Introduction
Breast cancer that occurs in women is a highly
malignant tumor with poor prognosis. As reported in the Global
Cancer Statistics 2020, breast cancer ranks first in cancer
incidence (number: 2261419; percentage: 11.7%) and fifth in
mortality (number: 684996; percentage: 6.9%) worldwide (1). Nowadays, surgery combined with
radiotherapy, chemotherapy, endocrine therapy and targeted therapy
is the main treatment for breast cancer (2). Despite the multiple treatment methods
available, a large proportion of patients will eventually die of
recurrence and metastasis of breast cancer (3,4).
Hence, it seems imperative to elaborate the molecular mechanisms
underlying the malignant progression of breast cancer and to
develop effective therapeutic targets against breast cancer
metastasis.
Nuclear receptor coactivator 5 (NCOA5), also known
as coactivator independent of activation function-2(AF-2) domain
(CIA), is a nuclear receptor coregulator (5). Recently, the abnormal expression of
NCOA5 in tumor tissues has attracted considerable attention.
Elevated NCOA5 in colorectal cancer is closely related to the
malignant biological behaviors of cancer cells and prognosis of
patients (6). Knockdown of NCOA5
can suppress hepatocellular carcinoma cell proliferation and
migration (7). Moreover, it has
been verified that NCOA5 is highly expressed in breast cancer
tissues, and is lowly expressed in adjacent non-cancerous tissues,
and NCOA5 is highly associated with the poor prognosis of breast
cancer patients (8). Furthermore,
NCOA5 was greatly correlated with lymph node metastasis in breast
cancer, and its expression could predict the overall survival time,
thus NCOA5 is considered as a promising novel management target for
breast cancer (9), whereas its
specific regulatory mechanism in the progression of breast cancer
has not been fully elucidated till now.
Protein interactors of NCOA5 were predicted by
querying the BioPlex database and BioPlex interaction data
presented that targeting protein for xenopus kinesin-like protein 2
(TPX2) could interact with NCOA5. TPX2 is a micro-associated
protein that associates with the formation and stability of the
mitotic spindle (10). High
expression of TPX2 in different human tissues can lead to
disordered phenomena including abnormal amplification of
centrosome, formation of aneuploidy, malignant cell transformation
(10,11). Recent researches have reported that
TPX2 is overexpressed in multiple malignancies and TPX2 expression
is highly associated with the occurrence, development and prognosis
of cancers (12,13). In addition, TPX2 expression is
remarkably elevated in breast cancer tissues compared with adjacent
non-cancerous tissues, and knockdown of TPX2 can suppress the
proliferation, invasion and migration of breast cancer cells
(14,15).
In general, the present work was formulated to
elucidate the functions of NCOA5 in the malignant biological
behaviors of breast cancer cells and to explore the molecular
mechanisms underlying the involvement of TPX2/NCOA5 in the
development of breast cancer.
Materials and methods
Cell culture
Human breast epithelial cell lines (MCF10A, MCF12A),
human breast cancer cell lines (MCF7, T47D) and human immortalized
umbilical vein endothelial cells (HUVEC/EAhy926) were obtained from
American Type Culture Collection (ATCC). MCF10A and MCF12A cells
were cultured in Dulbecco's modified Eagle's medium (DMEM)/F12
supplemented with 10% fetal bovine serum (FBS; Gibco; Thermo Fisher
Scientific, Inc.) and 1% penicillin/streptomycin solution (Gibco;
Thermo Fisher Scientific, Inc.). HUVECs, MCF7 and T47D cells were
cultured in DMEM medium supplemented with 10% FBS and 1%
penicillin/streptomycin solution. All cells were incubated at 37̊C
in a humidified 5% CO2 atmosphere.
Cell transfection
Small interfering RNA (siRNA) plasmid targeting
NCOA5 (si-NCOA5-1, si-NCOA5-2), siRNA plasmid targeting TPX2
(si-TPX2-1, si-TPX2-2) and empty siRNA plasmid (si-NC), NCOA5
overexpression plasmid (Ov-NCOA5) and the corresponding negative
control (Ov-NC) were purchased from GenePharma. Transfection was
conducted using Lipofectamine® 2000 (Invitrogen; Thermo
Fisher Scientific, Inc.) according to the manufacturer's protocol.
The NCOA5- and TPX2-specific siRNA sequences were listed as
follows: si-NCOA5-1, 5'-GACTTGATCTTCCTTAACACAGA-3', si-NCOA5-2,
5'-TTCTCCTTTTGCTATTGTCATCA-3', si-TPX2-1,
5'-CACAAGTTAAAAGCTCTTATTCC-3', and si-TPX2-2,
5'-AAGTTAAAAGCTCTTATTCCTAT-3'.
TNMplot analysis
Expressions of NCOA5 and TPX2 between paired
non-tumor and tumor tissues of breast cancer patients were compared
using TNMplot website (http://tnmplot.com/analysis/). Mann Whitney was used
for statistical test.
Cell counting kit-8 (CCK-8) assay
Cell viability was determined using CCK-8 assay. In
short, MCF7 cells (5x103 cells/well) were inoculated
into a 96-well plate and then incubated for 24, 48 or 72 h at 37̊C.
