Introduction
Wiskott-Aldrich Syndrome (WAS) is a rare X-linked
primary immunodeficiency disorder that affects males with a
frequency of one in 10,000,000(1).
It can lead to recurrent infection, thrombocytopenia, eczema, a
high incidence of malignancy and autoimmune complications (in
40-70% of patients) (2). These
complications can be fatal if not treated with bone marrow
transplantation or gene therapy (3). WAS is caused by a coding mutation in
the was gene, which is located on the short arm of the X
chromosome and was identified by positional cloning in
1994(4). The was gene
contains 12 exons and encodes a highly proline-rich protein of 502
amino acid residues, which is named WAS protein (WASp) (5).
The WASp protein contains multiple functional
domains and serves a key role in the regulation of branched actin
chain polymerization, primarily regulating the actin cytoskeleton
in hematopoietic cells (6). The
role of WASp deficiency in the development of autoimmunity in WAS
has been explored extensively (7-9).
It has been demonstrated that WASp is involved in regulation of
immunity, cell proliferation, differentiation, activation and
function in an actin-dependent manner (10,11).
WASP also influences the immune response by regulating the
transcription levels of CD19 receptor in B cells and inflammatory
factors (12). Moreover, it has
been demonstrated that WASp-deficient natural regulatory T cells
are defective in suppressing effector T cells and B lymphocyte
proliferation (13,14). However, the cellular and molecular
mechanisms in WASp underlying WAS, particularly the pathogenesis of
autoimmunity, remain unclear.
Studies have found that IL-10 production of
regulatory B cells is decreased in WASp-knockout (WAS-KO) mice
(15) and the abundance of Th1 and
Th17 cells is increased (16),
which leads to impairment of immune function. In addition,
WASP-deficient T cells in WAS-KO mice show markedly impaired
proliferation and antigen receptor cap formation in response to
anti-CD3ε stimulation, which is consistent with patients with WAS
(17,18). The spleen is the largest secondary
lymphoid organ in the body and serves a major role not only in
immunological functions but also in hematopoiesis and red blood
cell clearance (19,20). Therefore, the spleen may serve an
important role in the progression or treatment of patients with WAS
and the study of the role of the spleen in autoimmunity in WAS-KO
mice may inform understanding of the pathogenesis of autoimmune
manifestations in WAS.
The present study investigated the differences in
the spleen transcriptome of 10-week-old WAS-KO and wild-type (WT)
mice. Differentially expressed genes (DEGs) that were significantly
altered were identified and Gene Ontology (GO) analysis to
determine the specific functions of genes and Kyoto Encyclopedia of
Genes and Genomes (KEGG) analysis to evaluate the enrichment of
gene sets was performed. In addition, DisGeNET and Reactome
analyses were performed to find possible novel autoimmune
complications. To the best of our knowledge, the present study is
the first to investigate transcriptome sequences in the spleen of
WAS-KO mice, which may facilitate understanding of the cellular and
molecular mechanisms in WAS.
Materials and methods
Mice and tissue processing
Was -/-
(129S6/SvEvTac-Wastm1Sbs/J; WAS-KO) and 129S6/SvEvTac
(WT) mice were purchased from the Jackson Laboratory (Bar Harbor,
ME, USA). It is a mature model widely used in the study of WAS
(21,22). The total number of WAS-KO mice is 6
and WT mice is 6. All mice were male used in this study. All
animals were maintained with food and water ad libitum under
a 12/12-h light/dark cycle at 22±2˚C, with 50±10% relative
humidity. Ten-week-old male mice (about 25 g) were used and all
experiments were approved by the Medical Ethics Committee of The
Third People's Hospital of Shenzhen (approval no. 2021038). The
mice were anesthetized with pentobarbital sodium (1%, 30 mg/kg,
i.p.) and the spleen was isolated. Mice were euthanized by
CO2 gas (4.5 l/min) with 30% air displacement rate in an
individually caging system (KW-AL-G; Nanjing Calvin Biotechnology
Co., Ltd.). Total RNA was extracted from the right spleen using an
RNAqueous™ kit (cat. no. AM1912; Thermo Fisher Scientific, Inc.)
following the manufacturer's instructions. Purity was determined by
a NanoDrop 1000 (Thermo Fisher Scientific, Inc.).
RNA-sequencing (seq) analysis
RNA-seq library was prepared by a TruSeq RNA sample
preparation kit (Illumina, Inc.) and sequencing of libraries was
conducted on an Illumina HiSeq system. A total of three samples
were collected from each group. The read depth was
30-50x106 bp for each sample. Skewer (v0.2.2) software
(sourceforge.net) was used to dynamically remove
joint sequence fragments and low-quality fragments from the 3' end
of the sequencing data. FastQC (v0.11.5) software (Babraham
Bioinformatics) was used to conduct quality control analysis on the
preprocessed data. Sequence data were mapped to the mouse reference
genome GRCm38(23) with STAR
(v2.5.3a) software (github.com). StringTie (v1.3.1c)
software (ccb.jhu.edu) was used to calculate the
original sequence count of known genes for all samples and the
expression levels of known genes were calculated using fragments
per kilobase of transcript per million fragments mapped.
