Introduction
Melanoma, brought on by the malignant transformation
of melanocytes, has been reported as the deadliest skin cancer
(1). Melanoma easily metastasizes
and its 5-year survival rate is only 18% (2). The incidence of melanoma worldwide has
been rising year by year (3). Its
high mortality is related to its unresectability and easy
movability (3). Though present
therapeutic approaches are capable of curing early-stage melanoma
by surgical removal, immune-based methods and chemoprevention, the
prognosis of patients with advanced melanoma remains poor (4).
Long non-coding RNAs (lncRNAs), >200 nucleotides
in length, play key roles in several types of human cancer
(5). LncRNAs have been found to be
dysregulated in various types of cancer, including lung cancer,
breast cancer, liver cancer, colorectal cancer and melanoma
(6). There are growing numbers of
studies that have investigated melanoma-related lncRNAs, such like
lncRNA cancer susceptibility candidate 2(7), as a melanoma-suppressing lncRNA; and
homeobox A11-antisense RNA (8),
ZNFX1 antisense RNA 1(9) and H19
Imprinted Maternally Expressed Transcript (10), as melanoma-accelerating lncRNAs.
Taurine-upregulated gene 1 (TUG1) is a lncRNA that consists of
6.7-kb nucleotides and TUG1 has been shown to play important roles
in tumorigenesis (11).
MicroRNAs (miRNAs) are a large group of endogenous
RNAs consisting of 18-22 nucleotides, which are regarded as key
regulators of development and progression of various types of human
tumors (12). miRNAs perform
regulatory roles by directly binding to the 3' untranslated region
(3'UTR) of their target mRNAs to silence translation, which can
affect the proliferation, metastasis and apoptosis of tumor cells
(13). Previous studies have
indicated that miRNA (miR)-145-5p expression levels are decreased
in breast cancer (14), bladder
cancer (15) and melanoma (16) cell lines, which suggests that
miR-145-5p may function as a tumor-suppressor.
SOX2, a member of the SOX gene family, has been
shown to be associated with the regulation of certain biological
processes during tumor development (17). A previous study found that SOX2
contributes to oxidative metabolism, as well as elevates drug
resistance and metastatic capacity in melanoma cells (18). SOX2 is dispensable for primary
melanoma metastasis and formation (19). Nevertheless, whether miR-145-5p and
SOX2 are required for the mechanisms of action behind how TUG1
functions during the development of melanomas remains unclear.
In the present study, the expression levels of TUG1
were found to be elevated in melanoma tumor tissues and cell lines.
In addition, the effects of TUG1 on the proliferative, migratory
and invasive abilities of melanoma cells were investigated.
Downstream target and regulatory mechanism of TUG1 in melanoma
cells were also explored.
Materials and methods
Clinical specimens and cell
culture
The present study was approved by the Ethics
Committee of the First Affiliated Hospital of Anhui Medical
University (Hefei, China). Tumor tissues and adjacent noncancerous
tissues (normal tissues) were collected from 27 patients with
melanoma (aged 29-71 years; 15 male and 12 female), who were
diagnosed at the First Affiliated Hospital of Anhui Medical
University between March 2014 and January 2018. No patients
received chemotherapy or any other types of therapy before surgery.
Inclusion criteria included histologically confirmed melanoma.
Exclusion criteria included: i) Current or previous history of any
other severe or uncontrolled diseases, including previous history
of cancer, bleeding dyscrasia and immune system disease; ii)
currently on medications that could interfere with assessment of
biological outcomes, including estrogens, progestogens, androgens,
prednisone or psychoactive drugs; and iii) aged <16 years. All
patients signed a written informed consent. Fresh tissues were
immediately frozen and stored in liquid nitrogen.
Human melanoma cells (M14 and A375) and human
primary normal epidermal melanocytes HEMa-LP were purchased from
the American Type Culture Collection and cultured in DMEM (Gibco;
Thermo Fisher Scientific, Inc.) with 10% FBS (Invitrogen; Thermo
Fisher Scientific, Inc.) in a humidified atmosphere with 5%
CO2 at 37˚C, unless otherwise stated.
Transfection with oligonucleotides and
plasmids
Specific small interfering (si)-RNAs against TUG1
(si-TUG1#1, si-TUG1#2 and si-TUG1#3; final concentration, 20 nM),
the scrambled negative control (NC; si-NC; final concentration, 20
nM), miR-145-5p mimic (final concentration, 15 nM), miR-NC (final
concentration, 15 nM), pcDNA-TUG1 (final concentration, 1.5 µg/ml),
si-SOX2 (final concentration, 20 nM), miR-145-5p inhibitor
(in-miR-145-5p; final concentration, 25 nM), its negative control
(in-miR-NC; final concentration, 25 nM), pcDNA-SOX2 (final
concentration, 1 µg/ml), specific short hairpin (sh)RNA against
TUG1 (sh-TUG1) and its negative control sh-NC were all synthesized
by Shanghai GenePharma Co., Ltd. The pcDNA plasmid was purchased
from Thermo Fisher Scientific, Inc. The sequences for the siRNAs,
miRNA mimics and inhibitors used in the present study were shown in
Table I.
 | Table ISequences for the siRNAs, miRNA
mimics and inhibitors used in the present study. |
Table I
Sequences for the siRNAs, miRNA
mimics and inhibitors used in the present study.
| Name | Sequence
(5'-3') |
|---|
| si-TUG1#1 |
CAGUCCUGGUGAUUUAGACAGUCUU |
| si-TUG1#2 |
CCCAGAAGUUGUAAGUUCACCUUGA |
| si-TUG1#3 |
CAGCUGUUACCAUUCAACUUCUUAA |
| si-NC |
UUCUCCGAACGUGUCACGUTT |
| miR-145-5p
mimic |
GUCCAGUUUUCCCAGGAAUCCCU |
| miR-145-5p
inhibitor |
AGGGAUUCCUGGGAAAACUGGAC |
| miR-NC mimic |
ACUCUAUCUGCACGCUGACUU |
| miR-NC
inhibitor |
CAGUACUUUUGUGUAGUACAA |
| sh-TUG1 |
CCGGCTGTTGACCTTGCTGTGAGAACTCGA
GTTCTCACAGCAAGGTCAACAGTTTTTTG |
| sh-NC |
CCTAAGGTTAAGTCGCCCTCG |
| si-SOX2 sense |
UGAUGGAGACGGAGCUGAAUU |
| si-SOX2
antisense |
UUCAGCUCCGUCUCCAUCAUU |
The aforementioned oligonucleotides or plasmids were
transfected into M14 and A375 cells using Lipofectamine™
2000 reagent (Invitrogen; Thermo Fisher Scientific, Inc.) following
the manufacturer's instructions. Briefly, M14 and A375 cells were
cultured for 24 h, before the constructs were transfected into the
cells using 500 µl Opti-MEM containing 20 µl
Lipofectamine™ 2000 (1 mg/ml) in a humidified atmosphere
with 5% CO2 at 37˚C. After incubating for 6 h, culture
medium was replaced by fresh medium. Subsequently, 24, 48 or 72 h
after transfection, the cells were collected for further
analysis.
