Introduction
Cervical cancer is one of the most prevalent
malignant tumours and one of the most serious diseases threatening
the health of women worldwide (1).
According to the World Health Organization, there were an estimated
570,000 cases of cervical cancer and 311,000 deaths globally in
2018, making it the fourth most commonly diagnosed cancer and the
fourth leading cause of cancer-related deaths in women (2). Cervical cancer is caused by
persistent human papillomavirus (HPV) infections, particularly
HPV16 and HPV18(3). Surgery,
pharmaceutical treatment and radiotherapy are the currently
available therapeutic options for cervical cancer; however, they
are ineffective for treating advanced, metastatic, or recurrent
tumours. Multiple factors contribute to the development and
progression of cervical cancer. Therefore, the most promising
strategy for the treatment of cervical cancer is to analyse the
mechanism underlying cervical cancer development by identifying the
relevant signal transduction processes and targeted molecular
sites.
Transmembrane protein 121 (TMEM121) is a gene
product isolated from the chicken heart via subtractive
hybridisation (4). The human
TMEM121 protein is a 319-amino acid, six-transmembrane protein with
a proline-rich C-terminal motif and an N-terminal extracellular
signal-regulated kinase binding domain (D-domain) that is highly
conserved in the evolution of different species (4,5). A
previous study by the authors revealed that TMEM121 is
highly expressed in adult mouse hearts and acts as an inhibitor of
pathologic cardiac hypertrophy (6). However, there have been few reports
linking TMEM121 to cancer. In the present study, it was
determined that TMEM121 may play a role in the development
of cervical cancer.
Materials and methods
Tumour Immune Estimation Resource
(TIMER) 2.0
The Tumour Immune Estimation Resource (TIMER;
http://timer.cistrome.org/) is a
comprehensive resource for the systematic analysis of immune
infiltrates across various cancer types from The Cancer Genome
Atlas (TCGA) database, with 10,897 samples from 32 cancer types
(7). In the present study, the
Gene_DE module was used to assess the differential expression of
the TMEM121 gene in tumours and adjacent normal tissues
across all TCGA tumours. Statistical significance was calculated by
Wilcoxon rank sum test.
cBioPortal
The cBioPortal for Cancer Genomics (http://cbioportal.org) platform is a comprehensive web
resource containing cancer genomics data from multiple platforms
(8). The summary, plots and
co-expression of cancer types were used to analyse the
TMEM121 gene expression in cancer.
LinkedOmics analysis
LinkedOmics is a publicly accessible portal that
incorporates multi-omics and clinical data from the TCGA project
for 32 cancer types and 11,158 patients (9). In this study, the LinkFinder tab of
LinkedOmics was used to identify differentially expressed genes
(DEGs) associated with TMEM121. The search and target
datasets are derived from ribonucleic acid sequencing (RNA-SEQ).
The results were statistically analysed using Pearson's and
Spearman's correlation tests. The over-representation enrichment
analysis and gene set enrichment analysis (GSEA) were completed
using the LinkInterpreter tab.
Kaplan-Meier plotter
The Kaplan-Meier plotter (http://kmplot.com/analysis) can assess the impacts of
54K genes (mRNA, miRNA and protein) on survival in 21 cancer types.
The database sources include Gene Expression Omnibus, European
Genome-phenome Archive and TCGA. The main purpose of this tool is
to identify and validate survival biomarkers based on a
meta-analysis. The Kaplan-Meier plots were generated using the
‘survplot’ R package (http://www.cbs.dtu.dk/~eklund/survplot/) (10). Log rank testing was used to analyze
statistical significance.
UALCAN analysis
UALCAN is an interactive web portal that performs
in-depth analyses of TCGA gene expression data using TCGA level 3
RNA-seq and clinical data from 31 cancer types (11). In this study, UALCAN was used to
analyse the relative TMEM121 gene expression in normal as
well as cervical squamous cell carcinoma and endocervical
adenocarcinoma (CESE) tumour subgroups.
Cell cultures and transfection
HeLa cells were cultured in Dulbecco's modified
Eagle's medium (Gibco; Thermo Fisher Scientific, Inc.) containing
10% foetal bovine serum [Serana (WA) Pty, Ltd.] and incubated at
37˚C and 5% CO2. HeLa cells were transfected with
Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.),
according to the manufacturer's instructions, using 5 µg plasmid
per dish. TMEM121 overexpression was performed by inserting the CDS
sequence of human TMEM121 into pCMV-Tag2B (Agilent Technologies,
Inc.) to prepare pCMV-Tag2B-TMEM121. TMEM121-specific shRNA
sequence
5'-GATCCCCGCTTCTTCATCTCAGGAATTTCTAGAGAATTCCTGAGATGAAGAAGCTTTTTCTCGA-3'
was inserted into pSUPER (Oligoengine, Inc.) to produce
pSUPER-TMEM121 for TMEM121 knockdown. Negative controls for TMEM121
overexpression and knockdown were empty vectors of pCMV-Tag2B and
pSUPER, respectively. The temperature conditions for transfection
were the same as for cell culture for 6 h. Fresh medium was
replaced 6 h after transfection. CCK-8 and cell scratch assays were
performed 24 h after transfection, and western blotting was
performed 48 h after transfection.
Western blotting
Total cellular proteins were extracted with RIPA
lysis buffer (Elabscience Biotechnology, Inc.), and the protein
amount was quantified by BCA assay. Protein (10-25 µl of each
sample) was loaded on a 10% acrylamide gel. Electrophoresis was
conducted at a constant pressure of 80 V until the target protein
bands were separated. The protein was then transferred to a
nitrocellulose (NC) filter membrane under a constant current of 300
mA. The NC membrane was immersed in Tris-buffered saline with 0.05%
Tween-20 (TBST) containing 5% skim milk at room temperature for 1.5
h, and the excess milk was cleaned with TBST. The NC membrane was
incubated at room temperature for 90 min with a primary antibody
(diluted at a 1:1,000 by volume ratio), and then rinsed three times
with TBST for 5 min each time. Subsequently, the NC membrane was
incubated at room temperature for 90 min transferred with the
secondary antibody (diluted at a 1:5,000 by volume ratio), and then
rinsed with TBST three times for 5 min each time. Thereafter, a
developing solution was dripped onto the membrane for imaging.
