Introduction
Menstrual stem cells (MenSCs) are a novel population
of cells derived from human menstrual blood that possess typical
characteristics (self-renewal and multilineage differentiation
potential) of mesenchymal SCs (MSCs) (1). As an alternative source of MSCs,
MenSCs have competitive advantages such as non-invasive acquisition
procedure and high proliferation capability (2). However, they also share similar
safety concerns to MSCs, such as unpredictable pro-tumor effects
and tissue entrapment (3).
Cell-free therapies using cellular components, which exhibit the
benefits of cellular therapy while avoiding the disadvantages, have
been drawing increasing attention (4,5).
Previous studies have demonstrated the therapeutic value of
SC-derived exosomes for various diseases (6,7),
particularly cancer treatment (8).
Mitochondria are a popular target in cell-free therapy. Besides
being the primary energy supplier, mitochondria also work as
carriers of enriched genetic cargo participating in multiple
cellular functions, such as cellular stress responses, including
apoptosis and autophagy (9,10).
Alteration of energy metabolism in cancer cells was
first shown by Otto Warburg (11)
. In addition to the dominant aerobic glycolysis, mitochondrial
oxidative phosphorylation (OXPHOS) is also hypothesized to serve a
key role in the metabolic mode of cancer cells (12,13).
Mutations in mitochondria have been found in various tumors. For
example, variants in mitochondrial biogenesis genes have been shown
to influence the epithelial ovarian cancer risk (14). Previous research has revealed that
the transplantation of mitochondria from different sources has
adverse effects on different tumors. For example, the transfer of
MSC-derived mitochondria supports tumor progression by enhancing
proliferation and invasion of breast cancer and glioblastoma cells
(15,16). By contrast, transplantation of
mitochondria from normal human astrocytes inhibits malignant
proliferation of human glioma cells (17). However, to the best of our
knowledge, no studies have reported the effects of MenSC-derived
mitochondria on cancer.
Ovarian cancer is the most lethal gynecological
cancer, with a worldwide mortality rate of 1.9% (18). Molecular evidence suggests
dysregulation in mitochondria associated with biogenesis,
morphology, dynamics and apoptosis is involved in carcinogenesis of
ovarian cancer (19). Thus, the
defective mitochondria may be a potential therapeutic target. To
the best of our knowledge, however, there are no previous studies
on the transfer of SC-derived mitochondria to ovarian cancer. In
the present study, the possible effects of MenSC-derived
mitochondria on ovarian cancer were investigated from the
perspective of protein expression profiling. Human ovarian
carcinoma cell line SKOV3, isolated from the ascites of a patient
with ovarian cancer, is typically used for in vitro and
in vivo research of ovarian cancer (20). By comparing the proteome of
mitochondria from MenSCs and SKOV3 cells, it was hypothesized that
OXPHOS-related metabolic pathways may serve a key role in
enhancement of tumor progression after mitochondrial
transplantation.
Materials and methods
Ethics approval and consent to
participate
The research proposal for human menstrual blood
collection was approved by the Ethical Committee of The Second
Affiliated Hospital of Harbin Medical University (approval no.
KY2018-105). Participants signed informed consent.
Menstrual blood collection
Volunteers were from The Second Affiliated Hospital
of Harbin Medical University, Harbin, Heilongjiang Province, China
from January 2020 to December 2020. Donors were selected based on
the following criteria: i) 20-30 years old; ii) no sexual history;
iii) no history of infectious diseases; iv) regular menstrual
cycles and v) normal vaginal discharge. The research proposal for
human menstrual blood collection was approved by the Ethical
Committee of The Second Affiliated Hospital of Harbin Medical
University (approval no. KY2018-105). Participants signed informed
consent. In total, 10 females were enrolled (mean age 23.4, median
age, 24). In total, 5 ml menstrual blood was collected and
transferred into a collection tube containing 0.1 ml
penicillin/streptomycin (P/S) and 0.1 ml EDTA-Na2
(Sigma-Aldrich; Merck KGaA) in 5 ml PBS.
Cell isolation and culture
Mononuclear cells were isolated from the collected
menstrual blood by Ficoll-Paque (Sigma-Aldrich; Merck KGaA) density
gradient centrifugation according to the manufacturer's protocols.
The cells were cultured in a T-25 flask containing DMEM/F-12
(Invitrogen; Thermo Fisher Scientific, Inc.) supplemented with 1%
P/S and 10% FBS (Invitrogen; Thermo Fisher Scientific, Inc.), at
37˚C, 5%CO2 The medium was replaced with the complete
medium the next day. Once cells reached 80-90% confluence, the
adherent cells were trypsinized, resuspended [wash cells twice with
3 ml PBS; add 500 µl of trypsin digestion solution (0.25%) for 3-4
min gently shake the flask to dislodge the adherent cells from the
flask. Add equal volume of culture medium to terminate digestion,
collect cell suspension, discard the supernatant, add culture
medium to prepare cell suspension]and cultured at a density of
1.5x105 cells in a T-25 flask. The cells were passaged
twice/week. 3-5 passage cells were used as indicated for all
experiments. Human ovarian cancer cell line SKOV3 was obtained from
Procell Life Science & Technology Co., Ltd. SKOV3 cells were
maintained in DMEM (Invitrogen; Thermo Fisher Scientific, Inc.)
supplemented with 10% FBS (Invitrogen; Thermo Fisher Scientific,
Inc.), 100 U/ml penicillin and 100 mg/ml streptomycin. Cultures
were incubated at 37˚C in a humidified atmosphere containing 5%
CO2. The morphology of cultured cells was examined under
a phase contrast microscope (40X magnification) (AX 70; Olympus,
Japan).