Reverse transcription-quantitative
(RT-qPCR)
For the detection of mRNA expression levels, RNA was
isolated from melanoma tissues or cells using TRIzol®
Reagent (Invitrogen; Thermo Fisher Scientific, Inc.). RNA was
reverse-transcribed into cDNA using M-MLV Reverse Transcriptase
(cat. no. 28025021; Invitrogen; Thermo Fisher Scientific, Inc.)
with RNaseOUT™ Recombinant Ribonuclease Inhibitor (cat.
no. 10777; Thermo Fisher Scientific, Inc.), dNTPs (cat. no.
10297117; Thermo Fisher Scientific, Inc.) and the oligo (dT) 12-18
primer (cat. no. 18418012; Thermo Fisher Scientific, Inc.)
following the standard protocols: 65˚C for 5 min; 50 min at 37˚C;
and 70˚C for 15 min. Subsequent qPCR was conducted using
iQ™ SYBR® Green Supermix (cat. no. 1708882;
Bio-Rad Laboratories, Inc.). For the detection of miR-145-5p
expression levels, miRNA was extracted using a miRNeasy Mini Kit
(Qiagen, GmbH). Reverse transcription and RT-qPCR were conducted
using a TaqMan™ Reverse Transcription kit and
TaqMan™ MicroRNA Assays (Applied Biosystems; Thermo
Fisher Scientific, Inc.). β-actin was used as the internal control
for TUG1 and SOX2, whereas U6 was used as the internal control for
miR-145-5p. The primers for TUG1, SOX2, β-actin, miR-145-5p and U6
are as follows: TUG1 forward, 5'-GGACCTGGAACCCGAAAGAG-3' and
reverse, 5'-TGGTGGTAGTGCTTGCTCAG-3'; SOX2 forward,
5'-CAGCGCATGGACAGTTACG-3' and reverse, 5'-TTCATGTAGGTCTGCGAGCTG-3';
β-actin forward, 5'-TGGACTTCGAGCAAGAGATGG-3' and reverse,
5'-ACGTCACACTTCATGATGGAG-3'; miR-145-5p forward,
5'-CAGTCTTGTCCAGTTTTCCCAG-3' and reverse,
5'-TATGCTTGTTCTCGTCTCTGTGTC-3'; U6 forward, 5'-CTCGCTTCGGCAGCACA-3'
and reverse, 5'-AACGCTTCACGAATTTGCGT-3'. RT-qPCR was performed on a
StepOnePlus™ Real-time PCR Systems (Applied Biosystems;
Thermo Fisher Scientific, Inc.) with the following thermocycling
conditions: Initial denaturation for 10 min at 95˚C; 40 cycles of
95˚C for 15 sec and 60˚C for 1 min; followed by final elongation at
72˚C for 10 min. The expression levels of TUG1, SOX2 and miR-145-5p
were analyzed using the 2-ΔΔCq method (20).
Cell counting kit-8 (CCK-8) assay
For the detection of cell proliferative ability,
cells were seeded in 96-well plates containing DMEM with 10% FBS
(1,000 cells per well) for 0, 24, 48 and 72 h. The CCK-8 reagent
(MedChemExpress) was used for this assay. After collection, 10 µl
CCK-8 reagent was added into each well. In total, after incubation
at 37˚C for 2 h, the proliferative ability was determined through
measuring the absorbance at 450 nm of each well using a
spectrophotometer (NanoDrop Technologies; Thermo Fisher Scientific,
Inc.).
Transwell assay
Transwell migration and invasion assay was performed
using a transwell chamber (8 µm pore size; EMD Millipore). For
migration assays, 3x104 transiently transfected M14 and
A375 cells in serum-free DMEM were seeded into the upper chamber,
whilst DMEM supplemented with 10% FBS was added to the lower
chamber. Subsequently, the chamber was incubated at 37˚C. In total,
24 h later, the migrated cells were fixed with 4% paraformaldehyde
for 15 min at room temperature, before being stained with 0.1%
crystal violet for 30 mins at room temperature and counted under an
inverted light microscope (Nikon Eclipse TS100; Nikon Corporation)
at x20 magnification. For invasion assays, the protocol used was
identical, but the upper chamber was precoated with 50 µl Matrigel
(8 mg/ml; 1:8 dilution; Corning, Inc.) at 4˚C overnight.
Dual-luciferase reporter assay
The downstream target miRNAs of TUG1 were predicted
using the online website LncBase Predicted v.2 (http://carolina.imis.athenainnova-tion.gr/diana_tools/web/index.php?r=lncbasev2%2Findex-predicted).
Direct targets of miR-145-5p were predicted by online databse
miRTarBase (http://mirtarbase.mbc.nctu.edu.tw/php/detail.php?mirtid=MIRT000307).
The wild-type (WT) or mutant (MUT) sequences of TUG1 and SOX2 were
amplified and cloned into pGL3 luciferase promoter vector (Promega
Corporation) to synthesize TUG1 WT, TUG1 MUT, SOX2 WT and SOX2 MUT.
Renilla luciferase reporter plasmids were used as the
control for normalization. In total, 1x106 M14 and A375
cells were co-transfected with 8 µg recombinant luciferase reporter
plasmids, 0.16 µg pRL-TK plasmids and miR-145-5p mimics (final
concentration, 15 nM) or miR-NC (final concentration, 15 nM) using
LipofectamineTM 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer's instruction. The
luciferase activity was evaluated using the
Dual-Luciferase® reporter system (Beyotime Institute of
Biotechnology) according to the protocols supplied by the
manufacturer.
RNA immunoprecipitation (RIP)
assay
The RIP assay was performed using the extracts
obtained from the melanoma M14 and A375 cells using the EZ-Magna
RIP kit (cat. no. 17-701; EMD Millipore) according to the
manufacturer's instructions, as previously described (21). In brief, the extracted supernatant
(150 µg protein) was incubated with 1 mg magnetic beads that had
been pre-coated with human anti-argonaute2 (Ago2) antibodies (cat.
no. ab186733, 1:50, Abcam) or IgG (cat. no. PP64B, 1:20, EMD
Millipore) at 4˚C overnight, which was used as a negative control.
After overnight incubation at 4˚C, the RNA complex were incubate
with proteinase K (RIP wash buffer, 10% SDS and 10 mg/ml proteinase
K; EMD Millipore) for 30 min at 55˚C to digest the protein
remaining on the beads. The RNA was then purified from the samples
using phenol: chloroform: isoamyl alcohol separation. RNA detection
was conducted using RT-qPCR as aforementioned.
