Immunol.
19:417–432. 2019.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Hartman ML and Czyz M: MITF in melanoma:
Mechanisms behind its expression and activity. Cell Mol Life Sci.
72:1249–1260. 2015.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Grammatikakis I, Abdelmohsen K and Gorospe
M: Posttranslational control of HuR function. Wiley Interdiscip Rev
RNA. 8(e1372)2017.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Moutal A, White KA, Chefdeville A,
Laufmann RN, Vitiello PF, Feinstein D, Weimer JM and Khanna R:
Dysregulation of CRMP2 post-translational modifications drive its
pathological functions. Mol Neurobiol. 56:6736–6755.
2019.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Buuh ZY, Lyu Z and Wang RE: Interrogating
the roles of post-translational modifications of non-histone
proteins. J Med Chem. 61:3239–3252. 2018.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Schultz CW, Preet R, Dhir T, Dixon DA and
Brody JR: Understanding and targeting the disease-related RNA
binding protein human antigen R (HuR). Wiley Interdiscip Rev RNA.
11(e1581)2020.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Wang X, Zhou X, Li G, Zhang Y, Wu Y and
Song W: Modifications and trafficking of APP in the pathogenesis of
Alzheimer's disease. Front Mol Neurosci. 10(294)2017.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Han ZJ, Feng YH, Gu BH, Li YM and Chen H:
The post-translational modification, SUMOylation, and cancer. Int J
Oncol. 52:1081–1094. 2018.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Estevez A, Zhu D, Blankenship C and Jiang
J: Molecular interrogation to crack the case of O-GlcNAc.
Chemistry. 26:12086–12100. 2020.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Zhou H, Rao Y, Sun Q, Liu Y, Zhou X, Chen
Y and Chen J: MiR-4458/human antigen R (HuR) modulates PBX3 mRNA
stability in melanoma tumorigenesis. Arch Dermatol Res.
312:665–673. 2020.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Andrade D, Mehta M, Griffith J, Oh S,
Corbin J, Babu A, De S, Chen A, Zhao YD, Husain S, et al: HuR
reduces radiation-induced DNA damage by enhancing expression of
ARID1A. Cancers. 11(2014)2019.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Xu X, Song C, Chen Z, Yu CX, Wang Y, Tang
Y and Luo J: Downregulation of HuR inhibits the progression of
esophageal cancer through interleukin-18. Cancer Res Treat.
50:71–87. 2018.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Mostaan LV, Tabari A, Amiri P, Ashtiani
MK, Mahdkhah A, Yazdani N, Khaniki M, Tabari A, Tavakkoly-Bazzaz J
and Amoli MM: Survivin gene polymorphism association with tongue
squamous cell carcinoma. Genet Test Mol Biomarkers. 17:74–77.
2013.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Srikantan S and Gorospe M: HuR function in
disease. Front Biosci (Landmark Ed). 17:189–205. 2012.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Hinman MN and Lou H: Diverse molecular
functions of Hu proteins. Cell Mol Life Sci. 65:3168–3181.
2008.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Wang J, Wang B, Bi J and Zhang C:
Cytoplasmic HuR expression correlates with angiogenesis,
lymphangiogenesis, and poor outcome in lung cancer. Med Oncol. 28
(Suppl 1):S577–S585. 2011.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Zhou Y, Chang R, Ji W, Wang N, Qi M, Xu Y,
Guo J and Zhan L: Loss of scribble promotes snail translation
through translocation of HuR and enhances cancer drug resistance. J
Biol Chem. 291:291–302. 2016.PubMed/NCBI View Article : Google Scholar
|
|
20
|
To KK, Leung WW and Ng SS: Exploiting a
novel miR-519c-HuR-ABCG2 regulatory pathway to overcome
chemoresistance in colorectal cancer. Exp Cell Res. 338:222–231.
2015.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Zhang S, Wang W, Gu Q, Xue J, Cao H, Tang
Y, Xu X, Cao J, Zhou J, Wu J and Ding WQ: Protein and miRNA
profiling of radiation-induced skin injury in rats: The protective
role of peroxiredoxin-6 against ionizing radiation. Free Radic Biol
Med. 69:96–107. 2014.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Wang W, Furneaux H, Cheng H, Caldwell MC,
Hutter D, Liu Y, Holbrook N and Gorospe M: HuR regulates p21 mRNA
stabilization by UV light. Mol Cell Biol. 20:760–769.