Mitochondrial isolation
The mitochondria were extracted by a mitochondrial
isolation kit (Abkine, KTP4003, China). Briefly, MenSCs or ovarian
cancer cells were homogenized by 70 strokes using a Dounce Tissue
Grinder in isolation buffer A plus protease inhibitor cocktail.
Cell extracts were collected into equal volumes of isolation buffer
C with buffer A. Cell debris and nuclei were removed by
centrifugation at 700 x g for 10 min, 4˚C, and mitochondrial
fractions were collected by centrifugation at 3,000 x g for 15 min,
4˚C.
Flow cytometric analysis
MenSCs or MenSC-derived mitochondria were stained
and labeled with anti-human fluorophore-conjugated antibodies for
CD73-FITC (BioLegend, Inc.; Cat# 344015), CD90-FITC(BioLegend, Inc.
Cat#328107), CD34-FITC (BioLegend, Inc. Cat# 343503), CD45-FITC
(BioLegend, Inc. Cat# 304005), CD63-PE (BioLegend, Inc. Cat#
353003) and CD29-FITC (eBioscience; Thermo Fisher Scientific, Inc.
Cat# 11029942) and detected by flow cytometric analysis. Briefly,
trypsinized cells or mitochondria were washed then re-suspended in
ice-cold PBS containing 1% BSA (Invitrogen; Thermo Fisher
Scientific, Inc.). Fluorophore-conjugated antibodies were added at
concentrations recommended by the manufacturer's protocols (95 µl
staining buffer, 5 µl fluorophore-conjugated antibodies) and
incubated in the dark for 30 min, 25˚C. The cells or mitochondria
were washed twice in staining buffer (eBioscience, Thermo Fisher
Scientific, Inc.) and analyzed under a flow cytometer (LSR
Fortessa; BD Biosciences). Data analysis was performed using FlowJo
(BD Biosciences. v.7.6.5).
Protein extraction and trypsin
digestion
Protein was extracted from mitochondria in ice-cold
lysis buffer [8 M urea, 2 mM EDTA, 10 mM Dithiothreitol (Sigma,
Japan) and 1% Protease Inhibitor Cocktail III, Calbiochem, Germany]
using a high-intensity ultrasonic processor (Ningbo Scientz
Biotechnology Co., Ltd.) on ice. In total, 200 µg protein in
solution (0.1% formic acid, 2% acetonitrile/in water) was used for
each group to run liquid chromatography tandem mass spectrometry
(LC-MS/MS) analysis. After removing the debris, protein was
precipitated with cold 15% trichloroacetic acid for 2 h at -20˚C.
Following centrifugation at 12,000 g at 4˚C for 10 min, the
remaining precipitate was washed with cold acetone three times. The
protein was dissolved in a buffer at pH 8.0 comprising 8 M urea and
100 mM Tetraethylammonium bromide (TEAB). For digestion, protein
solution was reduced with 5 mM dithiothreitol for 30 min at 56˚C
and alkylated with 11 mM iodoacetamide for 15 min at room
temperature in darkness. The protein sample was diluted by adding
100 mM TEAB to urea concentration <2 M. Finally, trypsin was
added at 1:50 trypsin-to-protein mass ratio for the first digestion
overnight and 1:100 trypsin-to-protein mass ratio for a second 4 h
digestion. The pooling of individual samples is a cost-effective
approach for proteomic studies. Therefore, 10 samples with equal
amounts of protein from volunteers were mixed to obtain 3 pooled
samples (21).
LC-MS/MS analysis (4D label-free)
4D label-free is the new generation of quantitative
proteomics. With mobility as the fourth latitude added to 3D
label-free quantification, sensitivity and quality of the detection
are improved. Relative protein quantification is achieved by
comparing the number of identified MS/MS spectra of a sample's
proteolytic peptides (22). The
tryptic peptides were dissolved in solvent A (0.1% formic acid in
water) and directly loaded onto a home-made reversed-phase
analytical column. Peptides were separated with a gradient from 4
to 6% solvent B (0.1% formic acid in acetonitrile) in 2 min, 6 to
24% for 68 min, 24 to 32% in 14 min and climbing to 80% in 3 min,
then holding at 80% for 3 min, all at a constant flow rate of 300
nl/min on a nanoElute high-performance LC system (Bruker
Corporation). The peptides were subjected to Capillary source
followed by the timsTOF Pro (Bruker Daltonics) mass spectrometry.
The electrospray voltage was 1.60 kV. Precursors and fragments were
analyzed at the TOF detector, with a MS/MS scan range from 100 to
1,700 m/z. The timsTOF Pro was operated in parallel accumulation
serial fragmentation (PASEF) mode. Precursors with charge states
0-5 were selected for fragmentation and 10 PASEF-MS/MS scans were
acquired/cycle. Associated parameters are set to: ionization mode
positive, and we use nano-ESI (CaptiveSpray), no nebulizer; Dry
Gas: 3 l/min; Dry Temp: 180˚C. The dynamic exclusion was set to 30
sec.