Western blot assay
Cell lysates of melanoma cells (M14 and A375) were
prepared using the RIPA Reagent (Cell Signaling Technology, Inc.).
The protein concentration was quantified with bicinchoninic acid
reagent (Sigma-Aldrich; Merck KGaA). A total of 30 µg protein
samples were fractionated on 15% SDS-polyacrylamide gels and then
transferred onto PVDF membranes. Thereafter, the membranes were
blocked with 5% skimmed milk at room temperature for 2 h and probed
with primary antibodies targeting SOX2 (1:1,000, cat. no. cat. no.
ab171380; Abcam) or β-actin (1:1,000; cat. no. ab8227; Abcam) at
4˚C overnight. Subsequently, the membranes were incubated with
horseradish peroxidase-conjugated goat anti-rabbit secondary
antibodies (1:1,000; cat. no. ab205718, Abcam) at room temperature
for 2 h. The bands were visualized using an ECL detection kit
(Thermo Fisher Scientific, Inc.) and quantified using the ImageJ
1.8.0 software (National Institute of Health). The aforementioned
antibodies used for this investigation were purchased from Abcam
(Cambridge, MA, USA).
Xenograft mouse model
The animal experiments, which lasted 28 days, were
approved by the Animal Care and Scientific Committee of the First
Affiliated Hospital of Anhui Medical University (Hefei, China). A
total of 12 male BALB/c nude mice (5-week-old; 16-20 g weight
range) were obtained from Shanghai SLAC Laboratory Animal Co., Ltd.
(grouped into a sh-TUG1 group and sh-NC group, n=6 per group) and
maintained under temperature (20-22˚C) and humidity-controlled
(55±10%) pathogen-free specific conditions with a 12-h light/dark
cycle and given water and food ad libitum. The stable A375
cells expressing sh-NC or sh-TUG1 were produced and maintained in
medium containing 3 µg/ml puromycin (Sigma-Aldrich; Merck KGaA).
Prior to injection, the stable A375 cells were resuscitated and
grown for two passages. A total of 4x106 stable A375
cells expressing sh-NC or sh-TUG1 were collected and suspended in
100 µl sterile PBS solution and then subcutaneously inoculated into
the right flank of the nude mice. Tumor volume was measured every 7
days for 28 days and calculated according to the following
equation: Volume=(length x width2)/2. The health and
behavior of nude mice involved in the animal experiment were also
monitored every 7 days after inoculation. All mice were sacrificed
at day 28 following inoculation, for a pre-set 28-day test cycle.
The 12 nude mice were anesthetized using 2% methoxyflurane (an
inhalant anesthetic) (22).
Subsequently, the animals were sacrificed by cervical dislocation.
The xenograft tumor tissues were then collected for measurement of
weight and used for RT-qPCR, western blot analyses and
immunohistochemical assays.
Immunohistochemical (IHC) assay
The IHC assays were conducted to detect the
expression profile of SOX2 in resected tumors from the sh-NC group
and sh-TUG1 group, following the protocols from a previous report
(23). In brief, resected tumors
from the xenograft models were fixed in 3% formaldehyde overnight
at 4˚C and embedded in paraffin and then cut into 5-µm sections.
The slices were then incubated with 0.3% hydrogen peroxide
(H2O2) solution in methanol for 20 min at
room temperature to block the activity of endogenous peroxidases.
Sections were blocked in 10% horse serum at room temperature for 1
h and then incubated with goat anti-SOX2 antibodies (1:100; cat.
no. GT15098; Neuromics) at 4˚C overnight, followed by incubation
with peroxidase-conjugated horse anti-goat IgG (cat. no. PI-9500-1;
1:2,000; Vector Laboratories, Inc.) at room temperature for 30 min.
Immunoreactivity was measured using NovaRed peroxidase substrate
(Vector Laboratories, Inc.) under an Eclipse TS100 fluorescence
microscope (Nikon Corporation) at x20 magnification.
Statistical analysis
All data analyses were conducted using SPSS 22.0
software (IBM Corp.). Data from three independent experiments are
presented as the mean ± SD. Difference between means was analyzed
using paired Student's t-tests (for 2 groups) and one-way ANOVAs
(for ≥3 groups) followed by Tukey's post hoc tests. Pearson
correlation coefficient was used to analyze the correlation among
TUG1, miR-145-5p and SOX2. Differences were considered significant
at P<0.05.
Results
TUG1 expression is upregulated in
melanoma tissues and cells
To investigate the role of TUG1 in melanoma, the
expression levels of TUG1 were analyzed using RT-qPCR. As shown in
Fig. 1A, TUG1 expression was
significantly upregulated in melanoma tissues compared with that in
normal tissues. In addition, the expression levels of TUG1 were
significantly raised in the melanoma M14 and A375 cell lines
compared with that in normal melanocyte cell line, HEMa-LP cells
(Fig. 1B).
Silencing of TUG1 expression inhibits
the proliferative, migratory and invasive abilities of melanoma
cells
siRNAs against TUG1 (si-TUG1#1, si-TUG1#2 and
si-TUG1#3) were transfected into M14 and A375 cells to construct
TUG1-silenced melanoma cells to explore the effect of TUG1 on the
proliferative, migratory and invasive abilities of melanoma cells.
RT-qPCR assays were conducted to detect the silencing efficiency of
the 3 siRNAs (si-TUG1#1, si-TUG1#2 and si-TUG1#3) on the TUG1
expression in the transfected M14 and A375 cells. The mRNA
expression levels of TUG1 in M14 and A375 cells were decreased
after the indicated transfections, with the silencing efficiency of
si-TUG1#3 being the most significant (Fig. 2A and B). As such, si-TUG1#3 was used for
subsequent functional experiments. CCK-8 assays were employed to
detect the proliferative ability of transfected M14 and A375 cells,
which indicated that a reduced TUG1 expression inhibited the
proliferative ability of the two cell lines (Fig. 2C and D). Transwell assay indicated that
downregulation of TUG1 also repressed the migration and invasion of
M14 and A375 cells (Fig. 2E and
F).
 | Figure 2Silencing TUG1 expression inhibits
the proliferative, migratory and invasive abilities of melanoma
cells. (A) M14 and (B) A375 cells were transfected with si-NC,
si-TUG1#1, si-TUG1#2 or si-TUG1#3. The relative expression levels
of TUG1 in M14 or A375 cells were assessed using RT-qPCR assay. M14
and A375 cells were then transfected with si-TUG1. The
proliferative ability of transfected (C) M14 and (D) A375 cells
after 24, 48 and 72 h was tested using Cell Counting Kit-8 assays,
measuring the OD450. Transwell assays were performed to
detect cell (E) migratory and (F) invasive abilities of M14 and
A375 cells. Scale bars, 200 µm. *P<0.05 vs. si-NC
group. NC, negative control; OD, optical density; OD450,
OD at 450 nm; si, small interfering; TUG1, taurine-upregulated gene
1. |
miR-145-5p is a direct target of TUG1
and is negatively regulated by TUG1
LncBase Predicted v.2 was applied to seek the
downstream target of TUG1 and it was hypothesized that miR-145-5p
could bind to TUG1 (Fig. 3A).