2000.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Zhang J and Bowden GT: UVB irradiation
regulates Cox-2 mRNA stability through AMPK and HuR in human
keratinocytes. Mol Carcinog. 47:974–983. 2008.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Brand RM, Epperly MW, Stottlemyer JM,
Skoda EM, Gao X, Li S, Huq S, Wipf P, Kagan VE, Greenberger JS and
Falo LD Jr: A topical mitochondria-targeted redox-cycling nitroxide
mitigates oxidative stress-induced skin damage. J Invest Dermatol.
137:576–586. 2017.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Xue J, Yu C, Sheng W, Zhu W, Luo J, Zhang
Q, Yang H, Cao H, Wang W, Zhou J, et al: The Nrf2/GCH1/BH4 axis
ameliorates radiation-induced skin injury by modulating the ROS
cascade. J Invest Dermatol. 137:2059–2068. 2017.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Bellei B and Picardo M: Premature cell
senescence in human skin: Dual face in chronic acquired pigmentary
disorders. Ageing Res Rev. 57(100981)2020.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Toutfaire M, Bauwens E and
Debacq-Chainiaux F: The impact of cellular senescence in skin
ageing: A notion of mosaic and therapeutic strategies. Biochem
Pharmacol. 142:1–12. 2017.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Lee BY, Han JA, Im JS, Morrone A, Johung
K, Goodwin EC, Kleijer WJ, DiMaio D and Hwang ES:
Senescence-associated beta-galactosidase is lysosomal beta-
galactosidase. Aging Cell. 5:187–195. 2006.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Kiang JG and Olabisi AO: Radiation: A
poly-traumatic hit leading to multi-organ injury. Cell Biosci.
9(25)2019.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Soriano JL, Calpena AC, Souto EB and
Clares B: Therapy for prevention and treatment of skin ionizing
radiation damage: A review. Int J Radiat Biol. 95:537–553.
2019.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Wang Y, Tu W, Tang Y, Momeni A, Longaker
MT and Wan DC: Prevention and treatment for radiation-induced skin
injury during radiotherapy. Radiat Med Protect. 1:60–68. 2020.
|
|
33
|
Lal P, Cerofolini L, D'Agostino VG, Zucal
C, Fuccio C, Bonomo I, Dassi E, Giuntini S, Maio DD, Vishwakarma V,
et al: Regulation of HuR structure and function by
dihydrotanshinone-I. Nucleic Acids Res. 45:9514–9527.
2017.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Lv Z, He K, Shi L, Shi K, Jiang T and Chen
Y: Interaction between C2ORF68 and HuR in human colorectal cancer.
Oncol Rep. 41:1918–1928. 2019.PubMed/NCBI View Article : Google Scholar
|
|
35
|
de Silanes IL, Fan J, Yang X, Zonderman
AB, Potapova O, Pizer ES and Gorospe M: Role of the RNA-binding
protein HuR in colon carcinogenesis. Oncogene. 22:7146–7154.
2003.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Hack NJ, Wride MC, Charters KM, Kater SB
and Parks TN: Developmental changes in the subcellular localization
of calretinin. J Neurosci. 20(RC67)2000.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Schwaller B: Calretinin: From a ‘simple’
Ca(2+) buffer to a multifunctional protein implicated in many
biological processes. Front Neuroanat. 8(3)2014.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Boehning D, Patterson RL, Sedaghat L,
Glebova NO, Kurosaki T and Snyder SH: Cytochrome c binds to
inositol (1,4,5) trisphosphate receptors, amplifying
calcium-dependent apoptosis. Nat Cell Biol. 5:1051–1061.
2003.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Wu Y, Fabritius M and Ip C:
Chemotherapeutic sensitization by endoplasmic reticulum stress:
Increasing the efficacy of taxane against prostate cancer. Cancer
Biol Ther. 8:146–152. 2009.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Lautz K, Damm A, Menning M, Wenger J, Adam
AC, Zigrino P, Kremmer E and Kufer TA: NLRP10 enhances
Shigella-induced pro-inflammatory responses. Cell Microbiol.
14:1568–1583. 2012.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Providence KM, Higgins SP, Mullen A,
Battista A, Samarakoon R, Higgins CE, Wilkins-Port CE and Higgins
PJ: SERPINE1 (PAI-1) is deposited into keratinocyte migration
‘trails’ and required for optimal monolayer wound repair. Arch
Dermatol Res. 300:303–310. 2008.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Kortlever RM, Higgins PJ and Bernards R:
Plasminogen activator inhibitor-1 is a critical downstream target
of p53 in the induction of replicative senescence. Nat Cell Biol.