Next, 10 µl CCK-8 reagent (Beyotime) was added into each well for
another 4 h incubation. The optical density (OD450
nm) was measured using a microplate reader (Bio-Rad).
Wound healing assay
Cell migratory ability was evaluated using wound
healing assay. The transfected or untransfected MCF7 cells
(1x105 cells/well) were cultured in a 6-well plate and
grown to 90% confluence. Then, the wounds were created by
scratching the monolayer of cells with a sterile 200-µl pipette tip
and the detached cells were washed twice with PBS. Next, MCF7 cells
were incubated in fresh serum-free DMEM for 24 h. Images of the
wounds were captured at 0 and 24 h under a light microscope
(magnification, x100; Leica).
Transwell assay
Cell invasive ability was evaluated using transwell
assay. The transfected or untransfected MCF7 cells were suspended
in fresh serum-free DMEM. A total of 5x104 cells were
seeded into the upper chamber of transwell plates pre-coated with
matrigel (BD Biosciences) and 600 µl FBS-DMEM was added into the
lower chamber as a chemoattractant. After 24 h incubation,
non-invasive cells were gently removed and the invasive cells in
the lower chamber were fixed in 4% paraformaldehyde and stained
with 0.1% crystal violet solution. Stained cells were photographed
and counted under a light microscope (magnification, x200;
Leica).
Tube formation assay
In short, the conditioned media (CM) of MCF7 cells
was collected. HUVECs (2x104 cells/well) were seeded
into the 96-well plate pre-coated with Matrigel and then incubated
with CM at 37˚C for 24 h. Tube formation was observed and
photographed under a light microscope (magnification, x40; Leica,
Wetzlar, Germany).
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was extracted using TRIzol reagent
(Invitrogen; Thermo Fisher Scientific, Inc.) in compliance with the
manufacturer's standard procedures. RNA samples were reversely
transcribed into cDNA using a PrimeScript RT kit (Takara).
Subsequently, PCR reactions were performed on an ABI 7500 system
(Applied Biosystems) using SYBR Premix Ex Taq kit (Takara). The PCR
thermocycling conditions were as follows: Initial denaturation at
95˚C for 10 min; followed by 40 cycles of 95˚C for 15 sec and 64˚C
for 30 sec. Primer sequences were as follows: NCOA5 forward:
5'-TGCTATTGTCATCACCCAG-3', reverse: 5'-CTCATTCTTGTAACGCTCATA-3';
TPX2 forward: 5'-ATGGAACTGGAGGGCTTTTTC-3', reverse:
5'-TGTTGTCAACTGGTTTCAAAGGT-3'; GAPDH forward:
5'-GGTCTCCTCTGACTTCAACA-3', reverse: 5'-GTGAGGGTCTCTCTCTTCCT-3'.
GAPDH served as the endogenous control. The relative gene
expression was calculated using 2-∆∆Ct method (16).
Western blot assay
Total proteins were extracted using RIPA lysis
buffer (Beyotime) and protein concentration was determined using
BCA Protein Assay Kit (Beyotime). Equal amounts of protein samples
were separated by sodium dodecyl sulphate-polyacrylamide gel
electrophoresis (SDS-PAGE) and then transferred onto polyvinylidene
difluoride (PVDF) membranes (Millipore). Nonspecific binding
proteins were blocked with 5% non-fat milk for 1.5 h at room
temperature. Subsequently, membranes were incubated overnight at
4̊C with antibodies against NCOA5 (Bioworld, BS67243, 1:1,000),
Ki67 (Abcam, ab16667, 1:1,000), PCNA (Abcam, ab92552, 1:1,000),
MMP2 (Abcam, ab92536, 1:1,000), MMP9 (Abcam, ab76003, 1:1,000),
VEGFA (Thermo Fisher Scientific, OPA1-10110, 1:200), VEGFR2 (Abcam,
ab134191, 1:1,000), TPX2 (Abcam, ab252945, 1:1,000) and GAPDH
(Abcam, ab9485, 1:2,500). On the next day, membranes were incubated
with horseradish peroxidase (HRP)-conjugated secondary antibody
(Abcam, ab6721, 1:3,000) for 2 h at room temperature. GAPDH served
as the endogenous control. Enhanced chemiluminescence (ECL) kit was
applied to develop the protein bands and the blots were visualized
and analyzed by a Bio-Rad imaging system (Bio-Rad).
BioPlex network analysis
The general biological repository for interaction
data sets (https://bioplex.hms.harvard.edu/) was adopted to
explore the BioPlex interaction data and identify high-confidence
NCOA5 interactors.
Gene Expression Profiling Interactive
Analysis (GEPIA)
GEPIA (http://gepia.cancer-pku.cn) is a web-based data mining
platform with large RNA sequencing data from TCGA and GTEx. Gene
expression correlation analysis was performed in the ‘Correlation’
module. The correlation coefficient was conducted by Pearson's
correlation test.