Dual-luciferase assays were performed to validate the relationship
between TUG1 and miR-145-5p. RT-qPCR assay indicated that the
expression of miR-145-5p was significantly upregulated by
transfection of miR-145-5p mimic (Fig.
S1A), but was significantly suppressed more by transfection
with the miR-145-5p inhibitor (Fig.
S1B). In addition, both transfection of TUG1 wt or TUG1 mut
significantly elevated TUG1 expression (Fig. S1C). As depicted in Fig. 3B and C, the luciferase activity of M14 and A375
cells co-transfected with TUG1 WT and miR-145-5p mimics was
obviously lower than that of cells co-transfected with TUG1 WT and
miR-NC mimics. By contrast, the luciferase activity of M14 and A375
cells co-transfected with TUG1 MUT and miR-145-5p mimics or miR-NC
mimics showed no significant difference. A RIP assay was performed
to confirm the interaction between TUG1 and miR-145-5p, which
showed that TUG1 mRNA could be specifically recruited to the miRNA
complexes isolated using anti-Ago2 antibody following miR-145-5p
transfection, suggesting that TUG1 could interact with miR-145-5p
(Fig. 3D). RT-qPCR was performed to
analyze the expression level of miR-145-5p in melanoma tissues and
cells. miR-145-5p was found to be downregulated in melanoma tissues
compared with normal tissues (Fig.
3E). In addition, downregulation of miR-145-5p was also
detected in melanoma M14 and A375 cells compared with HEMa-LP cells
(Fig. 3F). Introduction of TUG1
repressed the expression levels of miR-145-5p in both M14 and A375
cells relative to cells transfected with pcDNA; while silencing of
TUG1 promoted miR-145-5p expression in the two cell lines compared
with cells transfected with si-NC, indicating that TUG1 inversely
regulated miR-145-5p expression levels (Fig. 3G).
 | Figure 3TUG1 targets miR-145-5p and
negatively regulates miR-145-5p. (A) The predicted binding site
between miR-145-5p and TUG1 and its MUT were predicted by LncBase
Predicted v.2. Dual-luciferase assays were conducted to measure the
luciferase activity of (B) M14 and (C) A375 cells that were
co-transfected with TUG1 WT + miR-NC mimics, TUG1 WT + miR-145-5p
mimics, TUG1 MUT + miR-NC mimics or TUG1 MUT + miR-145-5p mimics.
(D) RIP assays were conducted and the enrichment of TUG1 was
determined by the samples which had bound to the Ago2 antibody or
IgG in M14 and A375 cells. (E) RT-qPCR was used to measure TUG1
expression levels in melanoma tissues. (F) The expression levels of
TUG1 in melanoma cells were evaluated using RT-qPCR. (G) M14 and
A375 cells were transfected with pcDNA, pcDNA-TUG1, si-NC or
si-TUG1. RT-qPCR was used to measure miR-145-5p levels in
transfected melanoma cells. *P<0.05. Ago2,
argonaute2; miR, microRNA; MUT, mutant; NC, negative control; ns,
no significance; RT-qPCR, reverse transcription-quantitative PCR;
si, small interfering; TUG1, taurine-upregulated gene 1; WT,
wild-type. |
TUG1 regulates the proliferative,
migratory and invasive abilities of melanoma cells by targeting
miR-145-5p
To clarify whether TUG1 regulates the proliferative,
migratory and invasive abilities of melanoma cells by targeting
miR-145-5p, recovery experiments were performed. RT-qPCR assays
showed that miR-145-5p mimics significantly elevated miR-145-5p
expression levels in both M14 and A375 cells, while introduction of
TUG1 counteracted the promoting impact of miR-145-5p mimics on
miR-145-5p expression (Fig. 4A and
B). CCK-8 assays also indicated
that upregulation of miR-145-5p inhibited the proliferative ability
of M14 and A375 cells, whereas elevated TUG1 almost reversed the
repressive effect of miR-145-5p mimics on the proliferative ability
of the two cell lines (Fig. 4C and
D). Subsequently, the impact of
miR-145-5p alone or miR-145-5p combined with TUG1, on the migratory
and invasive abilities of melanoma cells was assessed using
Transwell assays. As displayed in Fig.
4E and F, overexpression of
miR-145-5p significantly inhibited the migratory and invasive
abilities of M14 and A375 cells, while raised expression levels of
TUG1 significantly weakened the repressive impact of miR-145-5p on
the migratory and invasive abilities of M14 and A375 cells.
miR-145-5p targets SOX2 and TUG1
regulates SOX2 expression by sponging miR-145-5p
The online software mirTarBase identified SOX2 as a
direct target of miR-145-5p. The putative binding site and its MUT
for miR-145-5p on the 3'-UTR of SOX2 mRNA are displayed in Fig. 5A. RT-qPCR assay indicated that SOX2
expression was significantly elevated by SOX2 wt or SOX2 mut
transfection (Fig. S1E).
Subsequently, the targeted relationship between miR-145-5p and SOX2
was validated using dual-luciferase reporter assays. The assay
indicated that upregulation of miR-145-5p significantly constrained
the luciferase activity of SOX2 WT reporter in co-transfected M14
and A375 cells, compared to the miR-NC transfected cells. However,
neither the miR-145-5p nor the miR-NC had little impact on the
luciferase activity of the SOX2 MUT reporter in co-transfected M14
and A375 cells (Fig. 5B and
C). Subsequently, RIP assays were
also implemented to confirm the direct interaction between
miR-145-5p and SOX2, and it was suggested that transfection with
miR-145-5p triggered SOX2 enrichment in the RIP-Ago2 group in
melanoma M14 and A375 cells compared with that in the RIP-IgG
group, supporting the notion that SOX2 was a direct target of
miR-145-5p (Fig. 5D). RT-qPCR
assays indicated that SOX2 was significantly upregulated in
melanoma tissues in comparison to normal tissues (Fig. 5E). Additionally, western blot assays
suggested that the protein expression levels of SOX2 were higher in
melanoma M14 and A375 cells than that in HEMa-LP cells (Fig. 5F). Western blot assays that the
revealed introduction of miR-145-5p reduced the protein expression
levels of SOX2 and that silencing of TUG1 also reduced the SOX2
protein expression levels (Fig. 5G
and H). Pearson analysis
illuminated a negative correlation between miR-145-5p and SOX2
expression levels (Fig. 5I), as
well as between TUG1 and miR-145-5p expression levels (Fig. 5J), while a positive correlation was
found between TUG1 and SOX2 expression levels (Fig. 5K) in melanoma tissues.