The anti-retinoblastoma (RB) (cat. no. 9313),
anti-B-cell lymphoma 2 (BCL-2) (cat. no. 4223), anti-cleaved
caspase-3 (cat. no. 9664), anti-cyclin D1 (cat. no. 2922),
anti-cyclin E1 (cat. no. 20808), anti-cyclin E2 (cat. no. 4132),
anti-light chain 3 α/β (LC3A/B) (cat. no. 4108),
anti-phosphorylated c-Jun N-terminal kinase (p-JNK) (cat. no.
4668), anti-JNK (cat. no. 9252), anti-p-p38 (cat. no. 4511),
anti-p38 (cat. no. 8690), anti-p-AKT (cat. no. 9271) and anti-AKT
(cat. no. 9272) antibodies (all (1:1,000) were purchased from Cell
Signaling Technology, Inc. The anti-p53 antibody was purchased from
ProteinTech Group, Inc. (1:1,000; cat. no. 10442-1-AP), and the
anti-E-cadherin antibody was purchased from Boster Biological
Technology (1:1,000; cat. no. PB9561). GAPDH and α-tubulin were
used as internal references, and antibodies against them were
purchased from ProteinTech Group, Inc. (1:1,000; cat. nos.
60004-1-Ig and 66031-1-Ig, respectively). Goat anti-mouse IgG
HRP-conjugated and goat anti-rabbit IgG HRP-conjugated secondary
antibodies were purchased from CWBio (1:5,000; cat. nos. CW0102 and
CW0103).
Cell Counting Kit-8 (CCK-8) assay
The cell viability assays were performed using Cell
Counting Kit-8 (CCK-8; Beyotime Institute of Biotechnology). In
brief, 20 µl CCK-8 reagent was applied per well at 24, 48 and 72 h
following plasmid vector transfection, and the cells were incubated
at 37˚C for 1 h in a humidified CO2 incubator. The
optical density of the medium was then measured as an absorbance at
a wavelength of 450 nm on a microplate reader (Bio-Rad
Laboratories, Inc.).
Cell scratch assay
Transfected cells were seeded on 6-well plates and
incubated to almost 100% confluence. A pipette tip perpendicular to
the cell plane was used to scratch the cell monolayer to create
scratches. After the scratches were completed, the cells were
incubated in 37˚C 5% CO2 with serum-free medium (using
the same reagents as aforementioned). Confluence on either side of
the wound at the start of the assay. The cells in the culture plate
were observed under an inverted microscope and images were captured
at 0, 24, 48 and 72 h. ImageJ 1.53c software (National Institutes
of Health) was then used for cell scratch area analysis. The
scratch healing rate was calculated according to the scratch area:
(Original scratch area-scratch area at a certain time)/original
scratch area x100.
Statistical analysis
Graphpad Prism 8.0 software (GraphPad Software,
Inc.) was used for statistical analysis. Data are expressed as the
mean ± SD. The unpaired t-test was used to evaluate the differences
between two groups. Wilcoxon rank-sum test was used in TIMER 2.0.
P<0.05 was used to indicate a statistically significant
difference. Each experiment was repeated at least three times. Log
rank testing was used to analyze survival curve significance.
Pearson's and Spearman's correlation tests were used in LinkedOmics
analysis.
Results
TMEM121 expression in tumours
TIMER was used to analyse and compare TMEM121
expression in various human tumour tissues with that in normal
tissues. As shown in Fig. 1,
TMEM121 expression was significantly downregulated in CESC,
kidney chromophobe, kidney renal papillary cell carcinoma, liver
hepatocellular carcinoma, and uterine corpus endometrial carcinoma
compared with that in normal tissues.
Subsequently, UALCAN was used to categorise CESC
samples derived from TCGA database based on sample type, cancer
stage, weight, age, tumour grade and lymph node metastatic status
in order to analyse the TMEM121 expression among various
categories. The results revealed that the TMEM121 expression
in the various CESC subgroups was lower than that in normal tissues
(Fig. 2). Furthermore, the
methylation levels of the TMEM121 gene promoter were higher
in cancerous tissues in various categories than those in normal
tissues (Fig. 3).