Co-immunoprecipitation (Co-IP)
The interaction between NCOA5 and TPX2 was validated
by employing Co-IP assay. Cells were lysed on ice for 30 min using
cell lysis buffer consisting of complete protease inhibitor. Then,
anti-NCOA5, anti-TPX2 or control IgG were added into the collected
supernatant of cell lysates and incubated for 12 h at 4̊C. Next, 20
µl of protein A/G agarose beads (Santa Cruz Biotechnology, Inc.)
were added to the mixture and incubated overnight at 4̊C. The
pelleted resin was washed three times by washing buffer and the
immuno-complexes were subjected to western blot analysis.
Statistical analysis
Data of three independent experiments were expressed
as means ± SD. Comparisons among multiple groups were carried out
using one-way analysis of variance (ANOVA) followed by Tukey's post
hoc test. *P<0.05 represented a statistically
significant difference.
Results
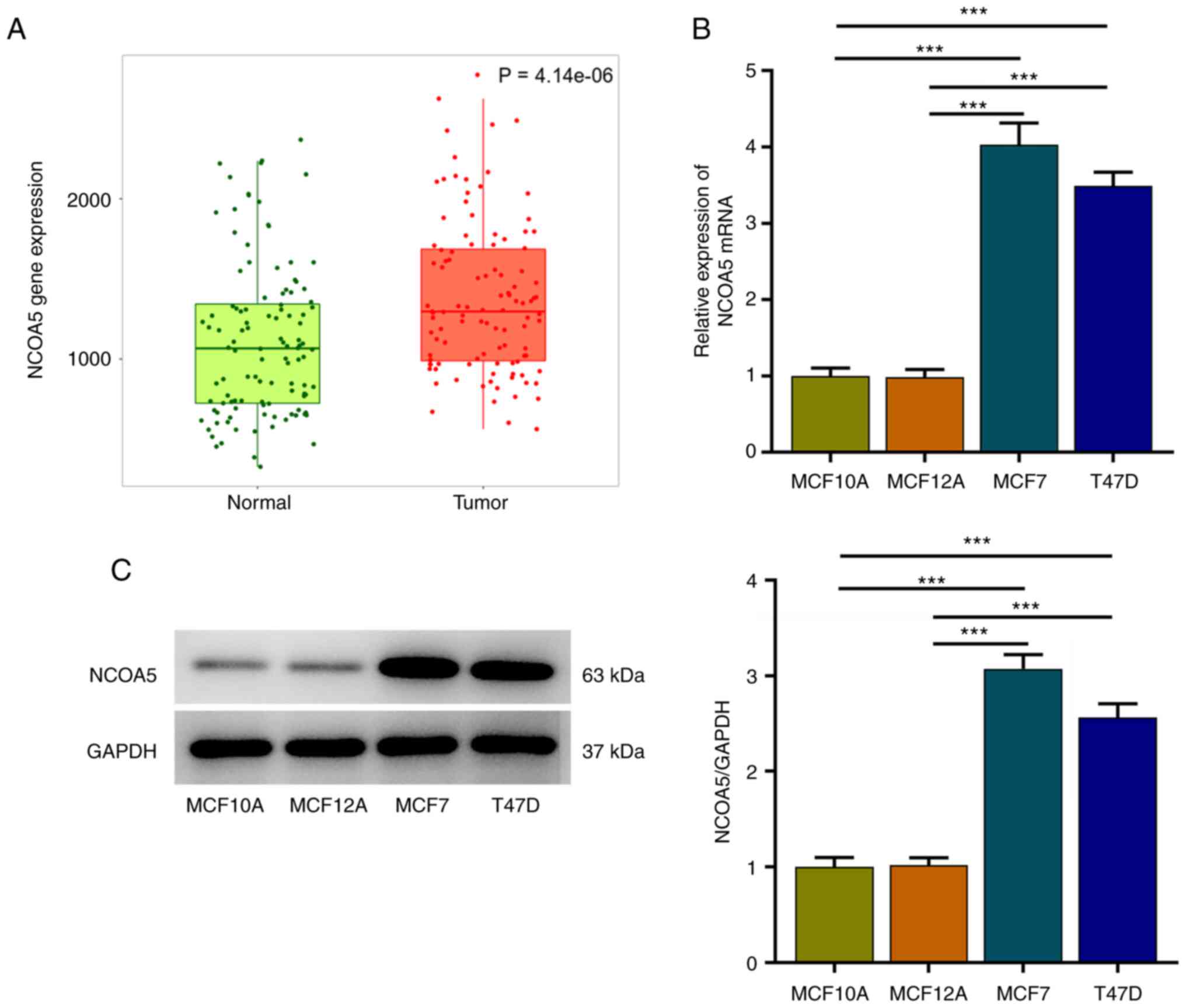
NCOA5 is highly expressed in breast
cancer
TNMplot was adopted to analyze NCOA5 expression in
paired non-tumor and tumor tissues of breast cancer patients. NCOA5
expression was markedly elevated in tumor tissues compared to the
adjacent non-tumor tissues (Fig.