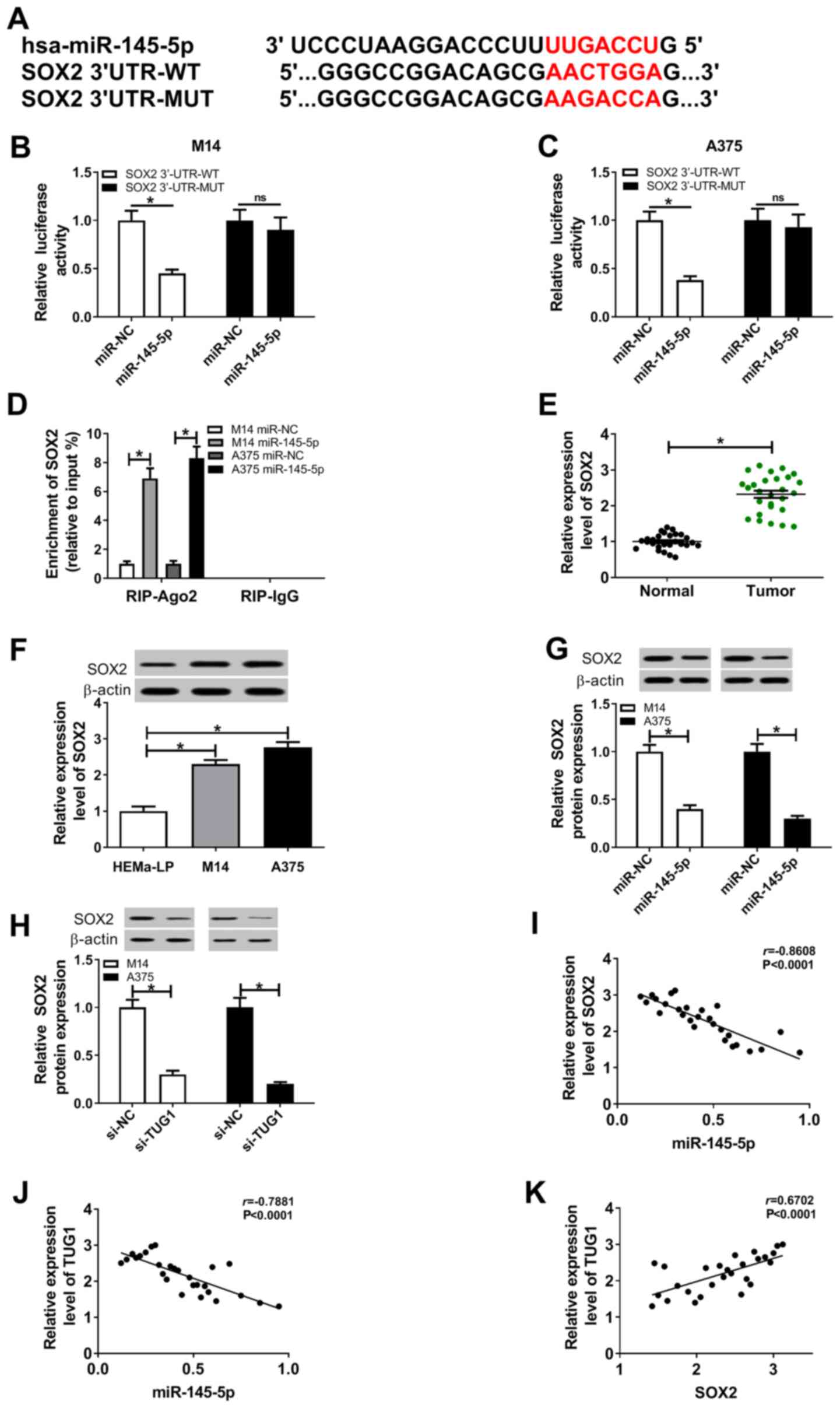 | Figure 5miR-145-5p targets SOX2 and TUG1
regulates SOX2 expression by sponging miR-145-5p. (A)
Bioinformatics analysis showed the potential binding sites of
miR-145-5p on SOX2, using mirTarBase. Dual-luciferase assays were
performed for the analysis of luciferase activity of the (B) M14
and (C) A375 cells co-transfected with SOX2 WT + miR-NC mimics,
SOX2 WT + miR-145-5p mimics, SOX2 MUT + miR-NC mimics or SOX2 MUT +
miR-145-5p mimics. (D) RIP assays were performed and SOX2
enrichment was detected in the samples bound to the Ago2 antibody
in M14 and A375 cells. (E) Reverse transcription-quantitative PCR
for SOX2 expression levels in melanoma tissues. (F) Western blot
assays for SOX2 expression levels in melanoma cells. (G) Western
blot assays for SOX2 expression levels in transfected M14 and A375
cells. (H) The protein expression levels of SOX2 in transfected M14
and A375 cells were analyzed using western blot assays. Correlation
analyses between (I) miR-145-5p and SOX2; (J) TUG1 and miR-145-5p2
and (K) TUG1 and SOX2. *P<0.05. Ago2, argonaute2;
miR, microRNA; MUT, mutant; NC, negative control; ns, no
significance; TUG1, taurine-upregulated gene 1; UTR, untranslated
region; WT, wild-type. |
Silencing of SOX2 restrains the cell
proliferative, migratory and invasive abilities of melanoma cells,
and TUG1 regulates the function of SOX2 by sponging miR-145-5p
To further clarify that TUG1 mediates the
proliferative, migratory and invasive abilities of melanoma cells
by targeting the miR-145-5p/SOX2 axis, restoration experiments were
conducted. As shown in Fig. S1D,
the expression of SOX2 was significantly by transfection with
si-SOX2. The effects of SOX2 on the proliferative ability of
melanoma cells were subsequently evaluated. CCK-8 assays found that
knockdown of SOX2 repressed the proliferative ability of M14 and
A375 cells; however, coinstantaneous knockdown of miR-145-5p
weakened the inhibitory impact (Fig.
6A and B). Similarly, knockdown
of SOX2 blocked the migratory and invasive capacities of M14 and
A375 cells, while simultaneous knockdown of miR-145-5p relieved the
inhibitory effect (Fig. 6C and
D). The CCK-8 assay also suggested
that introduction of SOX2 reversed the inhibitory impact of si-TUG1
on the proliferative ability of M14 and A375 cells (Fig. 6E and F). Upregulation of SOX2 also rescued the
suppressive effect of si-TUG1 on the migratory and invasive
capacities of M14 and A375 cells (Fig.