8:877–884. 2006.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Deotto ML, Spiller A, Sernicola A and
Alaibac M: Bullous pemphigoid: An immune disorder related to aging.
Exp Ther Med. 23(50)2022.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Bhatia E, Kumari D, Sharma S, Ahamad N and
Banerjee R: Nanoparticle platforms for dermal antiaging
technologies: Insights in cellular and molecular mechanisms. Wiley
Interdiscip Rev Nanomed Nanobiotechnol. 14(e1746)2022.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Burgio E, Piscitelli P and Migliore L:
Ionizing radiation and human health: Reviewing models of exposure
and mechanisms of cellular damage. An epigenetic perspective. Int J
Environ Res Public Health. 15(1971)2018.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Barzilai A and Yamamoto K: DNA damage
responses to oxidative stress. DNA Repair (Amst). 3:1109–1115.
2004.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Mehta M, Basalingappa K, Griffith JN,
Andrade D, Babu A, Amreddy N, Muralidharan R, Gorospe M, Herman T,
Ding WQ, et al: HuR silencing elicits oxidative stress and DNA
damage and sensitizes human triple-negative breast cancer cells to
radiotherapy. Oncotarget. 7:64820–64835. 2016.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Suzuki H, Miyagaki T, Otobe S, Nakajima R,
Oka T, Takahashi N, Kabasawa M, Suga H, Yoshizaki A, Asano Y, et
al: Increased endocan expression in lesional skin and decreased
endocan expression in sera in atopic dermatitis. J Dermatol.
44:1392–1395. 2017.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Zhang H, Yushkevich PA and Gee JC:
Deformable registration of diffusion tensor MR images with explicit
orientation optimization. Med Image Comput Comput Assist Interv.
8:172–179. 2005.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Roudnicky F, Poyet C, Wild P, Krampitz S,
Negrini F, Huggenberger R, Rogler A, Stöhr R, Hartmann A,
Provenzano M, et al: Endocan is upregulated on tumor vessels in
invasive bladder cancer where it mediates VEGF-A-induced
angiogenesis. Cancer Res. 73:1097–1106. 2013.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Moussion C, Ortega N and Girard JP: The
IL-1-like cytokine IL-33 is constitutively expressed in the nucleus
of endothelial cells and epithelial cells in vivo: A novel
‘alarmin’? PLoS One. 3(e3331)2008.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Kim J, Kim W, Le HT, Moon UJ, Tran VG, Kim
HJ, Jung S, Nguyen QT, Kim BS, Jun JB, et al: IL-33-induced
hematopoietic stem and progenitor cell mobilization depends upon
CCR2. J Immunol. 193:3792–3802. 2014.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Erdreich-Epstein A, Robison N, Ren X, Zhou
H, Xu J, Davidson TB, Schur M, Gilles FH, Ji L, Malvar J, et al:
PID1 (NYGGF4), a new growth-inhibitory gene in embryonal brain
tumors and gliomas. Clin Cancer Res. 20:827–836. 2014.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Bonala S, McFarlane C, Ang J, Lim R, Lee
M, Chua H, Lokireddy S, Sreekanth P, Leow MKS, Meng KC, et al: Pid1
induces insulin resistance in both human and mouse skeletal muscle
during obesity. Mol Endocrinol. 27:1518–1535. 2013.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Zhang CM, Chen XH, Wang B, Liu F, Chi X,
Tong ML, Ni YH, Chen RH and Guo XR: Over-expression of NYGGF4
inhibits glucose transport in 3T3-L1 adipocytes via attenuated
phosphorylation of IRS-1 and Akt. Acta Pharmacol Sin. 30:120–124.
2009.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Walia V, Ding M, Kumar S, Nie D, Premkumar
LS and Elble RC: hCLCA2 is a p53-inducible inhibitor of breast
cancer cell proliferation. Cancer Res. 69:6624–6633.
2009.PubMed/NCBI View Article : Google Scholar
|
|
57
|
Ramena G, Yin Y, Yu Y, Walia V and Elble
RC: CLCA2 interactor EVA1 is required for mammary epithelial cell
differentiation. PLoS One. 11(e0147489)2016.PubMed/NCBI View Article : Google Scholar
|
|
58
|
Bart G, Hämäläinen L, Rauhala L, Salonen
P, Kokkonen M, Dunlop TW, Pehkonen P, Kumlin T, Tammi MI,
Pasonen-Seppänen S and Tammi RH: rClca2 is associated with
epidermal differentiation and is strongly downregulated by
ultraviolet radiation. Br J Dermatol. 171:376–387. 2014.PubMed/NCBI View Article : Google Scholar
|