DEG analysis
DEG analysis was conducted between WT and WAS-KO
mice to identify significantly up- or downregulated genes. DEGs
were assessed by DESeq 2 (v1.16.1) software (bioconductor.org). Benjamini-Hochberg multiple test
correction method was used. DEGs were chosen according to
|log2 fold-change |≥1 and adjusted P-value <0.05.
Functional annotation clustering was performed using TopGO (v2.48)
software (bioconductor.org). GO (geneontology.org) and KEGG (genome.jp)
annotations were tested for enrichment and a
Benjamini-Hochberg-corrected P-value <0.05 was considered to
indicate a statistically significant difference. Functional
analysis of disease annotation was performed via functional
annotation and classification of disease types in the DisGeNET
disease database (disgenet.org) for DEGs. Reactome
metabolic pathway analysis was used to annotate DEGs for metabolic
pathways in the Reactome database (reactome.org/). Heatmaps were plotted by
Bioinformatics (bioinformatics.com.cn), a free online platform for
data analysis and visualization.
Results
DEG analysis
DEGs were selected according to the criteria |log2
(fold-change) |≥1 and P<0.05. DEGs are shown in Fig. 1A. A total of 1,964 genes were
differentially expressed in WAS-KO compared with WT mice, with 996
genes upregulated and 968 genes downregulated (Table SI). Cluster analysis of DEGs
between the WT and WAS-KO groups was performed and the results are
shown as a heatmap (Fig. 1B).
GO enrichment analysis
To determine the functions of the identified DEGs,
GO enrichment analysis was performed for significantly upregulated
and downregulated DEGs (Fig. 2).
The top 10 biological processes (BP), cellular components (CC) and
molecular functions (MF) in the GO analysis were selected. The GO
results showed that upregulated DEGs were involved in ‘heme
metabolic process’, ‘cell division’ and ‘cofactor biosynthetic
process’, according to BP. In terms of CC annotations, ‘cytoplasm’,
‘extracellular region part’ and ‘chromosome, centromeric region’
showed significant enrichment. In terms of the MF annotations,
‘cytoskeletal protein binding’, ‘ATPase regulator activity’ and
‘organic acid transmembrane transporter activity’ showed
significant enrichment (Fig. 2A).
To determine the association between GO clusters, network analysis
of the top 20 GO clusters in the upregulated DEGs was performed.
Genes were enriched in ‘erythrocyte development’, ‘cell cycle’ and
‘heme and pigment metabolic process’ (Fig. 2B).
GO analysis of downregulated DEGs showed that they
were involved in ‘microtubule bundle formation’, ‘anatomical
structure formation involved in morphogenesis’ and ‘cellular
component assembly involved in morphogenesis’ BPs. In terms of CC
annotation, ‘extracellular matrix component’ and ‘extrinsic
component of membrane’ showed significant enrichment. In terms of
the MF annotation, ‘ion binding’, ‘heparin binding’ and
‘glycosaminoglycan binding’ showed significant enrichment (Fig. 2C). The network analysis of the top
20 GO clusters in the downregulated DEGs yielded four clusters:
‘Anatomical structure morphogenesis’, ‘renal system’,
‘extracellular matrix’ and ‘cellular component assembly’ (Fig. 2D).