Database search
The resulting MS/MS data were processed using
Maxquant (Max-Planck-Institute of Biochemistry. v.1.6.6.0). Tandem
mass spectra were searched against the Uniprot human database
(20,600 sequences, accessed 03/2021) concatenated with a reverse
decoy database (maxquant.org/, SETTING PARAMETERS:
Database is Homosapiens9606SP20191115) Trypsin/P was specified as a
cleavage enzyme allowing up to 2 missing cleavages. The mass
tolerance for precursor ions was set as 40 parts/million in both
first and main search and the mass tolerance for fragment ions was
set as 0.04 Da. Carbamidomethyl on cysteine was specified as a
fixed modification. Acetylation on protein N-terminal and oxidation
on methionine were specified as variable modifications. Peptide and
protein false discovery rate were adjusted to <1%.
Bioinformatics annotation
analysis
Gene Ontology (GO) annotation and enrichment
analysis were derived from the UniProt-GOA database (ebi.ac.uk/interpro/). If some identified proteins were
not annotated, InterProScan (ebi.ac.uk/.
v.5.14-53.0) was used to annotate GO function based on protein
sequence alignment method. InterProScan was applied to predict the
domain functional descriptions of the differentially expressed
proteins. WoLF PSORT (wolfpsort.hgc.jp/. NAKAI Lab.) and iLoc-Animal
(http://www.jci-bioinfo.cn/iLoc-Animal) were used to
predict subcellular localization.
Protein-protein interaction (PPI)
analysis
PPI networks and pathways were assessed by STRING
(cn.string-db.org/. v.11.5) and Kyoto Encyclopedia
of Genes and Genomes (KEGG, genome.jp/kaas-bin/kaas_main) database,
respectively.
GO annotation
GO annotation proteomics was derived from the
UniProt-GOA database (ebi.ac.uk/GOA/). Firstly, identified protein IDs were
converted to UniProt IDs that were mapped to GO IDs by protein ID.
If identified proteins were not annotated by the UniProt-GOA
database, InterProScan software [InterProScan (https://www.ebi.ac.uk/. v.5.14-53.0)]was used to
annotate protein GO function based on the protein sequence
alignment method. Proteins were classified by GO annotation based
on three categories: i) biological process; ii) cellular component
and iii) molecular function.
Clusters of Orthologous Groups of
proteins
COG, Orthologs refer to proteins evolved from
vertical pedigrees (speciation) from different species and
typically retain the same function as the original protein. COG is
divided into two categories; one is prokaryotic and the other is
eukaryotic. Prokaryotes are called COG databases; eukaryotes are
called KOG databases. We performed COG/KOG functional
classification statistics for differentially expressed proteins by
database alignment analysis (ncbi.nlm.nih.gov/COG/).
Functional enrichment analysis
For each GO category, a two-tailed Fisher's exact
test was employed to test enrichment of the differentially
expressed protein against all identified proteins. GO with
corrected P<0.05 was considered to indicate statistical
significance. The KEGG database was used to identify enriched
pathways by two-tailed Fisher's exact test t. A pathway with
corrected P<0.05 was considered to indicate statistical
significance. These pathways were classified into hierarchical
categories (Metabolism,Genetic Information Processing,
Environmental Information Processing, Cellular Processes,
Organismal Systems, Human Diseases and Drug Development) according
to the KEGG website. For protein domain enrichment analysis, the
InterPro database was searched and a two-tailed Fisher's exact test
was employed to compare enrichment of the differentially expressed
protein. GO plot package (v.0.4) was employed for plotting GO or
pathway annotation and abundance (https://cran.r-project.org/web/packages/networkD3/).
Enrichment-based clustering
For further hierarchical clustering based on
differentially expressed protein functional classification (such as
GO, domain, pathway), categories obtained following enrichment
along with their P-values were filtered for those categories that
were least enriched in one of the clusters with P<0.05. This
filtered P-value matrix was transformed by the function
x=-log10(P). Finally, these values were z-transformed
for each functional category. The z-scores were then clustered by
one-way hierarchical clustering (Euclidean distance, average
linkage clustering) in Genesis (Genesis2000. v.9.1). Cluster
membership was visualized by a heat map using the heatmap.2
function from the gplots R package (cran.r-project.org/web/packages/cluster/,
v.2.0.3).
Parallel reaction monitoring (PRM)
analysis
PRM is a targeted quantitative MS approach,
generating high resolution precursor measurements and full scan
MS/MS data (23). The target
proteins suitable for PRM validation were decided by factors such
as the unique sequence coverage, intensity and MS/MS count. PRM MS
analysis was performed using MS/MS in Q Exactive™ Plus (Thermo
Fisher Scientific, Inc.). Sample processing and performance of PRM
analysis was performed by Jingjie PTM BioLab Co., Inc. LC
parameters, electrospray voltage, scan range and Orbitrap
resolution were the same as for the 4D label-free method. Automatic
gain control was set at 3x106 for full MS and
1x105 for MS/MS. The maximum Ion Trap was set at 20 msec
for full MS and was automated for MS/MS. The isolation window for
MS/MS was set at 2.0 m/z. After quantitative information was
normalized by the heavy isotope-labeled peptide, a relative
quantitative analysis, based on three biological replicates, was
performed on the target peptides.