Association between TMEM121 expression
and survival in patients with CESC
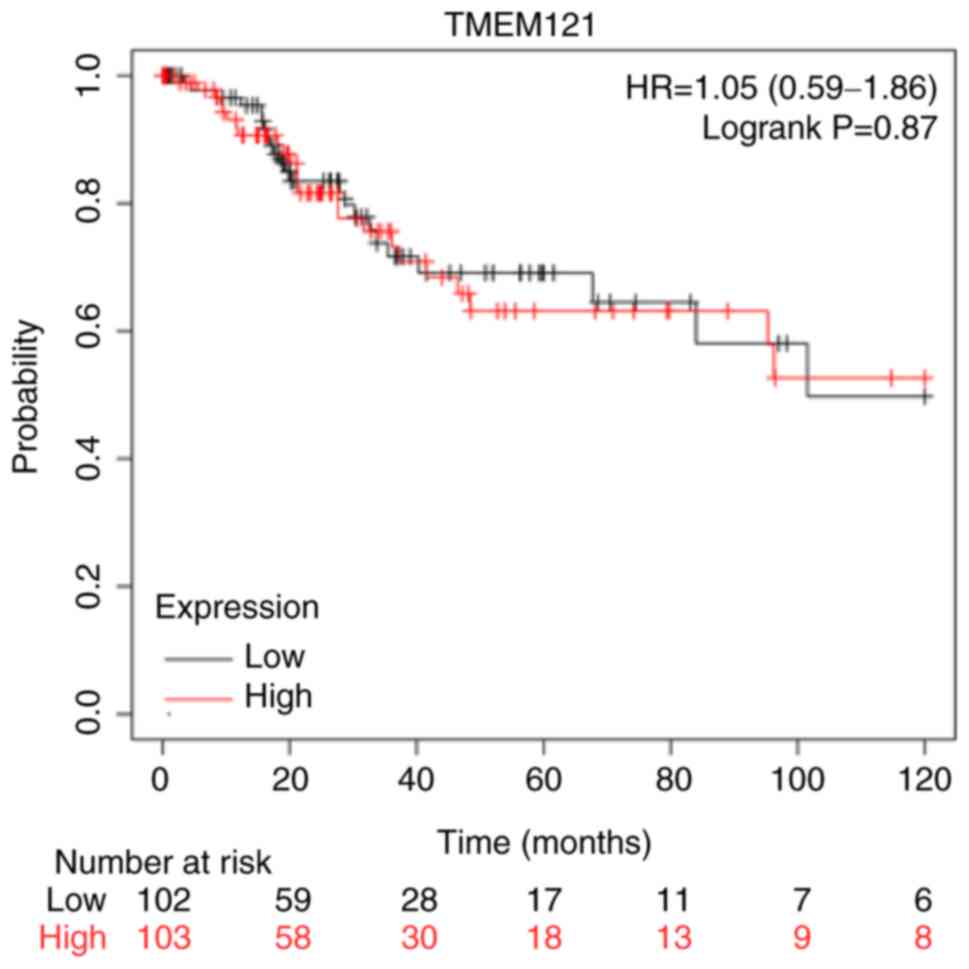
The Kaplan-Meier plotter was used to investigate the
prognostic significance of TMEM121 expression in patients
with CESC. The survival analysis revealed that there was no
significant difference in the overall survival between the low and
high TMEM121 expression groups of patients with CESC
(P=0.87) (Fig. 4).
DEGs associated with TMEM121
In CESC, DEGs associated with TMEM121 were
identified using LinkedOmics and displayed in heat maps (12). As illustrated in the volcano plot
(Fig. 5A), a total of 19,904 DEGs
were identified, with the top 50 significantly expressed, either
positively or negatively, and the related genes are displayed in
heat maps (Fig. 5B and C).
Functional enrichment analysis of
TMEM121
Next, the GSEA LinkInterpreter module was used for
Gene Ontology analysis to categorise TMEM121-related DEGs
into three categories based on their molecular functions,
biological processes they contribute to, and cellular locations
where they occur (13). In the
biological process category, the TMEM121-related DEGs were
primarily enriched in ‘monoamine transport’, nicotinamide adenine
dinucleotide/hydrogen dehydrogenase ‘(NADH) complex assembly’,
‘microvillus organisation’, and ‘cargo loading into the vesicle’
(Fig. 6A-a). In the category of
cellular components, they were primarily enriched in the
‘extracellular matrix’, ‘ribosome’, ‘endoplasmic reticulum exit
site’ and promyelocytic leukaemia ‘(PML) body’ (Fig. 6A-b). Furthermore, they were
primarily involved in molecular functions such as ‘structural
constituent of ribosome’, ‘extracellular matrix structural
constituent’, ‘ubiquitinyl hydrolase activity’ and ‘cysteine-type
peptidase activity’ (Fig.
6A-c).
Further analysis revealed the possible functional
pathways involved in the development and progression of CESC, which
include the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway
(‘ribosome’, ‘oxidative phosphorylation’, ‘glycosaminoglycan
biosynthesis’ and nucleotide-binding and oligomerisation domain
‘(NOD)-like receptor signalling pathway’; Fig. 6B-a), the PANTHER pathway (‘JAK/STAT
signalling pathway’, ‘phosphatidylinositol 3 (PI3)-kinase pathway’,
‘Toll receptor signalling pathway’ and platelet-derived growth
factor ‘(PDGF) signalling pathway’; Fig. 6B-b), and the WikiPathway
(‘cytoplasmic ribosomal proteins, ‘oxidative phosphorylation’,
epidermal growth factor/epidermal growth factor receptor
‘(EGF/EGFR) signalling pathway’, and the transforming growth factor
‘(TGF)-beta signalling pathway’; Fig.
6B-c). Therefore, the occurrence and progression of CESC
involve aberrant changes in multiple signalling pathways.