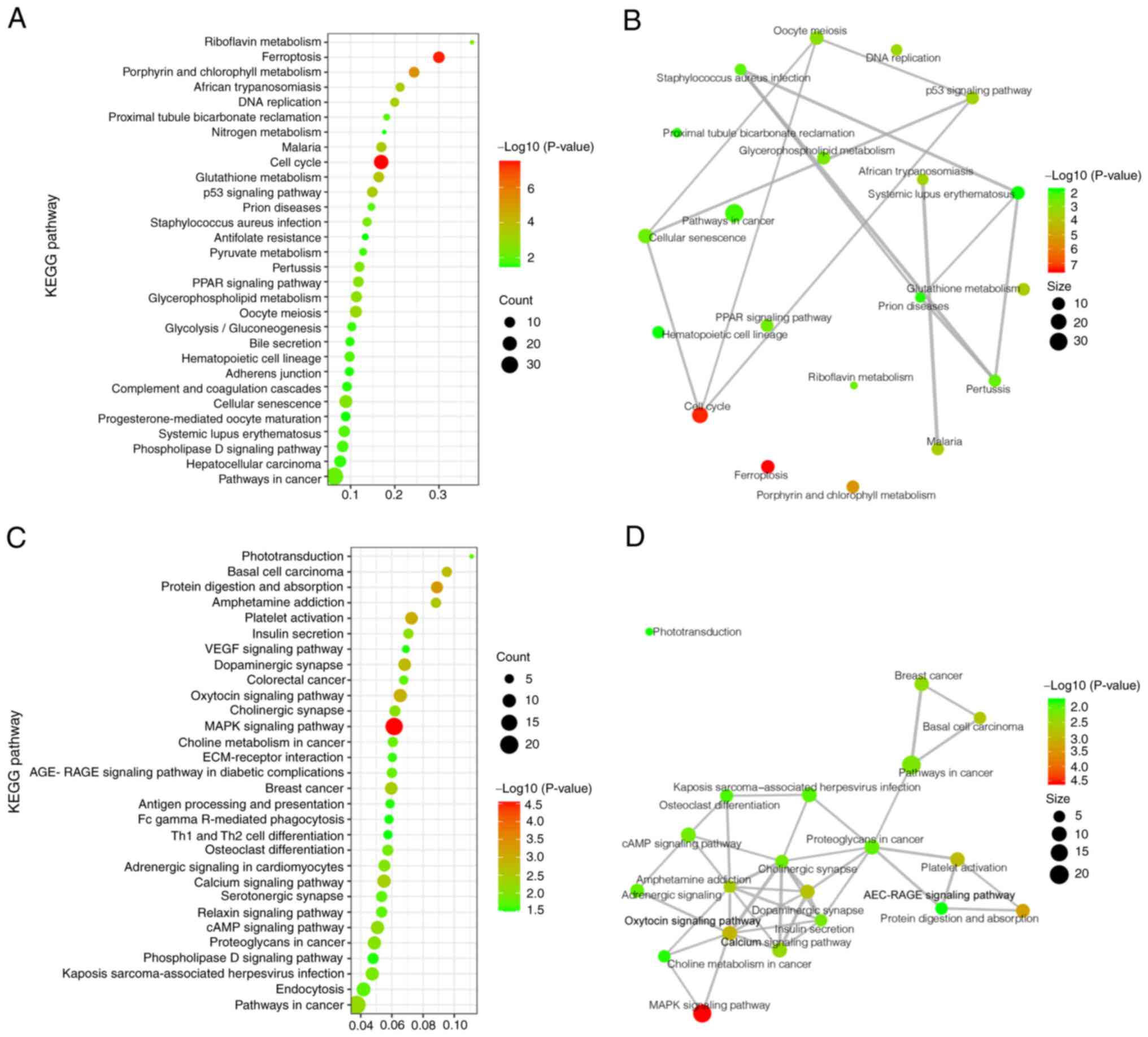
KEGG analysis
To determine which pathways may be directly affected
by gene deficiency in WAS-KO mice, the significantly upregulated
and downregulated DEGs were analyzed using the KEGG pathway
database. The significantly upregulated DEGs were primarily
involved in signaling pathways including ‘cellular senescence’,
‘p53 signaling pathway’ and ‘ferroptosis’ (a cell death pathway). A
number of other DEGs were found to participate in different
metabolic pathways, such as ‘glutathione metabolism’,
‘glycolysis/gluconeogenesis’ and ‘glycerophospholipid metabolism’
(Fig. 3A). To clarify the links
between these pathways, a network analysis of the top 20 KEGG
clusters in the upregulated DEGs was performed; the primary
upregulated pathway was cell cycle and cell death (Fig. 3B).
KEGG results of downregulated DEGs showed that these
genes were involved in ‘platelet activation’, ‘Th1 and Th2 cell
differentiation’ and ‘antigen processing and presentation’ for the
immune system and ‘MAPK signaling pathway’, ‘calcium signaling
pathway’ and ‘cAMP signaling pathway’ for signal transduction
(Fig. 3C). In addition, the
network analysis of the top 20 KEGG clusters in the downregulated
DEGs showed that the main downregulated pathway was signal
transduction.
DisGeNET and Reactome analysis
To investigate potential symptoms caused by DEGs, a
disease enrichment analysis was performed using the DisGeNET
database. Analysis of the top 30 clusters of disease terms showed
that ‘anemia, hemolytic’, ‘anemia, sickle cell’, ‘hereditary
spherocytic hemolytic’, ‘spherocytosis’, ‘thalassemia’ and
‘hemoglobin H disease’ were highly enriched (Fig. 4A). To determine the association
between these diseases, a network analysis of the top 20 clusters
was performed. The results showed that anemia and hemolysis were
primarily associated with ‘serum ferritin increased’, ‘hemoglobin
low’, ‘increased bilirubin level’ and ‘increased red cell osmotic
fragility’ (Fig. 4B).