1A). Besides, expression differences of NCOA5 in human breast
epithelial cell lines (MCF10A, MCF12A) and human breast cancer cell
lines (MCF7, T47D) were assessed via RT-qPCR and western blot
assay. In comparison with those in MCF10A and MCF12A cells, NCOA5
mRNA (Fig. 1B) and protein
(Fig. 1C) levels in MCF7 and T47D
cells were significantly increased, especially in MCF7 cells. Thus,
MCF7 cells were selected for the follow-up experiments.
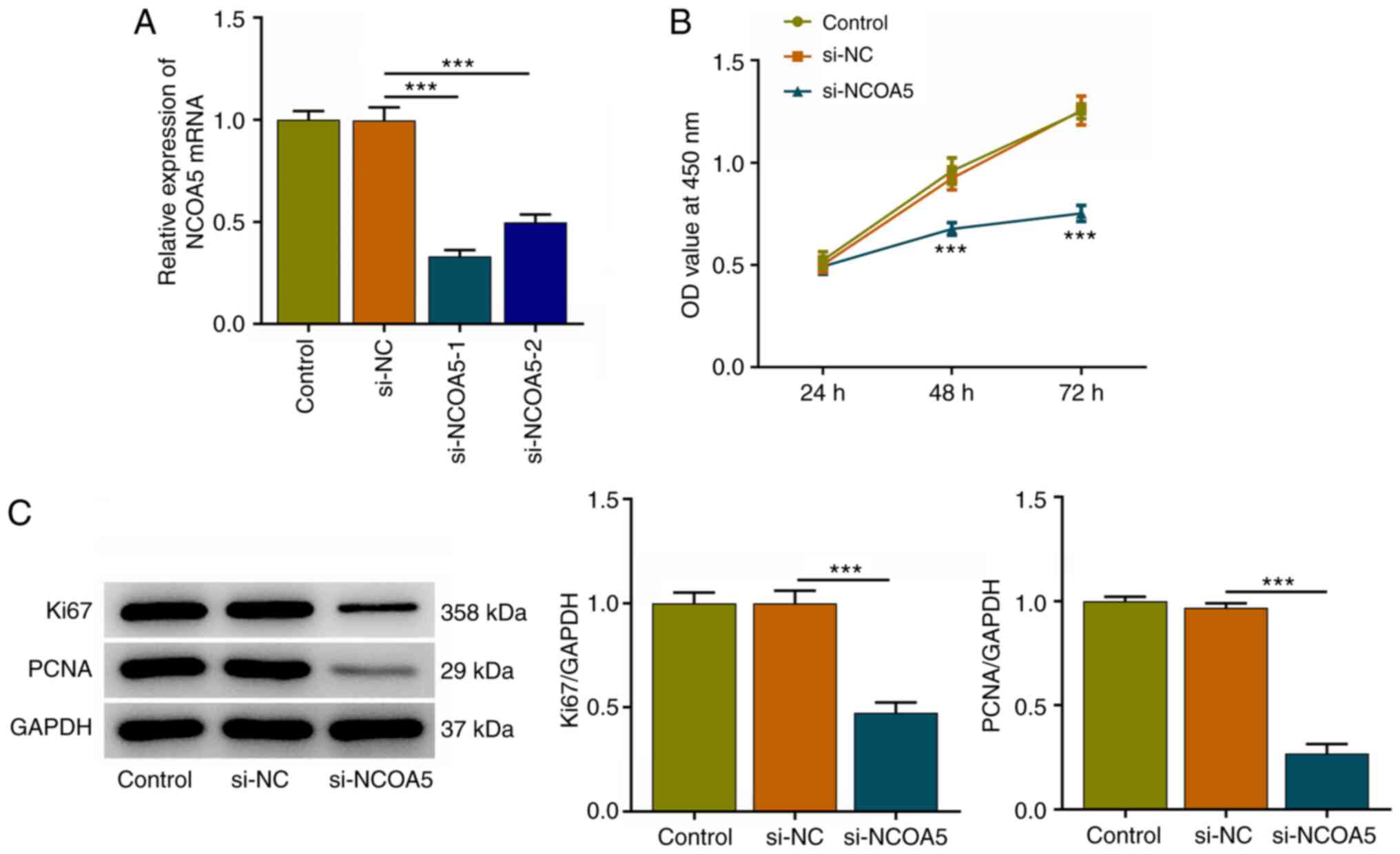
NCOA5 knockdown suppresses the
proliferation of breast cancer cells
In the current work, MCF7 cells were transfected
with si-NCOA5-1/2 or si-NC and transfection efficiency was
validated via RT-qPCR. NCOA5 expression was markedly downregulated
following transfection with si-NCOA5-1/2. Attributed a relative
high transfection efficacy, si-NCOA5-1 was selected for subsequent
research (Fig. 2A). Results of
CCK-8 assay indicated that NCOA5 knockdown inhibited the viability
of MCF7 cells (Fig. 2B).
Additionally, decreased expressions of proliferation markers (Ki67
and PCNA) following NCOA5 knockdown in MCF7 cells also demonstrated
that downregulation of NCOA5 could inhibit the proliferation of
breast cancer cells (Fig. 2C).