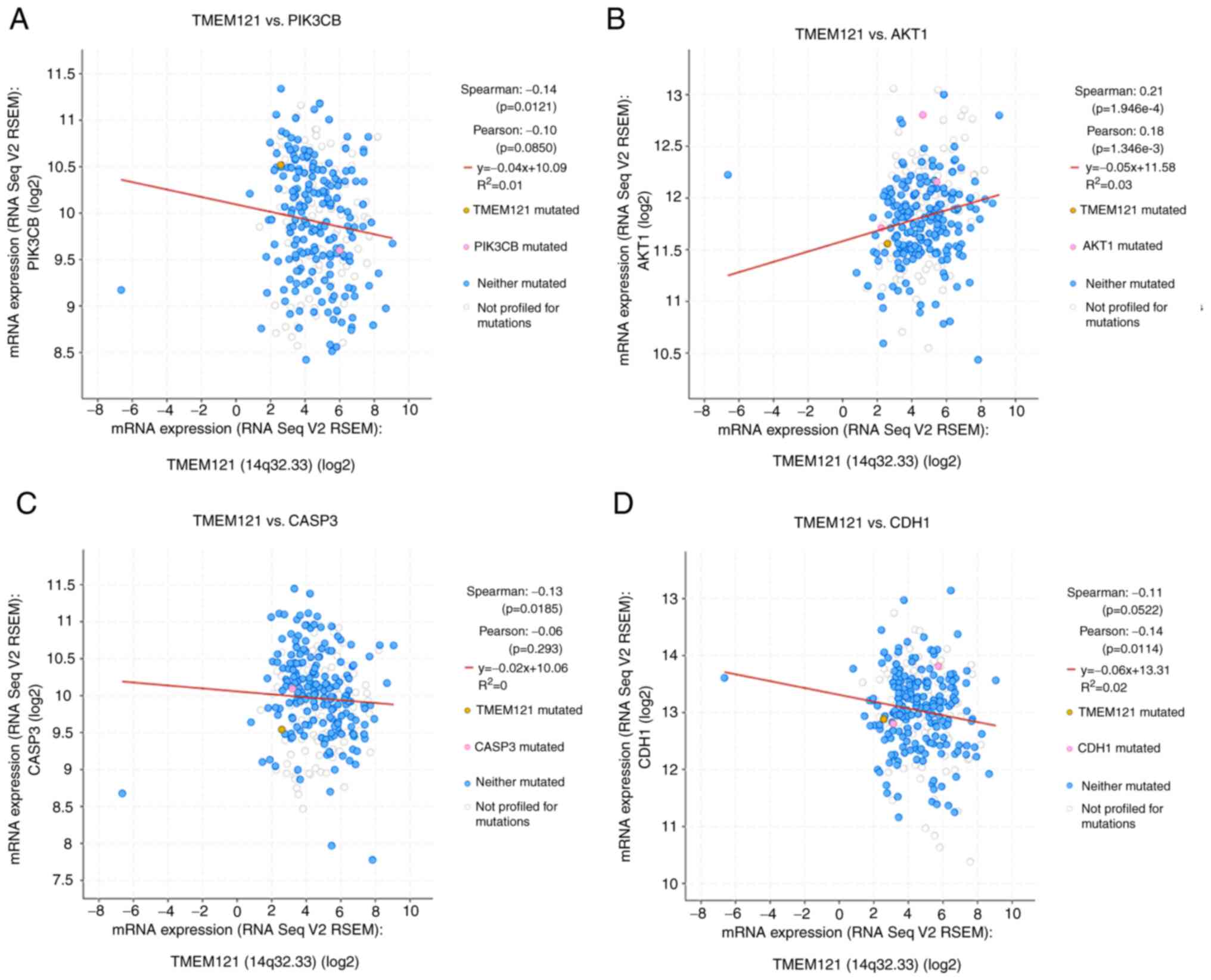
TMEM121 is associated with the
PI3K/AKT signalling pathway
To further identify TMEM121 as a putative
cancer regulator, the PI3K/AKT signalling pathway was investigated.
In CESC, the cBioPortal platform was used to correlate
TMEM121 expression levels with mutations of key molecules of
the PI3K/AKT signalling pathway, such as caspase-3 (CASP3) and
cadherin-1 (CDH1). Ultimately, PIK3CB, AKT1, CASP3 and CDH1 were
identified to be significantly correlated to TMEM121
expression in CESC (Fig. 7).
Pearson's correlation coefficient analysis revealed that the
expression level of TMEM121 was positively correlated with
the expression level of AKT1 and negatively correlated with the
expression levels of PIK3CB, CASP3 and CDH1. These results suggest
that TMEM121 influences cancer progression via the PI3K/AKT
signalling pathway and is closely associated to cell migration and
apoptosis.
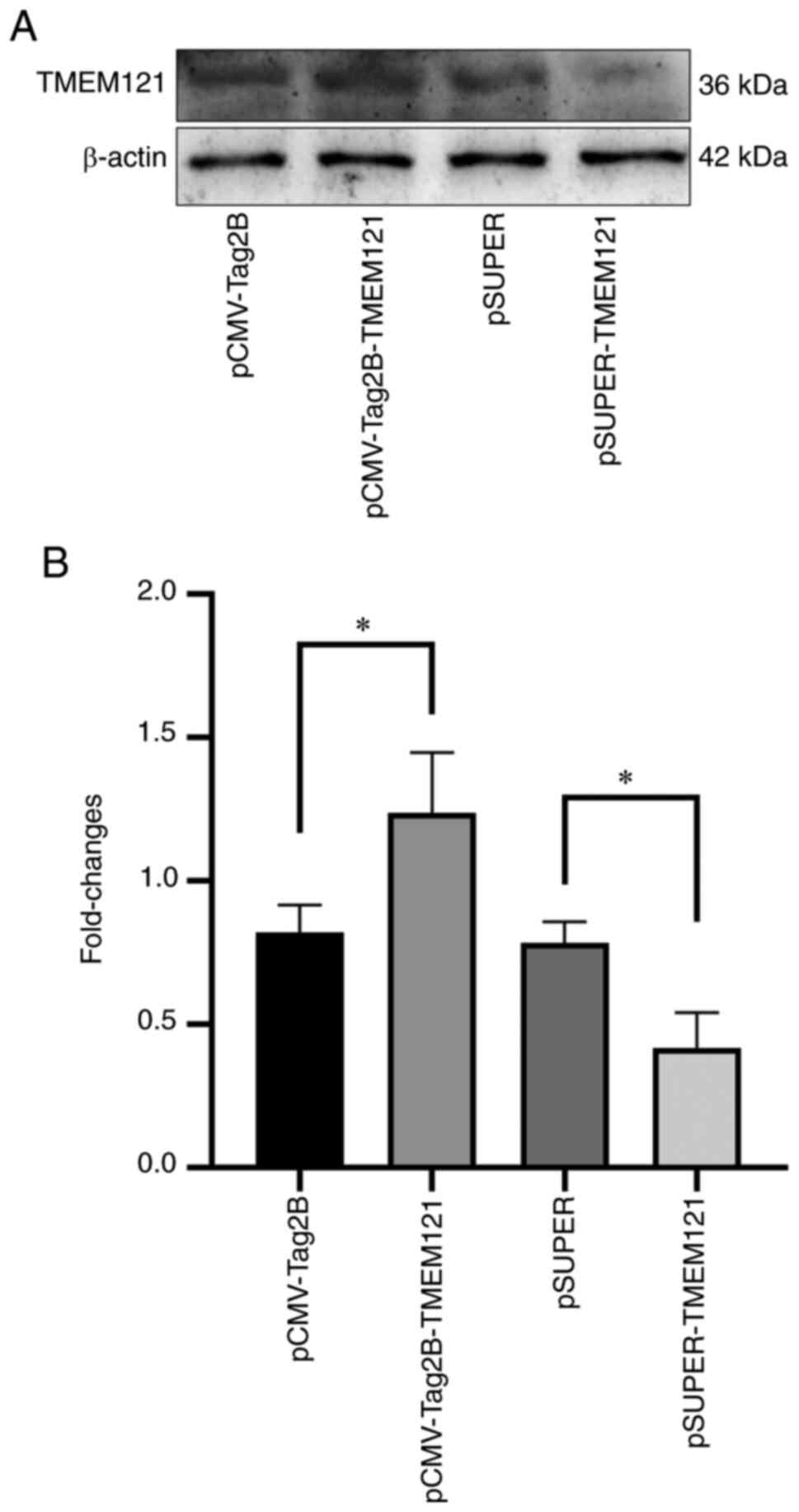
Validation of TMEM121 overexpression
and knockdown in the HeLa cell line
The overexpression and knockdown experiments were
used to explore the role of TMEM121 in cervical cancer
cells. The treated plasmids, pCMV-Tag2B, pCMV-Tag2B-TMEM121, pSUPER
and pSUPER-TMEM121, were transfected into the HeLa cell lines for
overexpression and RNA interference. The total proteins were
extracted for western blot analysis. As pCMV-Tag2B-TMEM121 and
pSUPER-TMEM121 were transfected into the HeLa cells, the