To elucidate the mechanisms by which these diseases
may occur in WAS-KO mice, enrichment analysis was performed using
the Reactome database. Analysis of the top 30 clusters of Reactome
terms showed that ‘heme biosynthesis’, ‘activation of ATR in
response to replication stress’, ‘collagen degradation’, ‘G1-TP53
regulates transcription of genes involved in G1 cell cycle arrest’
and ‘glutathione synthesis and recycling’ were highly enriched
(Fig. 4C). The network analysis of
the top 20 clusters showed that ‘collagen biosynthesis and
modifying enzymes’ and ‘erythrocytes take up oxygen and release
carbon dioxide’ were associated with these terms (Fig. 4D).
Prognostic implications of derived
markers genes in WAS
To validate the aforementioned signaling pathways
and determine potential novel marker genes in WAS, expression
levels of marker genes were compared between WT and WAS-KO mice
(Fig. 5). Marker genes involved in
CC assembly and morphogenesis were significantly decreased in
spleen of WAS-KO mice, such as Ccdc114, Ccdc136 and
Dnah1 (Fig. 5A). In
addition, expression of apoptosis marker genes such as Bcl2,
Fth, Pdcd and Maged increased significantly.
Furthermore, the marker genes involved in the immune system, such
as Mapk, Rasgrp2, Plag4b, COL1A2 and
Hspa were significantly downregulated. Moreover, the
expression levels of marker genes involved in signal transmission,
such as Camk2b, Camk4, Rac1 and Fos,
were significantly decreased in the spleen of WAS-KO mice. The
information of these marker genes is shown in Table SII.
Discussion
WAS has a wide clinical spectrum ranging from mild
with thrombocytopenia, recurrent infections and eczema to severe
presentation, which can include complications such as
life-threatening hemorrhage, immunodeficiency, atopy, autoimmunity
and cancer (4,6). The pathophysiology of features of WAS
is being elucidated by clinical and basic studies but remains
unclear, which hinders the application of targeted therapies. Gene
expression analysis is a useful tool in the study of disease. The
present study investigated the differences in spleen transcriptomes
of 10-week-old WAS-KO and WT mice. The processes of heme
metabolism, cell division and ATPase regulator activity were
enhanced, but microtubule bundle formation and anatomical structure
formation involved in morphogenesis were impaired in the spleens of
WAS-KO mice. The levels of cell senescence and apoptosis-associated
genes were increased, antigen processing and presentation
mechanisms involved in the immune response were damaged and signal
transduction processes were impaired in the spleens of WAS-KO mice.
Gene deletion may lead to anemia and hemolysis-associated disease,
mainly due to increased erythrocyte osmotic fragility, low
hemoglobin and increased bilirubin levels and serum ferritin. These
results indicated that the senescence and apoptosis of blood cells
play a key role in the occurrence of WAS. However, most studies
have focused only on the immune response (24-26).
Therefore, the present findings may provide a broader theoretical
basis for further study to improve the treatment of WAS.
The large amount of cell senescence and apoptosis in
the spleens of WAS-KO mice promoted cell division and cofactor
biosynthesis processes, but dysfunction in cell structure formation
and microtubule bundle formation may lead to functional defects in
newly generated cells and ultimately cause splenic damage spleen in
these mice. Rawlings et al (27) examined the susceptibility to
apoptosis of resting primary lymphocytes isolated from patients
with WAS in the absence of exogenous apoptogenic stimulation.
Rengan et al (28) also
found accelerated cell death in WAS lymphocytes, as evidenced by
increased caspase-3 activity. This suggests that was gene
deletion leads not only to lymphocyte apoptosis but also to
erythrocyte senescence and apoptosis; this topic requires more
research. Here, marker genes of apoptosis, such as Bcl2,
Fth, PD and Mage increased significantly in
spleen of WAS-KO mice. Deletion of was gene leads to the
differentiation defect of T cells by downregulating transcription
level of PD-1 and Bcl-6. (29,30).
Hashimoto et al (31) found
that PD-1 and Mage-a4 are involved in the aggressive
elements of soft tissue sarcomas (32). In addition, ferroptosis is an
iron-dependent programmed cell death event, which affected by
Fth gene (33). To the best
of our knowledge, there is no research on the correlation between
Fth and WAS. Thus, the marker genes PD, Mage
and Fth identified here may affect the progression of WAS.
In addition, reactive oxygen species (ROS) and inflammation may
induce cell death (34,35). Production of ROS is associated with
dynamic actin cytoskeleton reorganization (36) and NADPH oxidase-dependent
physiologically generated ROS negatively regulate actin
polymerization in stimulated neutrophils via driving reversible
actin glutathionylation (37).
Therefore, neutrophil dysfunction induced by ROS may accelerate
pathogenesis of WAS. Furthermore, inflammation is involved in
immunodeficiency (38). However,
no signaling pathways directly associated with ROS or inflammation
were enriched in the present study. The roles of ROS and
inflammation in WAS should be investigated in future.