Results
Isolation and characterization of
MenSCs and mitochondria
Adherent cells showed a fibroblast-like short
spindle-shaped morphology after 9-12 day primary culture and were
confirmed to express surface markers similar to MSCs (expression of
characteristic stem cell phenotypes, such as CD44, CD73, CD90,
CD146, but not hematopoietic stem cell phenotypes CD34, CD45, CD14,
CD11b); CD73 and CD90 were positively expressed and CD34 and CD45
were negatively expressed (Fig.
1A). The mitochondria were extracted from MenSCs and SKOV3
cells. Flow cytometry identified that mitochondria markers CD63 and
CD29 were positively expressed (Fig.
1B).
Differentially expressed protein
analysis
By LC-MS/MS, a quantitative analysis of the global
proteome in mitochondria of MenSCs and SKOV3 cells was performed.
In total, 5,597 proteins were identified, of which 3,856 proteins
were quantified. Proteins that displayed fold-change in expression
>2 were analyzed further. Compared with the mitochondria of
SKOV3 cells, 592 proteins showed increased expression in
mitochondria of MenSCs, while 583 proteins showed decreased
expression (Fig. 2A). GO
functional annotation was performed to classify candidate proteins
into three categories according to their respective level 2 GO
terms: i) biological process; ii) cellular component and iii)
molecular function (Fig. 2B). In
the category of biological process, metabolic process is the
important and predominant biological process, with 727 proteins
being involved.
Subcellular localization of
differentially expressed proteins and functional
classification
The cells of eukaryotic organisms are divided into
functionally distinct membrane-bound compartments (24). The subcellular localization of
differentially expressed proteins was characterized. The largest
subcellular localization categories of proteins that showed
increased expression were ‘cytoplasm’ (23.82%), ‘mitochondria’
(18.07%) and ‘plasma membrane’ (17.91%; Fig. 2C). The largest subcellular
localization categories of proteins that showed decreased
expression were ‘cytoplasm’ (41.51%), ‘nucleus’ (27.62%) and
‘plasma membrane’ (8.58%; Fig.
S1). The percentage of mitochondrial proteins with increased
expression was higher than those with decreased expression (18.07
vs. 7.89%). COG/EuKaryotic Orthologous Groups (COG/KOG) functional
classification was performed by database alignment analysis
(Fig. 2D). Most upregulated
proteins were classified as ‘signal transduction mechanism’,
‘post-translational modification’, ‘protein turnover, chaperone’,
‘general function prediction only’ and ‘energy production and
conversion’.
Functional enrichment analysis of
differentially expressed proteins
GO, KEGG pathway and protein domain analysis were
performed to determine functional classification of the
differentially expressed proteins.
GO functional enrichment analysis revealed the most
enriched functions in differentially expressed mitochondrial
proteins in the three GO enrichment classification categories
(Fig. 3). In proteins that showed
increased expression, the top three enriched functions in each
classification were ‘endomembrane system’, ‘membrane part’ and
‘endoplasmic reticulum’; ‘structural constituent of ribosome’,
‘cation-transporting ATPase activity’ and ‘hydrogen ion
transmembrane transporter activity’ and ‘protein localization to
endoplasmic reticulum’, ‘ATP hydrolysis coupled transmembrane
transport’ and ‘protein transport’ (Fig. 3A). Enrichment analysis of proteins
that showed decreased expression is shown in Fig. S2A. In proteins that showed
decreased expression, the top three enriched functions in each
classification were ‘extracellular exosome’, ‘extracellular
organelle’ and ‘extracellular vesicle’; ‘cell adhesion molecule
binding’, ‘cadherin binding’ and ‘nucleotide binding’; ‘actin
cytoskeleton organization’, ‘neutrophil mediated immunity’ and
‘myeloid leukocyte activation’.