6G and H).
 | Figure 6Silencing SOX2 restrains the
proliferative, migratory and invasive abilities of melanoma cells,
with TUG1 regulating the function of SOX2 by sponging miR-145-5p.
M14 and A375 cells were transfected with si-NC, si-SOX2, si-SOX2 +
in-miR-NC or si-SOX2 + in-miR-145-5p. (A) The proliferative
abilities of transfected (A) M14 and (B) A375 cells, were
determined using CCK-8 assays, through the measurement of OD450.
The (C) migratory and (D) invasive abilities of M14 and A375 cells
were measured using transwell assays. M14 and A375 cells were
transfected with si-NC, si-TUG1, si-TUG1 + pcDNA or si-TUG1 +
pcDNA-SOX2. CCK-8 assays were performed on the transfected (E) M14
and (F) A375 cells. The (G) migratory and (H) invasive abilities of
the transfected melanoma cells were measured using transwell
assays. *P<0.05. CCK, Cell Counting Kit; in-miR, miR
inhibitor; miR, microRNA; NC, negative control; OD, optical
density; OD450, OD at 450 nm; si, small interfering;
TUG1, taurine-upregulated gene 1. |
Downregulation of TUG1 impedes tumor
growth in nude mice
Finally, A375 cells stably transfected with sh-NC or
sh-TUG1 were constructed, with transfections being confirmed using
RT-qPCR (Fig. 7A). Downregulation
of TUG1 repressed the tumor volume (Fig. 7B) and weight (Fig. 7C) in nude mice compared with the
sh-NC group. In addition, the expression levels of TUG1 were
detected in tumor tissues of nude mice and the results showed that
TUG1 was significantly reduced in the sh-TUG1 group compared with
the sh-NC group (Fig. 7D). In
addition, TUG1 silencing significantly elevated miR-145-5p
expression in xenograft tumor tissues (Fig. 7E). IHC staining assay for SOX2
indicated that SOX2-positive cells in sh-TUG1 group were decreased
compared with that in the sh-NC group (Fig. 7F). Additionally, downregulation of
TUG1 also inhibited SOX2 protein expression levels in the tumors
from nude mice (Fig. 7G). In
summary, TUG1 regulated the proliferation, migration and invasion
of melanoma cells by modulating the miR-145-5p/SOX2 axis (Fig. 8).
Discussion
As the most common skin cancer, melanoma is
extremely aggressive and metastatic (24). A large number of studies on melanoma
have been conducted (2,3), but the mechanism of action behind
melanoma development and progression needs to be further
elucidated. The present study aimed to explore the functional roles
and potential mechanisms of action of TUG1 in melanoma
progression.
LncRNAs are a group of non-protein coding RNAs,
which participate in the modulation of cell growth, migration and
invasion, as well as other important cellular processes in various
types of tumor progression, including melanoma (25,26).
LncRNAs have also been shown to be linked to the development of
melanoma by regulating various mechanisms and signaling pathways
(27). Dysregulation of TUG1 has
been demonstrated in several types of cancer, for example, TUG1 is
upregulated in liver cancer, colorectal cancer, cervical cancer,
ovarian cancer and gastric cancer, while it is downregulated in
non-small cell lung cancer, breast cancer and glioma (11). Additionally, a former study revealed
that TUG1 expression was elevated in melanoma specimens and cell
lines (28), with similar
conclusions being drawn in the present study. TUG1 facilitates
melanoma development and metastasis through the
TUG1/miR-129-5p/Astrocyte-Elevated Gene-1 axis. Wang et al
(29) found that knockdown of TUG1
blocked melanoma cell growth and metastasis, and promoted cell
apoptosis by regulating miR-29c-3p and its target gene regulator of
G-protein signaling. In the present study, knockdown of TUG1
significantly inhibited the proliferative, migratory and invasive
abilities of melanoma cells.
A previous study has shown that lncRNAs can function
as miRNA sponges to decrease the expression levels of miRNAs
available to target mRNAs (30).
The bioinformatics analysis in the present study suggested that
TUG1 targeted miR-145-5p; and the luciferase and RIP assays also
confirmed this interaction between TUG1 and miR-145-5p.
Furthermore, this targeted relationship has been reported in
laryngocarcinoma, gastric cancer, hypertension and intrahepatic
cholangiocarcinoma (ICC) (31-34).
In laryngocarcinoma, TUG1 has been revealed to partake in
laryngocarcinoma progression by regulating the miR-145-5p/ROCK1
axis and activating the RhoA/ROCK/MMPs signaling pathway (31); Ren et al (32) showed that TUG1 augmented cell
proliferation and invasion of gastric cancer cells by directly
binding to miR-145-5p and repress miR-145-5p expression; the lncRNA
TUG1/miR-145-5p/FGF10 axis regulates proliferation and migration in
vascular smooth muscle cells during hypertension by activation of
the Wnt/β-catenin pathway (33);
and as for ICC, TUG1 sponges miR-145 to contribute to cancer
development and modulate glutamine metabolism via the sirtuin
3/glutamate dehydrogenase axis (34). The present study also examined the
downstream genes of the TUG1/miR-145-5p axis. Moreover, the effects
of the interaction between TUG1 and miR-145-5p were evaluated on
proliferation, migration and invasion of melanoma cells.
Upregulation of TUG1 enriched the endogenous levels of TUG1 in
melanoma cells, thereby negatively regulating the expression of
miR-145-5p and aggravating the inhibitory impact of TUG1 on
miR-145-5p expression.
The aforementioned results suggested that miR-145-5p
may play vital roles in the tumorigenesis of melanoma; therefore,
the present study explored the impact and potential mechanisms of
action of this miRNA. Jiang et al (14) claimed that miR-145-5p was
downregulated in breast cancer cell lines, and that it exerts an
important role in the linc01561/miR-145-5p/MMP11 regulation axis of
breast cancer cell proliferation and apoptosis. In bladder cancer,
it has been shown that miR-145-5p is downregulated and that
miR-145-5p inhibits cell proliferation and migration in bladder
cancer by directly targeting transgelin 2(15). Additionally, miR-145-5p represses
cell proliferation, invasion and migration, and induces apoptosis
of melanoma cells by restraining the MAPK and PI3K/AKT pathways
(16), highlighting that miR-145-5p
functions as a tumor-suppressor. Reduced expression levels of
miR-145-5p in melanoma tissues and cells was also detected in the
present study. Moreover, it was found that TUG1 performed its
inhibitory effects on the proliferative, migratory and invasive
abilities of melanoma cells by targeting miR-145-5p.