WASp is a cytoskeletal scaffolding adapter that
coordinates transmission of stimulatory signals to downstream
inducers of actin remodeling and cytoskeletal-dependent T cell
responses (39). To the best of
our knowledge, however, there is a lack of studies on the
interaction between marker genes in the immune system and WAS. The
present study found marker genes involved in immune system such as
Mapk, Rasgrp2, Plag4b, COL1A2 and
Hspa, which may contribute to the study of WAS. Heat shock
proteins serve an important cytoprotective role in cells exposed to
stressful conditions and are implicated in auto-immune disease
(40,41). In addition, MAPK signaling has
roles both in innate and adaptive immune responses, including
induction of pro-inflammatory mediators (42,43).
T-cell receptors, upon binding to specific ligands of major
histocompatibility complex (MHC) molecules on antigen-presenting
cells, initiate intracellular signaling that leads to extensive
actin polymerization (14,44). Thus, marker genes belonging to MHC
type, such as H2-Q6, H2-T10 and H2-T10, may be
associated with actin polymerization. Furthermore, WAS-interacting
protein influences the function of CD19 as a general hub for PI3K
signaling by regulating the cortical actin cytoskeleton in humans
with WAS (45). Consistent with
this, marker gene pik3r5, which is involved in PI3K
signaling pathway, was downregulated in WAS-KO mice, suggesting
that pik3r5 is a potential marker gene for WAS.
In addition, WASp serves an essential role in signal
transduction and effector functions of T cells; signal transduction
regulating the function of T cells in immune response is impaired
in WAS (14). In the present
study, the expression levels of marker genes involved in signal
transmission, such as Camk2b, Camk4, Rac1 and
Fos, were significantly decreased in the spleen of WAS-KO
mice, suggesting the signal transduction in WAS-KO mice was
impaired. For signal transduction, there are four pathways
required, of which MAPK pathway is the most important (46). The Ras pathway activates the
extracellular receptor-activated kinase (Erk) by phosphorylating
its substrate, Elk1. Phospho-Elk1 then stimulates the transcription
of c-Fos, a component of the transcription factor activation
protein 1(47). WASp is essential
for nuclear translocation of phospho-Erk, Elk1 phosphorylation and
expression of c-Fos (48). This
suggests that Ras1 and Fos may be used as clinical
markers to evaluate Ras signaling pathway in WAS. Furthermore, T
cell receptor engagement triggers mobilization of Ca2+,
which is required for T cell activation, gene expression, motility,
synapse formation, cytotoxicity, development and differentiation
(49,50). T cells from patients with WAS and
WAS-KO mice show defects in intracellular Ca2+
mobilization (51). Here, marker
genes Camk2b and Camk4 were significantly
downregulated in spleen of WAS-KO mice and may be involved in the
decreased intracellular Ca2+ mobilization. Therefore,
the present results may improve the knowledge of signal
transduction in WAS. Some of these marker genes have been confirmed
to be involved in the occurrence of WAS by clinical testing
(52,53), which indicates that the marker
genes detected in this study have the potential for clinical
application.
Studies have found hemolysis and thrombocytopenia in
WAS (54,55); for example, Burroughs et al
(56) found that hematopoietic
cell transplantation (HCT) significantly increases platelet levels
in patients with WAS and is associated with better outcomes when
performed at a younger age. The present study found that
thrombocytopenia may be associated with increased erythrocyte
osmotic fragility, low hemoglobin and increased bilirubin levels
and serum ferritin, which may be targeted by gene therapy.
Moreover, Rohrer et al (57) found WAS in a family with Fanconi
anemia, indicating the occurrence of these two rare genetic
disorders in a single family or the existence of an unusual variant
of Fanconi anemia. This is consistent with the present prediction
by disease enrichment analysis that was gene deletion may
lead to anemia- and hemolysis-associated disease.
The present study detected 1,964 DEGs between WAS-KO
and WT mice but only showed the top 30 enrichment results; other
results indicated that the role of the spleen in WAS needs more
attention in further studies (data not shown). In addition, the
present study did not verify expression changes of DEGs by other
methods. Further analysis of transcriptome data available may aid
in discovering novel mechanisms to improve therapies for WAS,
especially in the context of anemia and primary immunodeficiency.
Moreover, clinical samples are being collected from patients with
WAS; analysis of differences in transcriptome levels between WAS-KO
mice and patients with WAS in future studies and identification of
marker genes may provide the basis for clinical gene therapy.
Supplementary Material
Differentially expressed genes.
Relative expression of marker
genes.
Acknowledgements
Not applicable.
Funding
Funding: The present study was supported by Shenzhen Science and
Technology Innovation Commission Funds (grant no.
JCYJ20210324141011028) and Sanming Project of Medicine in Shenzhen
(grant no. SZSM201812002).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request. The RNA-seq data used in this manuscript are publicly
available in the Gene Expression Omnibus repository (accession no.
GSE214745; ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE214745).
Authors' contributions
FFL, JY, QG, YX, LLW, YYH and CP contributed to the
study design. FFL, JY and CP analyzed the data. QG, YX, LLW and YYH
collected data. FFL wrote the manuscript. CP revised the
manuscript. CP and FFL confirm the authenticity of all the raw
data. All authors have read and approved the final manuscript.