KEGG pathway enrichment analysis showed that
proteins that showed increased expression were enriched in 25
pathway entries; functions associated with ‘protein processing in
endoplasmic reticulum’, ‘ribosome’, ‘lysosome’, ‘protein export’
and ‘oxidative phosphorylation’ were most significantly enriched
(Fig. 3B). The components and
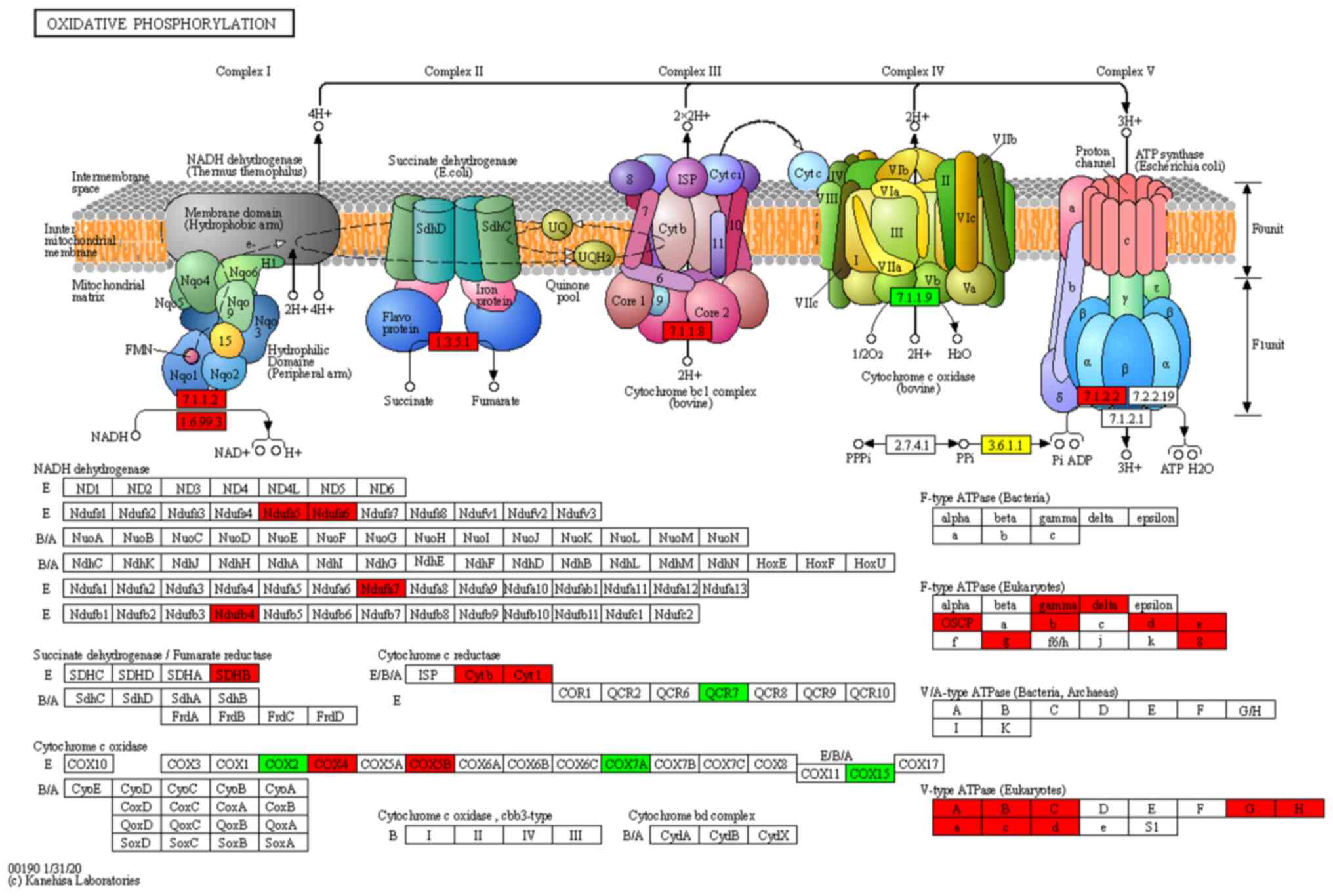
cascades of the OXPHOS pathway are shown in Fig. 4. The expression levels of
NADH:ubiquinone oxidoreductase subunit S5 (NDUFS5), NDUFS6),
succinate dehydrogenase complex iron sulfur subunit B (SDHB),
cytochrome c oxidase subunit 5B (COX5B), cytochrome c oxidase
subunit 4 (COX4), NDUFA7) and NDUFB4) were significantly increased
in MenSC-derived mitochondria and these proteins were identified in
the OXPHOS signaling pathway by KEGG pathway analysis (Fig. 4). This pathway has functions in
energy metabolism and driving ATP synthesis, which promote cell
proliferation. The pathways in which proteins that displayed
decreased expression were primarily enriched are shown in Fig S2B, such as ‘tight junction’,
‘regulation of actin cytoskeleton’, ‘adherens junction’ and
‘central carbon metabolism in cancer;
Protein domain enrichment analysis showed that
proteins that exhibited increased expression were primarily
enriched in domains such as ‘thioredoxin domain’, ‘thioredoxin-like
fold’, ‘band 7 domain’, ‘disulfide isomerase’ and ‘mitochondrial
carrier domain’ (Fig. 3C). Protein
domain enrichment analysis of proteins that showed decreased
expression is shown in Fig. S2C,
which included ‘GroEL-like apical domain’, ‘GroEL-like equatorial
domain’, ‘TCP-1-like chaperonin intermediate domain’ and
‘actin-depolymerising factor homology domain’.
Cluster based on function and
characteristics of protein
GO enrichment-based hierarchical clustering analysis
of results from GO, KEGG and protein domain functional enrichment
revealed that the differentially expressed proteins with P<0.05
were clustered into four groups, Q1 to Q4, based on the ratio of
mitochondria derived from MenSCs to SKOV3 cells. Q1 was defined as
a ratio between 0.0 and 0.5, which included 583 proteins. Q2 was
defined as a ratio between 0.500 and 0.667, which included 173
proteins. Q3 was defined as a ratio between 1.5 and 2.0, which
included 137 proteins. Q4 was defined as a ratio >2 and included
592 proteins. In the molecular function category, differentially
expressed proteins were primarily associated with ‘ATPase activity,
coupled to movement of substances’, ‘GTP binding’ and ‘ATP binding’
(Fig. 5A). ‘Oxoacid metabolic
process’, ‘regulation of cell motility’, ‘cellular chemical
homeostasis’ and ‘ATP hydrolysis coupled transmembrane transport’
were the subcategories differentially expressed proteins were most
associated with in the biological process category (Fig. 5B). Proteins associated with
‘ribosome’, ‘intrinsic component of organelle’ and
‘proton-transporting two-sector ATPase complex’ were upregulated in
cellular component category by enrichment analysis (Fig. 5C).