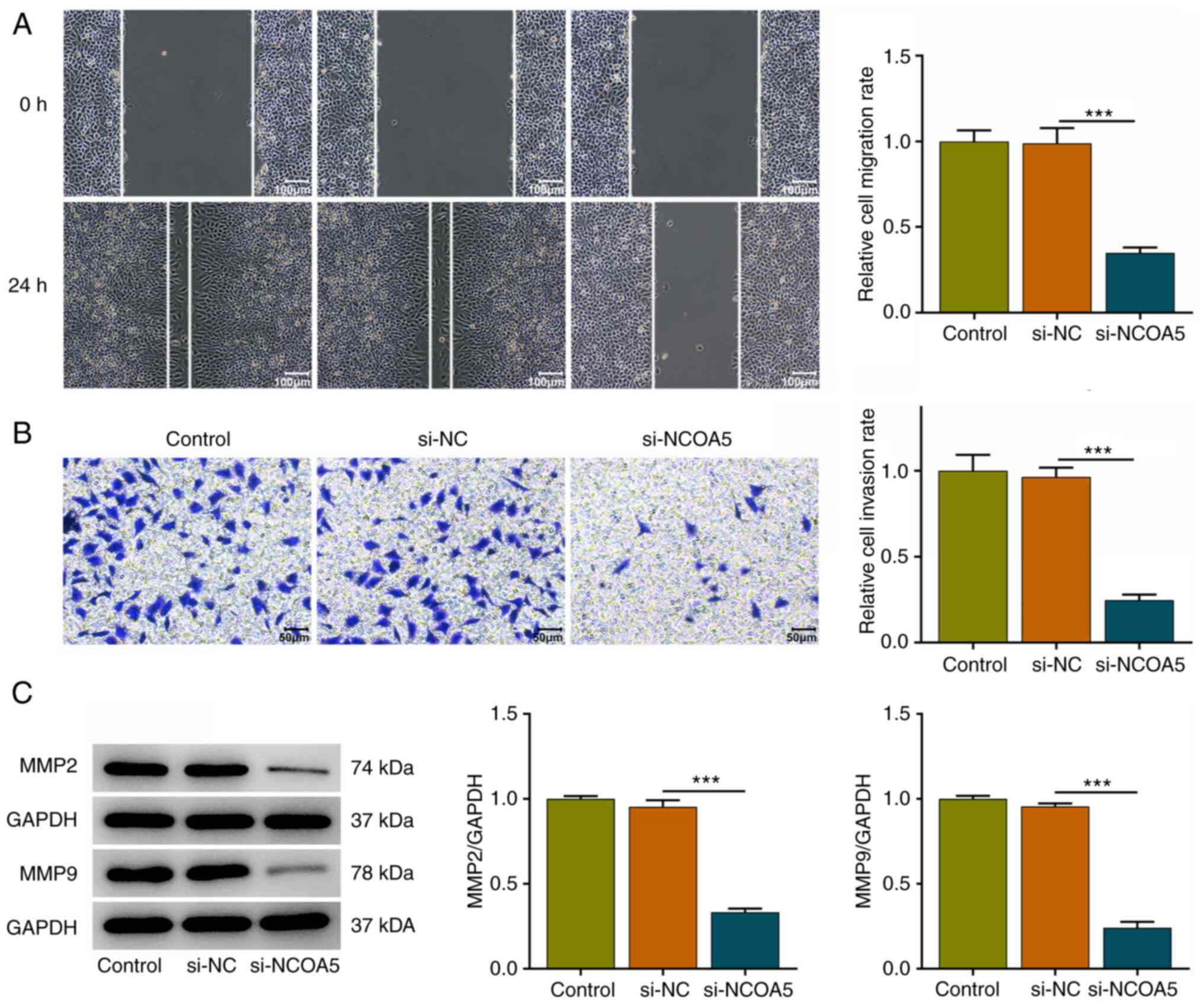
NCOA5 knockdown restrains the
migration and invasion of breast cancer cells
In addition, wound healing and transwell assays were
conducted to explore whether NCOA5 was functionally involved in the
migration and invasion of breast cancer cells. It was verified that
NCOA5 knockdown obviously repressed the migratory and invasive
abilities of MCF7 cells (Fig. 3A
and B). Besides, decreases in MMP2
and MMP9 expressions following NCOA5 knockdown also suggested that
downregulation of NCOA5 could suppress the migration and invasion
of breast cancer cells (Fig.
3C).