Ethics approval and consent to
participate
All experiments were approved by the Medical Ethics
Committee of The Third People's Hospital of Shenzhen (approval no.
2021038).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Peacocke M and Siminovitch KA:
Wiskott-Aldrich syndrome: New molecular and biochemical insights. J
Am Acad Dermatol. 27:507–519. 1992.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Yokoyama T, Yoshizaki A, Simon KL, Kirby
MR, Anderson SM and Candotti F: Age-dependent defects of regulatory
B cells in wiskott-aldrich syndrome gene knockout mice. PLoS One.
10(e0139729)2015.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Worth AJ and Thrasher AJ: Current and
emerging treatment options for Wiskott-Aldrich syndrome. Expert Rev
Clin Immunol. 11:1015–1032. 2015.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Ariga T: Wiskott-Aldrich syndrome; an
x-linked primary immunodeficiency disease with unique and
characteristic features. Allergol Int. 61:183–189. 2012.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Safaei S, Fazlollahi MR, Houshmand M,
Hamidieh AA, Bemanian MH, Alavi S, Mousavi F, Pourpak Z and Moin M:
Detection of six novel mutations in WASP gene in fifteen Iranian
Wiskott-Aldrich patients. Iran J Allergy Asthma Immunol.
11:345–348. 2012.PubMed/NCBI
|
|
6
|
Massaad MJ, Ramesh N and Geha RS:
Wiskott-Aldrich syndrome: A comprehensive review. Ann N Y Acad Sci.
1285:26–43. 2013.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Notarangelo LD, Notarangelo LD and Ochs
HD: WASP and the phenotypic range associated with deficiency. Curr
Opin Allergy Clin Immunol. 5:485–490. 2005.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Sasahara Y: WASP-WIP complex in the
molecular pathogenesis of Wiskott-Aldrich syndrome. Pediatr Int.
58:4–7. 2016.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Calle Y, Jones GE, Jagger C, Fuller K,
Blundell MP, Chow J, Chambers T and Thrasher AJ: WASp deficiency in
mice results in failure to form osteoclast sealing zones and
defects in bone resorption. Blood. 103:3552–3561. 2004.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Snapper SB and Rosen FS: The
Wiskott-Aldrich syndrome protein (WASP): Roles in signaling and
cytoskeletal organization. Annu Rev Immunol. 17:905–929.
1999.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Candotti F: Clinical manifestations and
pathophysiological mechanisms of the Wiskott-Aldrich Syndrome. J
Clin Immunol. 38:13–27. 2018.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Bai X, Zhang Y, Huang L, Wang J, Li W, Niu
L, Jiang H, Dai R, Zhou L, Zhang Z, et al: The early activation of
memory B cells from Wiskott-Aldrich syndrome patients is suppressed
by CD19 downregulation. Blood. 128:1723–1734. 2016.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Rey-Suarez I, Wheatley BA, Koo P, Bhanja
A, Shu Z, Mochrie S, Song W, Shroff H and Upadhyaya A: WASP family
proteins regulate the mobility of the B cell receptor during
signaling activation. Nat Commun. 11(439)2020.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Ngoenkam J, Paensuwan P, Wipa P, Schamel
WWA and Pongcharoen S: Wiskott-Aldrich syndrome protein: Roles in
signal transduction in T cells. Front Cell Dev Biol.
9(674572)2021.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Nguyen DD, Wurbel MA, Goettel JA, Eston
MA, Ahmed OS, Marin R, Boden EK, Villablanca EJ, Paidassi H, Ahuja
V, et al: Wiskott-Aldrich syndrome protein deficiency in innate
immune cells leads to mucosal immune dysregulation and colitis in
mice. Gastroenterology. 143:719–729.e2. 2012.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Du HQ, Zhang X, An YF, Ding Y and Zhao XD:
Effects of Wiskott-Aldrich syndrome protein deficiency on
IL-10-producing regulatory B cells in humans and mice. Scand J
Immunol. 81:483–493. 2015.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Snapper SB, Rosen FS, Mizoguchi E, Cohen
P, Khan W, Liu CH, Hagemann TL, Kwan SP, Ferrini R, Davidson L, et
al: Wiskott-Aldrich syndrome protein-deficient mice reveal a role
for WASP in T but not B cell activation. Immunity. 9:81–91.
1998.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Cleland SY and Siegel RM: Wiskott-Aldrich
Syndrome at the nexus of autoimmune and primary immunodeficiency
diseases. FEBS Lett. 585:3710–3714. 2011.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Lewis SM, Williams A and Eisenbarth SC:
Structure and function of the immune system in the spleen. Sci
Immunol. 4(eaau6085)2019.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Safeukui I, Buffet PA, Deplaine G, Perrot
S, Brousse V, Sauvanet A, Aussilhou B, Dokmak S, Couvelard A,
Cazals-Hatem D, et al: Sensing of red blood cells with decreased
membrane deformability by the human spleen. Blood Adv. 2:2581–2587.