TMEM121 protein expression level significantly increased or
decreased when compared to the empty vector groups (Fig. 8).
TMEM121 overexpression inhibits cell
proliferation in the CCK-8 assay
To elucidate the role of TMEM121 in the pathogenesis
of CESC, the CCK-8 assay was used to measure the proliferative
capacity of HeLa cells following TMEM121 overexpression or
knockdown. The CCK-8 assay measures cell viability and can be used
to assess cell proliferation. The results revealed that HeLa cells
with TMEM121 overexpression showed significantly reduced
cell viability at 24, 48 and 72 h when compared with the empty
vector control group, indicating that cell proliferation was
inhibited (Fig. 9A). By contrast,
there was no significant difference between the
TMEM121-knockdown group and the control group (Fig. 9B).
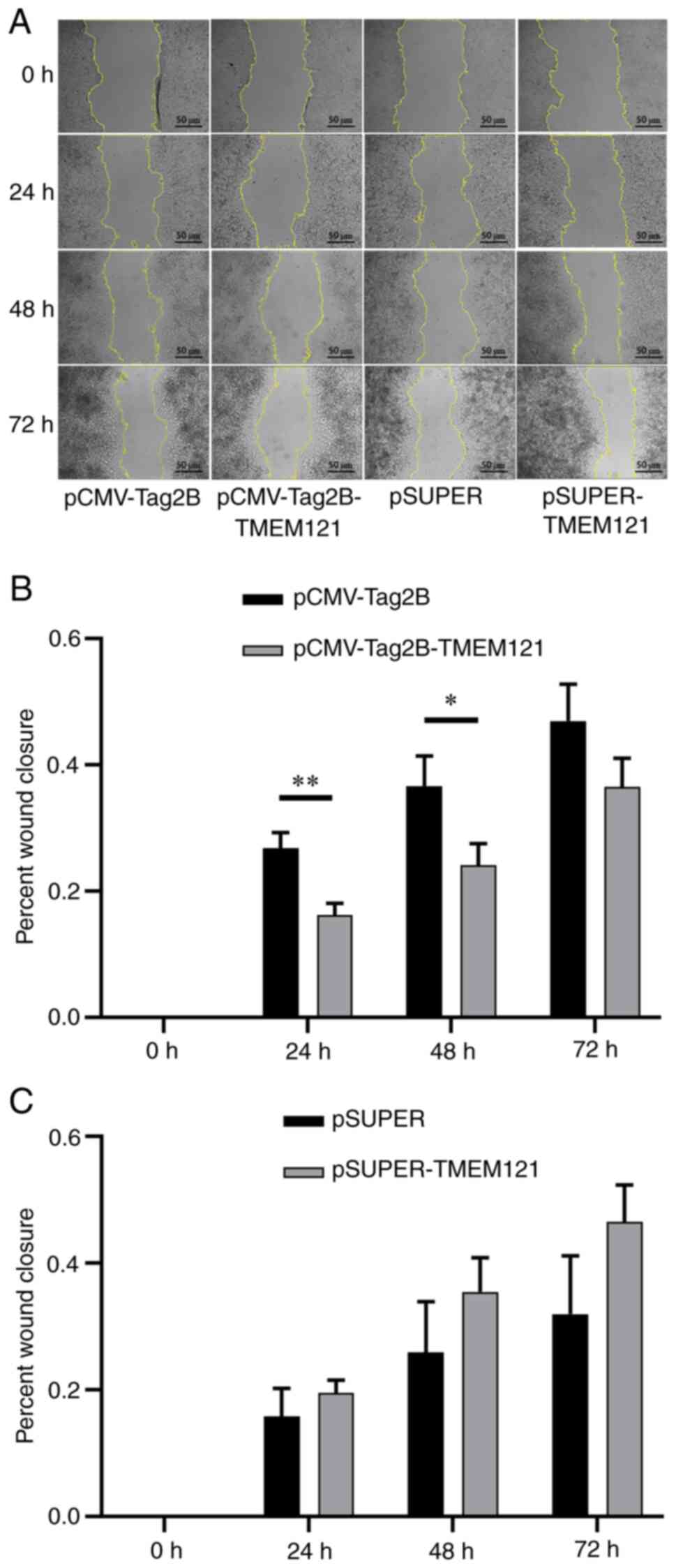
TMEM121 inhibits cervical cancer cell
migration in a cell scratch assay
A cell scratch experiment was also conducted to
assess cell migration through the wound closure rate of the HeLa
cells transfected with TMEM121-overexpression and TMEM121-knockdown
(Fig. 10A). The wound closure
rates were measured at 24, 48 and 72 h after the scratch, and
significant differences were found when compared with the empty
vector groups. The results revealed that the migratory ability of
HeLa cells with TMEM121 overexpression significantly
decreased at 24 and 48 h (Fig.