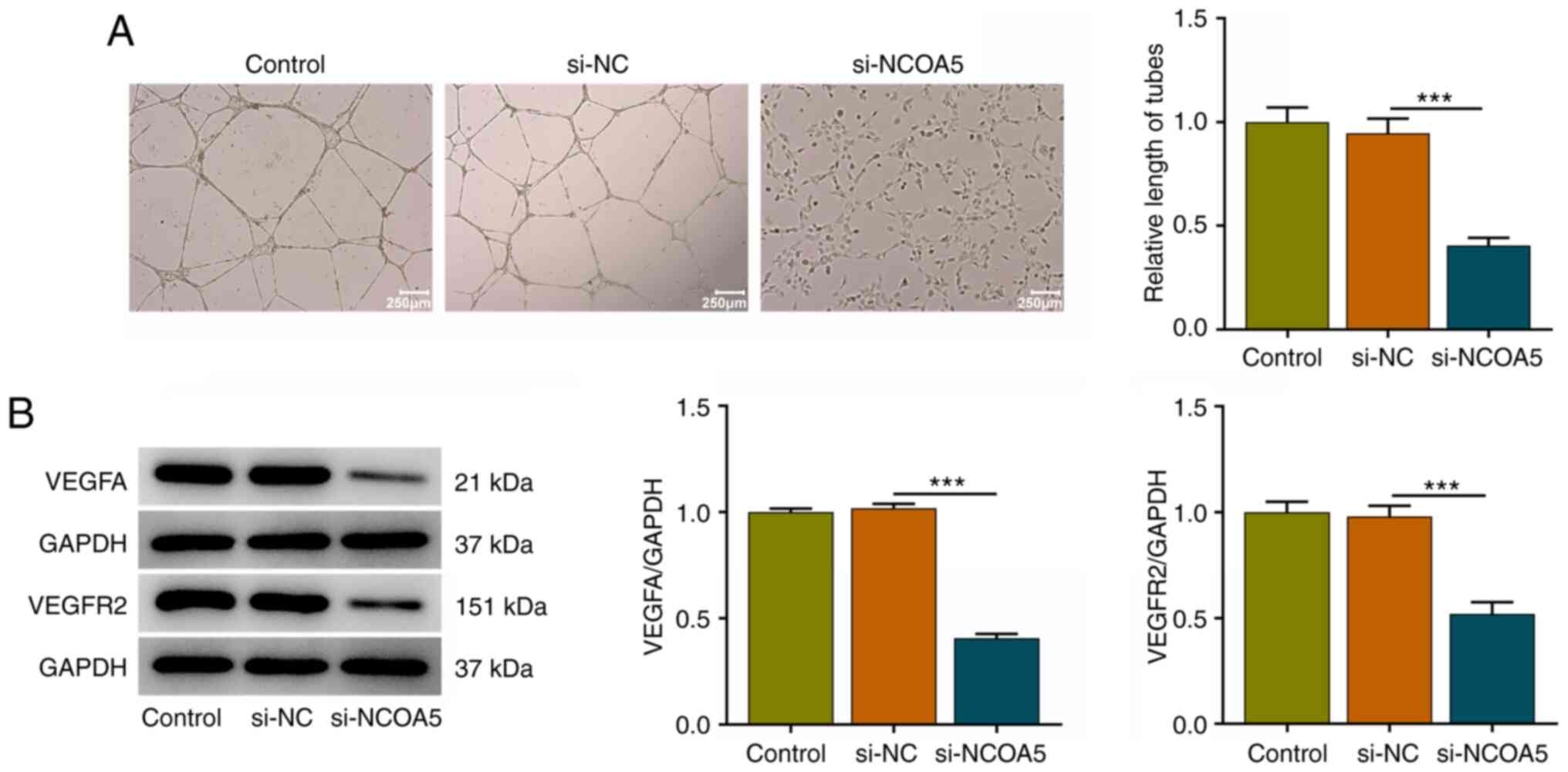
NCOA5 knockdown induces weaker in
vitro angiogenesis
Angiogenesis is a fundamental characteristic of
tumors. Tumor growth and metastasis need glorious angiogenesis for
nutrition provision. Vascular endothelial cell migration and tube
formation are important processes in tumor-related abnormal
angiogenesis. A tube formation assay of HUVECs revealed that
downregulation of NCOA5 induced a weaker in vitro
angiogenesis (Fig. 4A). Meanwhile,
decreased expressions of VEGFA and VEGFR2 in HUVECs together
demonstrated that NCOA5 was causally associated with in
vitro angiogenesis and NCOA5 knockdown could restrain in
vitro angiogenesis (Fig.
4B).