To identify the regulatory mechanism of action for
miR-145-5p in melanoma cells, bioinformatics analysis was also
performed to identify its downstream gene. SOX2 was identified as a
downstream gene of miR-145-5p. In prostate cancer, raising the
expression of miR-145-5p blocks proliferation and migration in
tumor cell lines, with miR-145-5p targeting SOX2 and inhibiting
SOX2 expression (35). Tang et
al (36) highlighted that
miR-145-5p inhibits breast cancer cell proliferation through
targeting SOX2, with both miR-145-5p and SOX2 being unfavorable
prognostic factors. SOX2 serves as downstream gene of the lncRNA
regulator of reprogramming/miR-145 axis, to maintain pluripotency
of human amniotic epithelial stem cells and regulate their directed
β islet-like cell differentiation efficiency (37). SOX2 has been validated to be
connected to the development and progression of lung cancer, breast
cancer, hepatocellular carcinoma, and ovarian cancer (38). SOX2 also accelerates the invasion
rate of melanoma cells (39). SOX2
has also been proven to reduce the suppressive effect of miR-625 on
the proliferation, migration and invasion in malignant melanoma
cells (40). In the present study,
the expression levels of SOX2 were upregulated, which was
negatively correlated with miR-145-5p expression, but positively
correlated with TUG1 expression. Silencing SOX2 hindered the
proliferative, migratory and invasive abilities of melanoma cells.
The present data manifested that SOX2 was involved in
TUG1/miR-145-5p-mediated melanoma cell proliferation and
metastasis. Additionally, TUG1 modulated SOX2 expression and
exerted its role in melanoma cells by sponging miR-145-5p.
In conclusion, TUG1 and SOX2 were expressed at
higher levels, while miR-145-5p expression was reduced, in melanoma
tissues and cell lines, compared with those in normal tissues and
cell lines. TUG1 inhibition elevated miR-145-5p expression while
repressed SOX2 expression. Moreover, silencing of TUG1 inhibited
the proliferative, migratory and invasive abilities of melanoma
cells, which was reversed by upregulating the expression levels of
miR-145-5p and downregulating the expression levels of SOX2. TUG1
regulated the expression and function of SOX2 by downregulating
miR-145-5p. In addition, knockdown of TUG1 inhibited tumor growth
in nude mice. Taken together, the TUG1/miR-145-5p/SOX2 axis
regulated the migration and invasion of melanoma cells.
Supplementary Material
Transfection efficiency of miR-145-5p
mimic, inhibitor, TUG1 wt, TUG1 mut, si-SOX2, SOX2 wt and SOX2 mut
in M14 and A375 cells. The expression of miR-145-5p in M14 and A375
cells transfected with (A) miR-145-5p mimic or (B) inhibitor.
*P<0.05 vs. miR-NC group or in-miR-NC. (C) TUG1
expression in M14 and A375 cells transfected with pcDNA, TUG1 wt or
TUG1 mut. *P<0.05 vs. pcDNA. (D) RT-qPCR assay for
SOX2 expression in M14 and A375 cells transfected si-NC or si-SOX2.
*P<0.05 vs. si-NC. (E) SOX2 expression in M14 and
A375 cells transfected with pcDNA, SOX2 wt or SOX2 mut.
*P<0.05 vs. pcDNA. miR, microRNA; in-miR, miR
inhibitor; RT-qPCR, reverse transcription-quantitative PCR; wt,
wild type; mut, mutant type; TUG1, taurine-upregulated gene 1;
SOX2, SRY (sex determining region Y)-box 2.
Acknowledgements
Not applicable.
Funding
Funding: This work was supported by Anhui Provincial Institute
of Translational Medicine Research Funded Project (grant no.
2017zhyx07).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
Conceptualization of the study was performed by JD
and YL. JD, YL and FZ designed the investigation. JD, YL and JS
developed the method. JS and FZ performed the project
administration. JD and JS collected and analyzed the data. YL
provided the resources. JD and FZ supervised the study. JD and FZ
wrote, reviewed and edited the manuscript. JD and FZ confirm the
authenticity of all the raw data. All authors read and approved the
final version of the manuscript.
Ethics approval and consent to
participate
The current study was permitted by the Ethics
Committee of the First Affiliated Hospital of Anhui Medical
University. All patients signed written informed consent. The
current animal experiment was approved by the Animal Care and
Scientific Committee of the First Affiliated Hospital of Anhui
Medical University.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Ali Z, Yousaf N and Larkin J: Melanoma
epidemiology, biology and prognosis. EJC Suppl. 11:81–91.
2013.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2017. CA Cancer J Clin. 67:7–30. 2017.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Robert C, Long GV, Brady B, Dutriaux C,
Maio M, Mortier L, Hassel JC, Rutkowski P, McNeil C,
Kalinka-Warzocha E, et al: Nivolumab in previously untreated
melanoma without BRAF mutation. N Engl J Med. 372:320–330.
2015.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Abildgaard C and Guldberg P: Molecular
drivers of cellular metabolic reprogramming in melanoma. Trends Mol
Med. 21:164–171. 2015.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Fang Y and Fullwood MJ: Roles, functions,
and mechanisms of long non-coding RNAs in cancer. Genomics
Proteomics Bioinformatics. 14:42–54. 2016.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Li X, Wu Z, Fu X and Han W: lncRNAs:
Insights into their function and mechanics in underlying disorders.
Mutat Res Rev Mutat Res. 762:1–21. 2014.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Yu X, Zheng H, Tse G, Zhang L and Wu WKK:
CASC2: An emerging tumour-suppressing long noncoding RNA in human
cancers and melanoma. Cell Prolif. 51(e12506)2018.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Lu CW, Zhou DD, Xie T, Hao JL, Pant OP, Lu
CB and Liu XF: HOXA11 antisense long noncoding RNA (HOXA11-AS): A
promising lncRNA in human cancers. Cancer Med. 7:3792–3799.
2018.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Liang L, Zhang Z, Qin X, Gao Y, Zhao P,
Liu J and Zeng W: Long noncoding RNA ZFAS1 promotes tumorigenesis
through regulation of miR-150-5p/RAB9A in melanoma. Melanoma Res.
29:569–581. 2019.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Shi G, Li H, Gao F and Tan Q: lncRNA H19
predicts poor prognosis in patients with melanoma and regulates
cell growth, invasion, migration and epithelial-mesenchymal
transition in melanoma cells. Onco Targets Ther. 11:3583–3595.
2018.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Ghaforui-Fard S, Vafaee R and Taheri M:
Taurine-upregulated gene 1: A functional long noncoding RNA in
tumorigenesis. J Cell Physiol. 234:17100–17112. 2019.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Di Leva G, Garofalo M and Croce CM:
MicroRNAs in cancer. Annu Rev Pathol. 9:287–314. 2014.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Kloosterman WP and Plasterk RH: The
diverse functions of microRNAs in animal development and disease.