10B), whereas there was no significant change in the migratory
ability of HeLa cells with knocked down TMEM121 expression
(Fig. 10C).
Overexpression and knockdown of
TMEM121 regulate cancer-related factors in CESC
To verify the impact of TMEM121
overexpression and knockdown on CESC progression at the protein
expression level, western blotting was conducted to assess markers
associated with cancer cell proliferation and apoptosis (Fig. 11A). The quantitative results
revealed that when TMEM121 was overexpressed in HeLa cells,
the protein expression levels of BCL-2, cyclin D1, and cyclin E2
significantly decreased, whereas those of p27 significantly
increased (Fig. 11B).
TMEM121 knockdown significantly inhibited RB, p53 and p27
protein expression, and promoted the expression of cyclin E1. In
addition, neither treatment had a significant effect on cleaved
caspase-3 protein expression (Fig.
11B).
 | Figure 11Western blot detection of tumour cell
proliferation and apoptosis markers. (A) Western blot results of
RB, p53, BCL-2, cleaved caspase-3, p27, cyclin D1, cyclin E1,
cyclin E2 and β-tubulin. (B) Quantitative analysis of the western
blot results (n=3). *P<0.05 and
**P<0.01. RB, retinoblastoma protein; BCL-2, B-cell
lymphoma 2; TMEM121, transmembrane protein 121. |
Subsequently, the expression levels of proteins
related to cancer cell migration and autophagy were determined
(Fig. 12A), and the results
revealed that the expression level of E-cadherin significantly
increased when TMEM121 was overexpressed, indicating that
cell migration was inhibited, which was consistent with the results
of the cell migration scratch assay (Fig. 12B). When the expression of TMEM121
was decreased, the expression of E-cadherin was downregulated
(Fig. 12B), even though there was
no significant change in cell migration. However, when
TMEM121 was overexpressed or inhibited, LC3, a marker of
autophagy, was not significantly altered (Fig. 12B).
 | Figure 12Western blot detection of tumour cell
proliferation and apoptosis markers. (A) Western blot results of
E-cadherin, LC3A/B, p-JNK, JNK, p-p38, p38, p-AKT, AKT and GAPDH.
(B) Quantitative analysis of western blot results of p-JNK, p-p38
and p-AKT (n=3). (C) Quantitative analysis of the western blot
results of E-cadherin and LC3 (n=3). *P<0.05,
**P<0.01 and ***P<0.001.. LC3A/B light
chain 3 α/β; p-, phosphorylated; JNK, c-Jun N-terminal kinase;
GAPDH, glyceraldehyde 3-phosphate dehydrogenase. |
In addition, the phosphorylation levels of several
key factors in cancer-related signalling pathways were estimated.
The results revealed that when TMEM121 was overexpressed,
P38 phosphorylation was promoted, whereas AKT phosphorylation was
inhibited. When TMEM121 was knocked down, the expression of
both p-P38 and p-JNK was decreased (Fig. 12C).
Discussion
The identification of new regulatory factors and
novel mechanisms underlying cervical cancer development and
metastasis is crucial to develop more effective cancer diagnostics
and treatment. A previous study by the authors revealed that the
TMEM121 gene is highly expressed in the adult heart and may
inhibit cardiac hypertrophy (6).
However, it has not been reported whether TMEM121 is
implicated in cancer development.
The present study is the first, to the best of our
knowledge, to focus on the association between TMEM121 and
cancer. Bioinformatics analysis revealed that TMEM121
expression was significantly downregulated in human cervical cancer
tissues compared with that in normal tissues in various
classification groups. As an epigenetic phenomenon, aberrant gene
silencing in cancer cells has been linked to CpG island
hypermethylation of DNA in gene promoter regions (14-16).
Abnormally high methylation levels of TMEM121 gene promoter
were detected in a number of CESC subgroups, suggesting that the
epigenetic downregulation of TMEM121 may contribute to CESC
development. Notably, in the bioinformatics analysis, it was
determined that the survival curves of patients with high
expression of TMEM121 were not significantly different. TMEM121 is
a transmembrane protein (5), so we
speculate that it receives and transmits signals on the cell
membrane to regulate intracellular cancer development. Therefore,
it is hypothesised that the mutation of TMEM121 will affect the
early stage of cervical cancer, and will affect the occurrence and
metastasis of tumour cells. The mutation of TMEM121 will assist
cervical cancer metastasis. A limitation of this study is the lack
of significant differences in the survival curves of patients with
high and low TMEM121 expression. This may be due to the
insufficient number of patients with cervical cancer included in
the TCGA database or as the differential expression of this gene is
not sufficient to be significantly reflected in the survival rate.
Furthermore, TMEM121 has been linked to PI3K/AKT signalling
as well as other cancer-related factors. Genetic events leading to
the aberrant activation of the PI3K/AKT pathway are one of the most
common drivers of human cancer (17). These results suggest that
TMEM121 is involved in CESC regulation. Cell function
analysis revealed that TMEM121 overexpression reduced not
only the migratory ability of cervical cancer cells but also their
proliferation ability. However, the reverse was not evident in the
HeLa cell lines with TMEM121 knockdown. These findings
suggest that TMEM121 can inhibit cervical cancer cell
migration and proliferation but is not required for either of these
processes to occur.