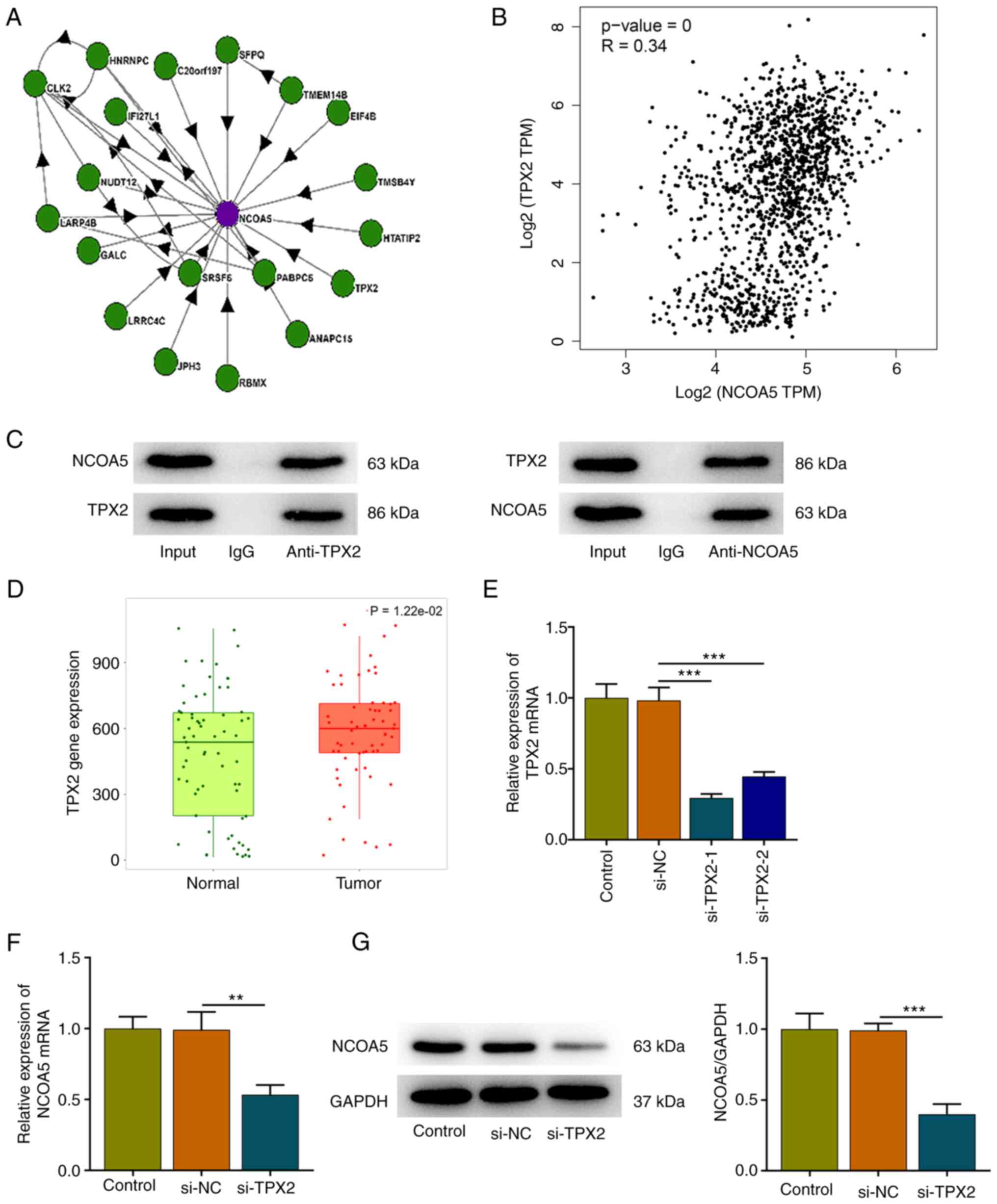
TPX2 interacts with NCOA5
High-confidence NCOA5 interactors were identified
based on BioPlex network data sets (Fig. 5A). Then, the correlation expression
of NCOA5 and TPX2 was analyzed by the Correlation Analysis module
of GEPIA. NCOA5 was moderately correlated with TPX2 in breast
cancer (Fig. 5B). Furthermore, the
interaction between TPX2 and NCOA5 was validated by employing Co-IP
assay. NCOA5 protein existed in the anti-TPX2 group and TPX2
protein existed in the anti-NCOA5 group, suggesting that TPX2 and
NCOA5 could interact with each other (Fig. 5C). In addition, TNMplot presented
that TPX2 expression was markedly elevated in tumor tissues of
breast cancer patients in comparison with that in the adjacent
non-tumor tissues (Fig. 5D). Then,
MCF7 cells were transfected with si-TPX2 or si-NC. TPX2 expression
was markedly downregulated following transfection and si-TPX2-1
with a relatively high transfection efficiency was selected for
subsequent research (Fig. 5E). It
was observed that TPX2 knockdown downregulated NCOA5 expression,
indicating a positive correlation between TPX2 and NCOA5 expression
in breast cancer cells (Fig. 5F
and G).
TPX2 knockdown suppresses the
proliferation of breast cancer cells by downregulating NCOA5
Subsequently, Ov-NCOA5 was introduced into MCF7
cells for rescue experiments, aiming to investigate whether TPX2
mediated the malignant biological behaviors of breast cancer cells
by regulating NCOA5 expression. Transfection efficiency was
validated via RT-qPCR and transfection of Ov-NCOA5 markedly
upregulated NCOA5 expression (Fig.
6A). Results of CCK-8 assay indicated that TPX2 knockdown
inhibited the viability of MCF7 cells, which was reversed by NCOA5
overexpression (Fig. 6B).
Additionally, increased expressions of Ki67 and PCNA in MCF7 cells
also demonstrated that upregulation of NCOA5 reversed the
suppressive effect of TPX2 knockdown on the proliferation of breast
cancer cells (Fig. 6C). In a word,
these evidences implied that TPX2 knockdown could inhibit the
proliferative capacity of breast cancer cells by downregulating
NCOA5 expression.