2018.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Keszei M, Kritikou JS, Sandfort D, He M,
Oliveira MMS, Wurzer H, Kuiper RV and Westerberg LS:
Wiskott-Aldrich syndrome gene mutations modulate cancer
susceptibility in the p53(±) murine model. Oncoimmunology.
7(e1468954)2018.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Li S, Huang J, Zhang YL, Zhu Y, An YF, Du
J, Zhang ZL, Xia Y, Liu L, Wang L and Luo XH: Wiskott-Aldrich
syndrome protein may be critical for CD8(+) T cell function
following MCMV infection. Cell Immunol. 338:43–50. 2019.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Sarsani VK, Raghupathy N, Fiddes IT,
Armstrong J, Thibaud-Nissen F, Zinder O, Bolisetty M, Howe K,
Hinerfeld D, Ruan X, et al: The Genome of C57BL/6J ‘Eve’, the
mother of the laboratory mouse genome reference strain. G3
(Bethesda). 9:1795–1805. 2019.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Yang H and Hultmark D: Drosophila muscles
regulate the immune response against wasp infection via
carbohydrate metabolism. Sci Rep. 7(15713)2017.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Thrasher AJ: WASp in immune-system
organization and function. Nat Rev Immunol. 2:635–646.
2002.PubMed/NCBI View
Article : Google Scholar
|
|
26
|
Blundell MP, Worth A, Bouma G and Thrasher
AJ: The Wiskott-Aldrich syndrome: The actin cytoskeleton and immune
cell function. Dis Markers. 29:157–175. 2010.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Rawlings SL, Crooks GM, Bockstoce D,
Barsky LW, Parkman R and Weinberg KI: Spontaneous apoptosis in
lymphocytes from patients with Wiskott-Aldrich syndrome:
Correlation of accelerated cell death and attenuated bcl-2
expression. Blood. 94:3872–3882. 1999.PubMed/NCBI
|
|
28
|
Rengan R, Ochs HD, Sweet LI, Keil ML,
Gunning WT, Lachant NA, Boxer LA and Omann GM: Actin cytoskeletal
function is spared, but apoptosis is increased, in WAS patient
hematopoietic cells. Blood. 95:1283–1292. 2000.PubMed/NCBI
|
|
29
|
Raes L, Pille M, Harizaj A, Goetgeluk G,
Van Hoeck J, Stremersch S, Fraire JC, Brans T, de Jong OG,
Maas-Bakker R, et al: Cas9 RNP transfection by vapor nanobubble
photoporation for ex vivo cell engineering. Mol Ther Nucleic Acids.
25:696–707. 2021.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Zhang X, Dai R, Li W, Zhao H, Zhang Y,
Zhou L, Du H, Luo G, Wu J, Niu L, et al: Abnormalities of
follicular helper T-cell number and function in Wiskott-Aldrich
syndrome. Blood. 127:3180–3191. 2016.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Hashimoto K, Nishimura S, Ito T, Kakinoki
R and Akagi M: Immunohistochemical expression and
clinicopathological assessment of PD-1, PD-L1, NY-ESO-1, and
MAGE-A4 expression in highly aggressive soft tissue sarcomas. Eur J
Histochem. 66(3393)2022.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Hashimoto K, Nishimura S, Sakata N, Inoue
M, Sawada A and Akagi M: Characterization of PD-1/PD-L1 immune
checkpoint expression in the pathogenesis of musculoskeletal
Langerhans cell histiocytosis: A retrospective study. Medicine
(Baltimore). 100(e27650)2021.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Fuhrmann DC, Mondorf A, Beifuß J, Jung M
and Brüne B: Hypoxia inhibits ferritinophagy, increases
mitochondrial ferritin, and protects from ferroptosis. Redox Biol.
36(101670)2020.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Oliveira JB and Gupta S: Disorders of
apoptosis: Mechanisms for autoimmunity in primary immunodeficiency
diseases. J Clin Immunol. 28 (Suppl 1):S20–S28. 2008.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Fuchs TA, Abed U, Goosmann C, Hurwitz R,
Schulze I, Wahn V, Weinrauch Y, Brinkmann V and Zychlinsky A: Novel
cell death program leads to neutrophil extracellular traps. J Cell
Biol. 176:231–241. 2007.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Park SJ, Kim YT and Jeon YJ: Antioxidant
dieckol downregulates the Rac1/ROS signaling pathway and inhibits
Wiskott-Aldrich syndrome protein (WASP)-family verprolin-homologous
protein 2 (WAVE2)-mediated invasive migration of B16 mouse melanoma
cells. Mol Cells. 33:363–369. 2012.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Sakai J, Li J, Subramanian KK, Mondal S,
Bajrami B, Hattori H, Jia Y, Dickinson BC, Zhong J, Ye K, et al:
Reactive oxygen species-induced actin glutathionylation controls
actin dynamics in neutrophils. Immunity. 37:1037–1049.