The RB gene is the first discovered tumour
suppressor gene. The RB protein binds to various cellular proteins
during the G0 and G1 phases, such as the E2F family and ELF-1, and
inhibits their activated transcription function, thereby blocking
the G1 phase and preventing S-phase entry (18). Therefore, RB is considered an
important checkpoint in controlling cell proliferation (19). p27 belongs to the Cip/Kip family of
cyclin-dependent kinase inhibitors (20). As an important recently discovered
tumour suppressor gene, the reduction of p27 protein expression
level is closely related to the development, progression, and
prognosis of malignancies (21,22).
When TMEM121 expression was knocked down, RB and p27 expression was
downregulated, which may predict increased cancer risk or
progression. The cell cycle progression in mammalian cells from the
G1 to S phases is driven by cyclins D-type (including cyclin D1)
and E-type (including cyclins E1 and E2) (23,24).
In contrast to RB and p27, when TMEM121 was overexpressed, the
protein expression levels of cyclin D1 and cyclin E2 decreased, and
cells were prevented from entering the S phase, inhibiting cell
proliferation. Conversely, when TMEM121 is knocked down, an
increase in cyclin E2 protein may promote cell proliferative
activity, although this increase does not seem to be apparent.
LC3A and LC3B are autophagosome markers (25), and their expression was not altered
by TMEM121 overexpression or knockdown. E-cadherin
expression has been linked to tumour metastasis. As a key molecule
mediating the intercellular adhesion, E-cadherin acts as the ‘glue’
between cells and a marker for cell migration (26). When E-cadherin expression is
upregulated, cell migration is inhibited. The internal molecular
mechanism underlying E-cadherin upregulation may be attributed to a
reduction in AKT phosphorylation following TMEM121
overexpression. Previous research has revealed that inhibiting the
PI3K/AKT signalling pathway by decreasing the p-AKT expression can
effectively prevent cancer cell migration (27,28).
It is hypothesised that endogenous TMEM121 inhibits cervical cancer
cell migration via downregulation of p-AKT. The mechanism by which
TMEM121 inhibits p-AKT activation is not well understood.
Nevertheless, the structural characteristics of TMEM121 suggest
that it interacts with kinases on the membrane and disrupts the
kinase signal transmission. Upstream of the AKT signal activation,
there is a protein-protein interaction on the membrane; however, it
is unknown whether TMEM121 is involved in this interaction. This
should be verified in future experiments.
The present study mainly focused on the
identification of novel candidate genes regulating carcinogenesis
through bioinformatics and experiments for preliminary verification
of its functions. In fact, previous laboratory work has
demonstrated the function of TMEM121 in colorectal cancer cells,
gastric cancer cells, liver cancer HepG2 cells, lung cancer A549
cells and cervical cancer HeLa cells, but it was found that TMEM121
only played a role in HeLa cells. Therefore, it can only be
confirmed that TMEM121 is involved in the regulation of cervical
cancer. However, the Hela cell line was only used throughout the
study. Considering the heterogeneity of different cell lines even
within the same type of cancer, and the complexity of further
molecular mechanism studies, in the future, further validation with
the use of other cell lines will be performed. TMEM121 is a triple
transmembrane protein. Questions as to how it sorts the signals on
the membrane for cancer cells, and how it affects the
phosphorylation of AKT through a protein interaction mechanism will
be addressed in a follow-up study.
In conclusion, the results of the present study
demonstrated for the first time, to the best of our knowledge, that
TMEM121 may be a novel inhibitor of cervical cancer migration that
is linked to multiple signalling pathways. The present study offers
unique theoretical insights into the development of cervical cancer
and the pursuit of novel diagnostic and interventional
approaches.
Acknowledgements
Not applicable.
Funding
Funding: The present study was supported by grants from the
National Natural Science Foundation of China (grant nos. 81670290,
81801392, 32071175, 31572349, 81370230, 81570279, 81974019 and
81600320), the National Key Research and Development Program of
China (grant nos. 2018YFA0108700 and 2017YFA0105602), the NSFC
Projects of International Cooperation and Exchanges (grant no.
81720102004), The Research Team Project of Natural Science
Foundation of Guangdong Province of China (grant no.
2017A030312007), the Science and Technology Planning Project of
Guangdong Province (grant no. 2022B1212010010), the Key Program of
Guangzhou Science Research Plan (grant no. 201904020047), The
Special Project of Dengfeng Program of Guangdong Provincial
People's Hospital (grant nos. DFJH201812, KJ012019119 and
KJ012019423) and the Hunan Provincial Natural Science Foundation of
China (grant no. 2020JJ5354).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
XF, YL, XW and PZ made significant contributions to
the conception or design of the study. YWa, ZJ, QZ, ZP, JC, YWe,
YS, XZ, XY, JZ, WY, YCh and FL performed experiments. BY, YCa and
JA were responsible for the acquisition, analysis or interpretation
of the data and drafted this work. BY and YCa confirm the
authenticity of all the raw data. All authors have read and
approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Liu Y, Zhu H, Mo L, Xu R, Li X, Li T, Zhao
L, Ren Y, Ou R and Xu Y: Flt3L and GM-CSF enhance anti-tumor effect
of HPV16/18 vaccine via increasing immune response. Am J Transl
Res. 12:6027–6042. 2020.PubMed/NCBI
|
|
2
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Forman D, de Martel C, Lacey CJ,
Soerjomataram I, Lortet-Tieulent J, Bruni L, Vignat J, Ferlay J,
Bray F, Plummer M and Franceschi S: Global burden of human
papillomavirus and related diseases. Vaccine. 5:F12–F23.