Dev Cell. 11:441–450. 2006.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Jiang R, Zhao C, Gao B, Xu J, Song W and
Shi P: Mixomics analysis of breast cancer: Long non-coding RNA
linc01561 acts as ceRNA involved in the progression of breast
cancer. Int J Biochem Cell Biol. 102:1–9. 2018.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Zhang H, Jiang M, Liu Q, Han Z, Zhao Y and
Ji S: miR-145-5p inhibits the proliferation and migration of
bladder cancer cells by targeting TAGLN2. Oncol Lett. 16:6355–6360.
2018.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Jin C, Wang A, Liu L, Wang G, Li G and Han
Z: miR-145-5p inhibits tumor occurrence and metastasis through the
NF-κB signaling pathway by targeting TLR4 in malignant melanoma. J
Cell Biochem. 120:11115–11126. 2019.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Grimm D, Bauer J, Wise P, Kruger M,
Simonsen U, Wehland M, Infanger M and Corydon TJ: The role of SOX
family members in solid tumours and metastasis. Semin Cancer Biol.
67:122–153. 2019.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Andreucci E, Pietrobono S, Peppicelli S,
Ruzzolini J, Bianchini F, Biagioni A, Stecca B and Calorini L: SOX2
as a novel contributor of oxidative metabolism in melanoma cells.
Cell Commun Signal. 16(87)2018.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Schaefer SM, Segalada C, Cheng PF, Bonalli
M, Parfejevs V, Levesque MP, Dummer R, Nicolis SK and Sommer L:
Sox2 is dispensable for primary melanoma and metastasis formation.
Oncogene. 36:4516–4524. 2017.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Wang SH, Zhang WJ, Wu XC, Weng MZ, Zhang
MD, Cai Q, Zhou D, Wang JD and Quan ZW: The lncRNA MALAT1 functions
as a competing endogenous RNA to regulate MCL-1 expression by
sponging miR-363-3p in gallbladder cancer. J Cell Mol Med.
20:2299–2308. 2016.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Aldred AJ, Cha MC and Meckling-Gill KA:
Determination of a humane endpoint in the L1210 model of murine
leukemia. Contemp Top Lab Anim Sci. 41:24–27. 2002.PubMed/NCBI
|
|
23
|
Laga AC, Zhan Q, Weishaupt C, Ma J, Frank
MH and Murphy GF: SOX2 and nestin expression in human melanoma: An
immunohistochemical and experimental study. Exp Dermatol.
20:339–345. 2011.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Singh S, Zafar A, Khan S and Naseem I:
Towards therapeutic advances in melanoma management: An overview.
Life Sci. 174:50–58. 2017.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Peng WX, Koirala P and Mo YY:
LncRNA-mediated regulation of cell signaling in cancer. Oncogene.
36:5661–5667. 2017.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Yu X, Zheng H, Tse G, Chan MT and Wu WK:
Long non-coding RNAs in melanoma. Cell Prolif.
51(e12457)2018.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Richtig G, Ehall B, Richtig E,
Aigelsreiter A, Gutschner T and Pichler M: Function and clinical
implications of long non-coding RNAs in melanoma. Int J Mol Sci.
18(715)2017.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Long J, Menggen Q, Wuren Q, Shi Q and Pi
X: Long noncoding RNA taurine-upregulated gene1 (TUG1) promotes
tumor growth and metastasis through
TUG1/Mir-129-5p/astrocyte-elevated gene-1 (AEG-1) axis in malignant
melanoma. Med Sci Monit. 24:1547–1559. 2018.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Wang Y, Liu G, Ren L, Wang K and Liu A:
Long non-coding RNA TUG1 recruits miR-29c-3p from its target gene
RGS1 to promote proliferation and metastasis of melanoma cells. Int
J Oncol. 54:1317–1326. 2019.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Thomson DW and Dinger ME: Endogenous
microRNA sponges: Evidence and controversy. Nat Rev Genet.
17:272–283. 2016.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Zhuang S, Liu F and Wu P: Upregulation of
long noncoding RNA TUG1 contributes to the development of
laryngocarcinoma by targeting miR-145-5p/ROCK1 axis. J Cell
Biochem. 120:13392–13402. 2019.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Ren K, Li Z, Li Y, Zhang W and Han X: Long
noncoding RNA taurine-upregulated gene 1 promotes cell
proliferation and invasion in gastric cancer via negatively
modulating miRNA-145-5p. Oncol Res. 25:789–798. 2017.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Shi L, Tian C, Sun L, Cao F and Meng Z:
The lncRNA TUG1/miR-145-5p/FGF10 regulates proliferation and
migration in VSMCs of hypertension. Biochem Biophys Res Commun.
501:688–695. 2018.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Zeng B, Ye H, Chen J, Cheng D, Cai C, Chen
G, Chen X, Xin H, Tang C and Zeng J: LncRNA TUG1 sponges miR-145 to
promote cancer progression and regulate glutamine metabolism via
Sirt3/GDH axis. Oncotarget. 8:113650–113661. 2017.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Ozen M, Karatas OF, Gulluoglu S, Bayrak
OF, Sevli S, Guzel E, Ekici ID, Caskurlu T, Solak M, Creighton CJ
and Ittmann M: Overexpression of miR-145-5p inhibits proliferation
of prostate cancer cells and reduces SOX2 expression. Cancer
invest. 33:251–258. 2015.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Tang W, Zhang X, Tan W, Gao J, Pan L, Ye
X, Chen L and Zheng W: miR-145-5p suppresses breast cancer
progression by inhibiting SOX2. J Surg Res. 236:278–287.
2019.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Zou G, Liu T, Guo L, Huang Y, Feng Y,
Huang Q and Duan T: miR-145 modulates lncRNA-ROR and Sox2
expression to maintain human amniotic epithelial stem cell
pluripotency and β islet-like cell differentiation efficiency.
Gene. 591:48–57. 2016.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Zeng H, Wang L, Wang J, Chen T, Li H,
Zhang K, Chen J, Zhen S, Tuluhong D, Li J and Wang S:
microRNA-129-5p suppresses Adriamycin resistance in breast cancer
by targeting SOX2. Arch Biochem Biophys. 651:52–60. 2018.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Girouard SD, Laga AC, Mihm MC, Scolyer RA,
Thompson JF, Zhan Q, Widlund HR, Lee CW and Murphy GF: SOX2
contributes to melanoma cell invasion. Lab Invest. 92:362–370.
2012.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Fang W, Fan Y, Fa Z, Xu J, Yu H, Li P and
Gu J: microRNA-625 inhibits tumorigenicity by suppressing
proliferation, migration and invasion in malignant melanoma.
Oncotarget. 8:13253–13263. 2017.PubMed/NCBI View Article : Google Scholar
|






