Differentially expressed protein
interaction network
To understand the potential PPIs of the
differentially expressed proteins, PPI proteomics network analysis
was performed using string. The robust and cross-talking signaling
interactions among the top 100 upregulated proteins sorted by
P-value are mapped in Fig. S3.
STRING 11.5 online software was used for PPI analysis with a
confidence score >0.4 (medium confidence). According to this
network, proteins associated with OXPHOS and ribosome were
significantly enriched.
PRM analysis of differentially
expressed proteins
To validate the results from 4D label-free data
analysis, PRM was performed on significantly differentially
expressed proteins involved in the OXPHOS pathway. Due to
limitations associated with low relative abundance, 6 proteins of
interest, namely T cell immune regulator 1 (TCIRG1), ATP5PO,
ATP6V1B2, ATP5F1C, ATP5PB and ATP5PD, were selected for PRM
analysis (Fig. S4). Two unique
peptides with anticipated chemical stability were selected for each
protein and the relative protein abundance was expressed as the
mean of the two normalized peptide peak areas. A total of four
(TCIRG1, ATP5PO, ATP6V1B2 and ATP5PD) of the six candidate proteins
displayed similar trends to 4D label-free results, which supported
the reliability of quantitative proteomics analysis.
Discussion
In the present study, proteomic analysis was
performed to identify the differentially expressed proteins between
mitochondria derived from MenSCs and the SKOV3 cell line.
Subcellular localization of differentially expressed proteins
showed that more mitochondrial proteins had increased expression in
mitochondria of MenSCs. Functional enrichment analysis revealed
that proteins in the OXPHOS pathway were significantly enriched,
which was validated by PRM analysis.
Abnormal metabolism is a hallmark of cancer
(25). Unlike normal cells, cancer
cells depend on aerobic glycolysis to produce the energy needed for
rapid proliferation (26).
Mitochondrial OXPHOS increases electron leak from the
electron-transport chain and mitochondrial superoxide production to
stimulate pro-tumoral signaling pathways and antioxidant defenses
(27). Increased activity of the
OXPHOS pathway has been demonstrated to contribute to cancer
metastasis, whereby chemo-resistant cells undergo a shift towards a
more active oxidative metabolism (28). In ovarian cancer, increased OXPHOS
has been reported to enhance IL-6 production, promoting cancer cell
survival and proliferation and impairing responsiveness to
chemotherapy (29,30). KEGG pathway enrichment analysis
showed that proteins involved in the OXPHOS pathway were
significantly enriched in MenSC-derived mitochondria. This
indicated that, by facilitating the energy metabolism of the cancer
cells, MenSC-derived mitochondria may promote progression of
ovarian cancer. Moreover, protein domain analysis showed that
‘thioredoxin domain’ was significantly enriched. Thioredoxin is a
small redox-regulating protein domain that serves key roles in both
oxidative stress and hypoxia in the tumoral microenvironment, which
is hypothesized to contribute to carcinogenesis (31,32).
Therefore, enrichment in ‘thioredoxin domain’ and ‘thioredoxin-like
fold domain’ in the mitochondria of MenSCs indicated that the
transplantation of MenSC-derived mitochondria may enhance ovarian
cancer progression.
Therapeutic regimens targeting mitochondria are a
potential treatment for cancer in the future (29,33,34).
There is in vivo and in vitro evidence to suggest
that mitochondrial transplantation inhibits proliferation of tumor
cells or promotes tumor progression (17,35).
SCs are optimal donors because of ‘high proliferation rate’, ‘low
immunogenicity’ and ‘low carcinogenicity’ to provide mitochondria
for transplantation. However, MenSC-derived mitochondria have, to
the best of our knowledge, not yet been investigated. By comparing
the protein expression profile of the mitochondria from MenSCs and
the ovarian cancer cell line SKOV3, it was found that the
mitochondria from MenSCs may serve as an enhancer for ovarian
cancer.
The present study had limitations. Mitochondria from
MenSCs were not transferred into ovarian cancer cells and changes
in the biological properties of cancer cells were not observed;
experimental results and conclusions were derived from comparative
proteomics. Mitochondria derived from MenSCs were proposed and
explored for cancer treatment. The present findings indicated that
transfer of mitochondria of MenSCs may promote ovarian cancer
progression by activating OXPHOS. Although our hypothesis suggests
that transplantation of MenSCs -derived mitochondria into ovarian
cancer cells does not inhibit tumor development, previous studies
have shown that stem cell-derived mitochondria transplantation is
helpful for cancer treatment. This may be related to the source of
stem cells, the mode of metastasis, etc. Despite of the promise of
cell-free therapy by foreign mitochondria transfer, there is
uncertainty regarding SC-derived mitochondrial transplantation and
more research is needed in future. In next studies, we will try to
select mitochondria or cellular vesicles from other sources of stem
cells, transplant them into ovarian cancer cells and explore the
status of cancer cells; other research directions also include the
selection of mitochondria from MenSCs for the treatment of other
malignant tumors.