2012.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Nesset AL and Bader DM: Hole is a novel
gene product expressed in the developing heart and brain. Mech Dev.
117:347–350. 2002.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Zhou J, Li Y, Liang P, Yuan W, Ye X, Zhu
C, Cheng Y, Wang Y, Li G, Wu X and Liu M: A novel six-transmembrane
protein hhole functions as a suppressor in MAPK signaling pathways.
Biochem Biophys Res Commun. 333:344–352. 2005.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Xu W, Wang Y, Zhou J, Zhu X, Zhang S, Yuan
W, Liu X, Shi Y, Cao L, Zeng Q, et al: Cardiac specific
overexpression of hHole attenuates isoproterenol-induced
hypertrophic remodeling through inhibition of extracellular
signal-regulated kinases (ERKs) signalling. Curr Mol Med.
16:515–523. 2016.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Li T, Fan J, Wang B, Traugh N, Chen Q, Liu
JS, Li B and Liu XS: TIMER: A web server for comprehensive analysis
of tumor-infiltrating immune cells. Cancer Res. 77:e108–e110.
2017.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Gao J, Aksoy BA, Dogrusoz U, Dresdner G,
Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, et al:
Integrative analysis of complex cancer genomics and clinical
profiles using the cBioPortal. Sci Signal. 6(pl1)2013.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Vasaikar SV, Straub P, Wang J and Zhang B:
LinkedOmics: Analyzing multi-omics data within and across 32 cancer
types. Nucleic Acids Res. 46:D956–D963. 2017.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Nagy Á, Munkácsy G and Győrffy B:
Pancancer survival analysis of cancer hallmark genes. Sci Rep.
11(6047)2021.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Chandrashekar DS, Bashel B, Balasubramanya
SA, Creighton CJ, Ponce-Rodriguez I, Chakravarthi BV and Varambally
S: UALCAN: A portal for facilitating tumor subgroup gene expression
and survival analyses. Neoplasia. 19:649–658. 2017.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Gu Z, Eils R and Schlesner M: Complex
heatmaps reveal patterns and correlations in multidimensional
genomic data. Bioinformatics. 32:2847–2849. 2016.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Gene Ontology Consortium: Going forward.
Nucleic Acids Res. 43:D1049–D1056. 2015.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Esteller M: CpG island hypermethylation
and tumor suppressor genes: A booming present, a brighter future.
Oncogene. 21:5427–5440. 2002.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Herman JG and Baylin SB: Gene silencing in
cancer in association with promoter hypermethylation. N Engl J Med.
349:2042–2054. 2003.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Ushijima T and Asada K: Aberrant DNA
methylation in contrast with mutations. Cancer Sci. 101:300–305.
2010.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Hoxhaj G and Manning BD: The PI3K-AKT
network at the interface of oncogenic signalling and cancer
metabolism. Nat Rev Cancer. 20:74–88. 2020.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Kato J, Matsushime H, Hiebert SW, Ewen ME
and Sherr CJ: Direct binding of cyclin D to the retinoblastoma gene
product (pRb) and pRb phosphorylation by the cyclin D-dependent
kinase CDK4. Genes Dev. 7:331–342. 1993.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Knudsen ES and Knudsen KE: Tailoring to
RB: Tumour suppressor status and therapeutic response. Nat Rev
Cancer. 8:714–724. 2008.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Pérez-Luna M, Aguasca M, Perearnau A,
Serratosa J, Martínez-Balbas M, Pujol MJ and Bachs O: PCAF
regulates the stability of the transcriptional regulator and
cyclin-dependent kinase inhibitor p27 Kip1. Nucleic Acids Res.
40:6520–6533. 2012.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Perearnau A, Orlando S, Islam A,
Gallastegui E, Martínez J, Jordan A, Bigas A, Aligué R, Pujol MJ
and Bachs O: p27Kip1, PCAF and PAX5 cooperate in the
transcriptional regulation of specific target genes. Nucleic Acids
Res. 45:5086–5099. 2017.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Roilo M, Kullmann MK and Hengst L:
Cold-inducible RNA-binding protein (CIRP) induces translation of
the cell-cycle inhibitor p27Kip1. Nucleic Acids Res. 46:3198–3210.
2018.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Liu L, Michowski W, Inuzuka H, Shimizu K,
Nihira NT, Chick JM, Li N, Geng Y, Meng AY, Ordureau A, et al: G1
cyclins link proliferation, pluripotency and differentiation of
embryonic stem cells. Nat Cell Biol. 19:177–188. 2017.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Hydbring P, Wang Y, Fassl A, Li X, Matia
V, Otto T, Choi YJ, Sweeney KE, Suski JM, Yin H, et al:
Cell-cycle-targeting micrornas as therapeutic tools against
refractory cancers. Cancer Cell. 31:576–590.e8. 2017.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Wu J, Dang Y, Su W, Liu C, Ma H, Shan Y,
Pei Y, Wan B, Guo J and Yu L: Molecular cloning and
characterization of rat LC3A and LC3B-two novel markers of
autophagosome. Biochem Biophys Res Commun. 339:437–442.
2006.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Bruner HC and Derksen PWB: Loss of
e-cadherin-dependent cell-cell adhesion and the development and
progression of cancer. Cold Spring Harb Perspect Biol.
10(a029330)2018.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Wang D, Yang T, Liu J, Liu Y, Xing N, He
J, Yang J and Ai Y: Propofol inhibits the migration and invasion of
glioma cells by blocking the PI3K/AKT pathway through miR-206/ROCK1
axis. Onco Targets Ther. 13:361–370. 2020.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Sheng XY, Wang CH, Wang CF and Xu HY:
Long-chain non-coding SOX21-AS1 promotes proliferation and
migration of breast cancer cells through the PI3K/AKT signaling
pathway. Cancer Manag Res. 12:11005–11014. 2020.PubMed/NCBI View Article : Google Scholar
|