2012.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Perelygina L, Icenogle J and Sullivan KE:
Rubella virus-associated chronic inflammation in primary
immunodeficiency diseases. Curr Opin Allergy Clin Immunol.
20:574–581. 2020.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Zhang J, Dong B and Siminovitch KA:
Contributions of Wiskott-Aldrich syndrome family cytoskeletal
regulatory adapters to immune regulation. Immunol Rev. 232:175–194.
2009.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Zininga T, Ramatsui L and Shonhai A: Heat
shock proteins as immunomodulants. Molecules.
23(2846)2018.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Grundtman C, Kreutmayer SB, Almanzar G,
Wick MC and Wick G: Heat shock protein 60 and immune inflammatory
responses in atherosclerosis. Arterioscler Thromb Vasc Biol.
31:960–968. 2011.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Arthur JS and Ley SC: Mitogen-activated
protein kinases in innate immunity. Nat Rev Immunol. 13:679–692.
2013.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Liu Y, Shepherd EG and Nelin LD: MAPK
phosphatases-regulating the immune response. Nat Rev Immunol.
7:202–212. 2007.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Matalon O, Reicher B and Barda-Saad M:
Wiskott-Aldrich syndrome protein-dynamic regulation of actin
homeostasis: From activation through function and signal
termination in T lymphocytes. Immunol Rev. 256:10–29.
2013.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Keppler SJ, Gasparrini F, Burbage M,
Aggarwal S, Frederico B, Geha RS, Way M, Bruckbauer A and Batista
FD: Wiskott-Aldrich syndrome interacting protein deficiency
uncovers the role of the Co-receptor CD19 as a generic Hub for PI3
kinase signaling in B cells. Immunity. 43:660–673. 2015.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Conley JM, Gallagher MP and Berg LJ: T
cells and gene regulation: The switching on and turning up of genes
after T cell receptor stimulation in CD8 T cells. Front Immunol.
7(76)2016.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Smith-Garvin JE, Koretzky GA and Jordan
MS: T cell activation. Annu Rev Immunol. 27:591–619.
2009.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Sossey-Alaoui K, Ranalli TA, Li X, Bakin
AV and Cowell JK: WAVE3 promotes cell motility and invasion through
the regulation of MMP-1, MMP-3, and MMP-9 expression. Exp Cell Res.
308:135–145. 2005.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Feske S: Calcium signalling in lymphocyte
activation and disease. Nat Rev Immunol. 7:690–702. 2007.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Veytia-Bucheli JI, Alvarado-Velázquez DA,
Possani LD, González-Amaro R and Rosenstein Y: The Ca2+
channel blocker verapamil inhibits the in vitro activation and
function of T lymphocytes: A 2022 Reappraisal. Pharmaceutics.
14(1478)2022.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Menotti M, Ambrogio C, Cheong TC, Pighi C,
Mota I, Cassel SH, Compagno M, Wang Q, Dall'Olio R, Minero VG, et
al: Wiskott-Aldrich syndrome protein (WASP) is a tumor suppressor
in T cell lymphoma. Nat Med. 25:130–140. 2019.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Wang Y, Kang W, Shang L, Song A and Ge S:
N-WASP knockdown upregulates inflammatory cytokines expression in
human gingival fibroblasts. Arch Oral Biol.
110(104605)2020.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Ferrua F, Marangoni F, Aiuti A and
Roncarolo MG: Gene therapy for Wiskott-Aldrich syndrome: History,
new vectors, future directions. J Allergy Clin Immunol.
146:262–265. 2020.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Lee WI, Yang CY, Jaing TH, Huang JL, Chien
YH and Chang KW: Clinical aspects and molecular analysis of Chinese
patients with Wiskott-Aldrich syndrome in Taiwan. Int Arch Allergy
Immunol. 145:15–23. 2008.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Notarangelo LD, Miao CH and Ochs HD:
Wiskott-Aldrich syndrome. Curr Opin Hematol. 15:30–36.
2008.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Burroughs LM, Petrovic A, Brazauskas R,
Liu X, Griffith LM, Ochs HD, Bleesing JJ, Edwards S, Dvorak CC,
Chaudhury S, et al: Excellent outcomes following hematopoietic cell
transplantation for Wiskott-Aldrich syndrome: A PIDTC report.
Blood. 135:2094–2105. 2020.PubMed/NCBI View Article : Google Scholar
|
|
57
|
Rohrer J, Ribeiro RC, Auerbach AD, Mirro B
and Conley ME: Wiskott-Aldrich syndrome in a family with Fanconi
anemia. J Pediatr. 129:50–55. 1996.PubMed/NCBI View Article : Google Scholar
|