Supplementary Material
Subcellular localization of proteins
with decreased expression. The majority of proteins with decreased
expression were distributed in ‘cytoplasm’ (41.51%), ‘nucleus’
(27.62%) and ‘plasma membrane’ (8.58%).
Functional enrichment analysis of
proteins with decreased expression. (A) Gene Ontology enrichment
analysis of biological process, cellular component and molecular
function. (B) Kyoto Encyclopedia of Genes and Genomes pathway
enrichment analysis. (C) Protein domain enrichment analysis.
PPI network analysis of proteins with
increased expression. Network nodes represent proteins; edges
represent protein-protein associations. Line thickness indicates
the strength of supporting data. The hub shows proteins with
functions in relation to oxidative phosphorylation. PPI,
protein-protein interaction.
Validation of six selected proteins by
PRM. A total of four candidate proteins displayed similar trends
with 4D label-free quantitative proteomics analysis. PRM, parallel
reaction monitoring; TCIRG1, T cell immune regulator 1.
Acknowledgements
Not applicable.
Funding
Funding: The present study was supported by China Postdoctoral
Science Foundation Grant (grant no. 2018M641854), National Natural
Science Foundation of China (grant no. 81802592), National Natural
Science Foundation of China (grant no. 81801402), Natural Science
Foundation of Heilongjiang Province (grant no. LH2020H049).
Availability of data and materials
The datasets generated and/or analyzed during the
current study are available in the Proteomics Identification
Database repository (accession no. PXD036148), ebi.ac.uk/pride/archive/projects/PXD036148.
Authors' contributions
XC and WZ confirm the authenticity of all the raw
data. XC, YC and XK designed the study. QK contributed to
conception and design. WZ and BL collected menstrual blood samples,
extraction of menstrual blood stem cells and their mitochondria and
cultured tumor cells. TS and WZ analyzed the data and wrote the
manuscript. All authors have read and approved the final
manuscript.
Ethics approval and consent to
participate
The research proposal for human menstrual blood
collection was approved by the Ethics Committee of The Second
Affiliated Hospital of Harbin Medical University (approval no.
KY2018-105). All volunteers participating in the experiment signed
the informed consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Chen L, Qu J, Cheng T, Chen X and Xiang C:
Menstrual blood-derived stem cells: Toward therapeutic mechanisms,
novel strategies, and future perspectives in the treatment of
diseases. Stem Cell Res Ther. 10(406)2019.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Chen L, Qu J and Xiang C: The
multi-functional roles of menstrual blood-derived stem cells in
regenerative medicine. Stem Cell Res Ther. 10(1)2019.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Zhou J, Tan X, Tan Y, Li Q, Ma J and Wang
G: Mesenchymal stem cell derived exosomes in cancer progression,
metastasis and drug delivery: A comprehensive review. J Cancer.
9:3129–3137. 2018.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Raik S, Kumar A and Bhattacharyya S:
Insights into cell-free therapeutic approach: Role of stem cell
‘soup-ernatant’. Biotechnol Appl Biochem. 65:104–118.
2018.PubMed/NCBI View
Article : Google Scholar
|
|
5
|
Foo JB, Looi QH, Chong PP, Hassan NH, Yeo
GEC, Ng CY, Koh B, How CW, Lee SH and Law JX: Comparing the
therapeutic potential of stem cells and their secretory products in
regenerative medicine. Stem Cells Int. 2021(2616807)2021.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Huang-Doran I, Zhang CY and Vidal-Puig A:
Extracellular vesicles: Novel mediators of cell communication in
metabolic disease. Trends Endocrinol Metab. 28:3–18.
2017.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Xu HK, Chen LJ, Zhou SN, Li YF and Xiang
C: Multifunctional role of microRNAs in mesenchymal stem
cell-derived exosomes in treatment of diseases. World J Stem Cells.
12:1276–1294. 2020.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Parfejevs V, Sagini K, Buss A, Sobolevska
K, Llorente A, Riekstina U and Abols A: Adult stem cell-derived
extracellular vesicles in cancer treatment: Opportunities and
challenges. Cells. 9(1171)2020.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Liu K, Zhou Z, Pan M and Zhang L: Stem
cell-derived mitochondria transplantation: A promising therapy for
mitochondrial encephalomyopathy. CNS Neurosci Ther. 27:733–742.
2021.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Nakamura Y, Park JH and Hayakawa K:
Therapeutic use of extracellular mitochondria in CNS injury and
disease. Exp Neurol. 324(113114)2019.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Warburg O, Wind F and Negelein E: The
metabolism of tumors in the body. J Gen Physiol. 8:519–530.
1927.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Viale A, Corti D and Draetta GF: Tumors
and mitochondrial respiration: A neglected connection. Cancer Res.
75:3685–3686. 2015.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Jia D, Lu M, Jung KH, Park JH, Yu L,
Onuchic JN, Kaipparettu BA and Levine H: Elucidating cancer
metabolic plasticity by coupling gene regulation with metabolic
pathways. Proc Natl Acad Sci U S A. 116:3909–3918. 2019.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Permuth-Wey J, Chen YA, Tsai YY, Chen Z,
Qu X, Lancaster JM, Stockwell H, Dagne G, Iversen E, Risch H, et
al: Inherited variants in mitochondrial biogenesis genes may
influence epithelial ovarian cancer risk. Cancer Epidemiol
Biomarkers Prev. 20:1131–1145. 2011.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Caicedo A, Fritz V, Brondello JM, Ayala M,
Dennemont I, Abdellaoui N, de Fraipont F, Moisan A, Prouteau CA,
Boukhaddaoui H, et al: MitoCeption as a new tool to assess the
effects of mesenchymal stem/stromal cell mitochondria on cancer
cell metabolism and function. Sci Rep. 5(9073)2015.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Mombo NB, Gerbal-Chaloin S, Bokus A,
Daujat-Chavanieu M, Jorgensen C, Hugnot JP and Vignais ML:
MitoCeption: Transferring isolated human msc mitochondria to
glioblastoma stem cells. J Vis Exp. 22(55245)2017.PubMed/NCBI View
Article : Google Scholar
|
|
17
|
Gomzikova MO, James V and Rizvanov AA:
Mitochondria donation by mesenchymal stem cells: Current
understanding and mitochondria transplantation strategies. Front
Cell Dev Biol. 9(653322)2021.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Signorile A, De Rasmo D, Cormio A, Musicco
C, Rossi R, Fortarezza F, Palese LL, Loizzi V, Resta L, Scillitani
G, et al: Human ovarian cancer tissue exhibits increase of
mitochondrial biogenesis and cristae remodeling. Cancers (Basel).
11(1350)2019.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Ke X, Li L, Li J, Zheng M and Liu P:
Anti-oncogenic PTEN induces ovarian cancer cell senescence by
targeting P21. Cell Biol Int. 46:118–128. 2022.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Li F, Wang Y, Li Y, Yang H and Wang H:
Quantitative analysis of the global proteome in peripheral blood
mononuclear cells from patients with new-onset psoriasis.
Proteomics. 18(e1800003)2018.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Lin H, Zhang W, Xu Y, You Z, Zheng M, Liu
Z and Li C: 4D label-free quantitative proteomics analysis to
screen potential drug targets of Jiangu Granules treatment for
postmenopausal osteoporotic rats. Front Pharmacol.
13(1052922)2022.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Heil LR, Remes PM and MacCoss MJ:
Comparison of unit resolution versus high-resolution accurate mass
for parallel reaction monitoring. J Proteome Res. 20:4435–4442.
2021.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Dacks JB, Field MC, Buick R, Eme L,
Gribaldo S, Roger AJ, Brochier-Armanet C and Devos DP: The changing
view of eukaryogenesis-fossils, cells, lineages and how they all
come together. J Cell Sci. 129:3695–3703. 2016.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Pavlova NN and Thompson CB: The emerging
hallmarks of cancer metabolism. Cell Metab. 23:27–47.
2016.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Vaupel P, Schmidberger H and Mayer A: The
Warburg effect: Essential part of metabolic reprogramming and
central contributor to cancer progression. Int J Radiat Biol.
95:912–919. 2019.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Payen VL, Zampieri LX, Porporato PE and
Sonveaux P: Pro- and antitumor effects of mitochondrial reactive
oxygen species. Cancer Metastasis Rev. 38:189–203. 2019.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Zampieri LX, Grasso D, Bouzin C, Brusa D,
Rossignol R and Sonveaux P: Mitochondria participate in
chemoresistance to cisplatin in human ovarian cancer cells. Mol
Cancer Res. 18:1379–1391. 2020.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Emmings E, Mullany S, Chang Z, Landen CN
Jr, Linder S and Bazzaro M: Targeting mitochondria for treatment of
chemoresistant ovarian cancer. Int J Mol Sci.
20(229)2019.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Matassa DS, Amoroso MR, Lu H, Avolio R,
Arzeni D, Procaccini C, Faicchia D, Maddalena F, Simeon V,
Agliarulo I, et al: Oxidative metabolism drives
inflammation-induced platinum resistance in human ovarian cancer.
Cell Death Differ. 23:1542–1554. 2016.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Ghareeb H and Metanis N: The thioredoxin
system: A promising target for cancer drug development. Chemistry.
26:10175–10184. 2020.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Karlenius TC and Tonissen KF: Thioredoxin
and cancer: A role for thioredoxin in all states of tumor
oxygenation. Cancers (Basel). 2:209–232. 2010.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Pustylnikov S, Costabile F, Beghi S and
Facciabene A: Targeting mitochondria in cancer: Current concepts
and immunotherapy approaches. Transl Res. 202:35–51.
2018.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Wang J, Li H, Yao Y, Zhao T, Chen YY, Shen
YL, Wang LL and Zhu Y: Stem cell-derived mitochondria
transplantation: A novel strategy and the challenges for the
treatment of tissue injury. Stem Cell Res Ther.
9(106)2018.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Kheirandish-Rostami M, Roudkenar MH,
Jahanian-Najafabadi A, Tomita K, Kuwahara Y, Sato T and Roushandeh
AM: Mitochondrial characteristics contribute to proliferation and
migration potency of MDA-MB-231 cancer cells and their response to
cisplatin treatment. Life Sci. 244(117339)2020.PubMed/NCBI View Article : Google Scholar
|