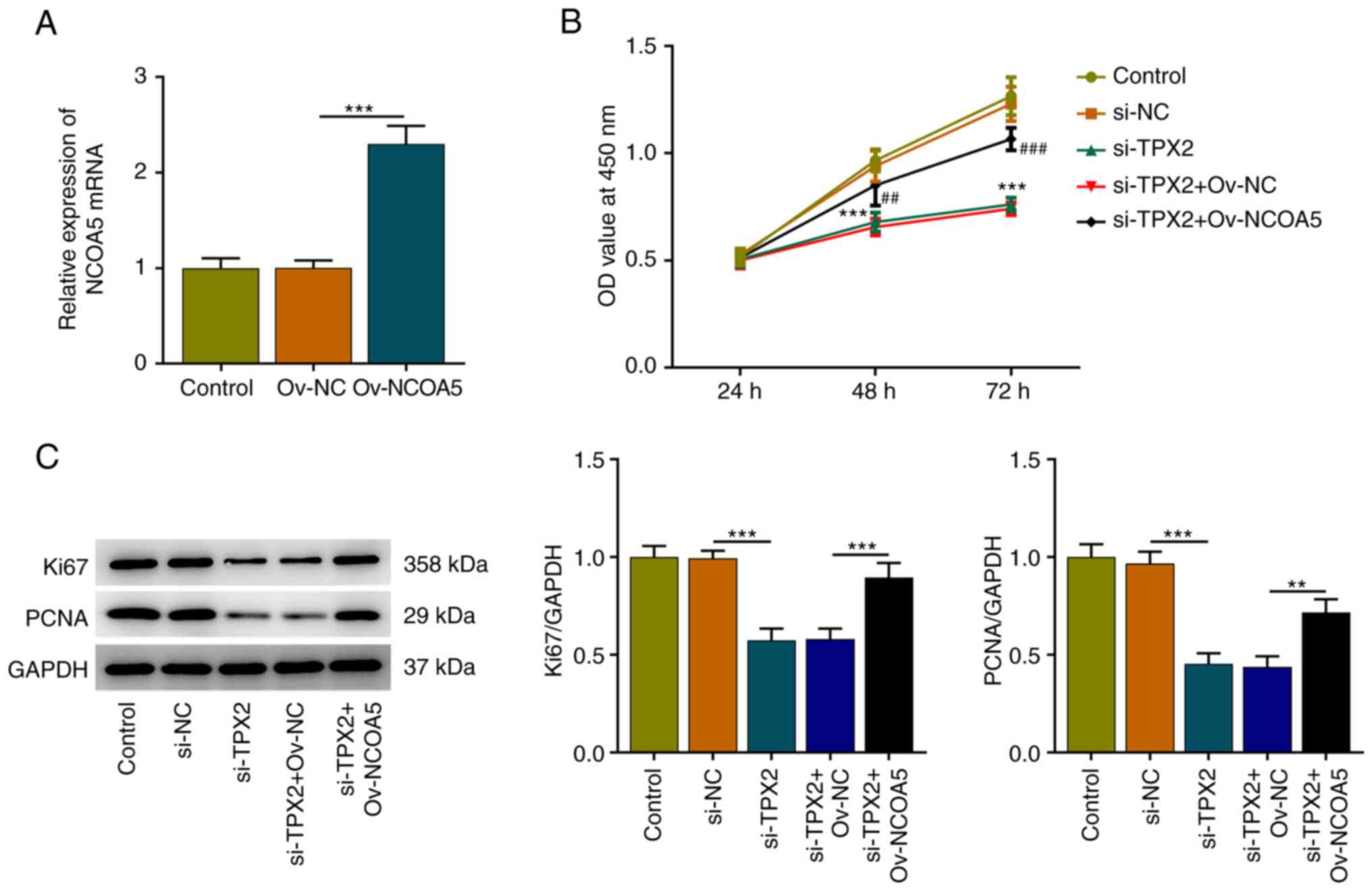 | Figure 6TPX2 knockdown suppresses the
proliferation of breast cancer cells by downregulating NCOA5. (A)
MCF7 cells were transfected with Ov-NCOA5 or Ov-NC. The
transfection efficiency of si-NCOA5 in MCF7 cells was validated via
RT-qPCR. ***P<0.001. (B) MCF7 cells were transfected
with si-TPX2 or co-transfected with si-TPX2 and Ov-NCOA5. Cell
Counting Kit-8 assay for determination of the viability of MCF7
cells. ***P<0.001 vs. si-TPX2.
##P<0.01, ###P<0.001 vs. si-TPX2 +
Ov-NC. (C) MCF7 cells were transfected with si-TPX2 or
co-transfected with si-TPX2 and Ov-NCOA5. Western blot analysis for
determination of Ki67 and PCNA protein expression in MCF7 cells.
**P<0.01, ***P<0.001. NC, negative
control; NCOA5, nuclear receptor coactivator 5; OD, optical
density; Ov, overexpression plasmid; PCNA, proliferating cell
nuclear antigen; RT-qPCR, reverse transcription-quantitative PCR;
si, small interfering RNA; TPX2, targeting protein for xenopus
kinesin-like protein 2. |
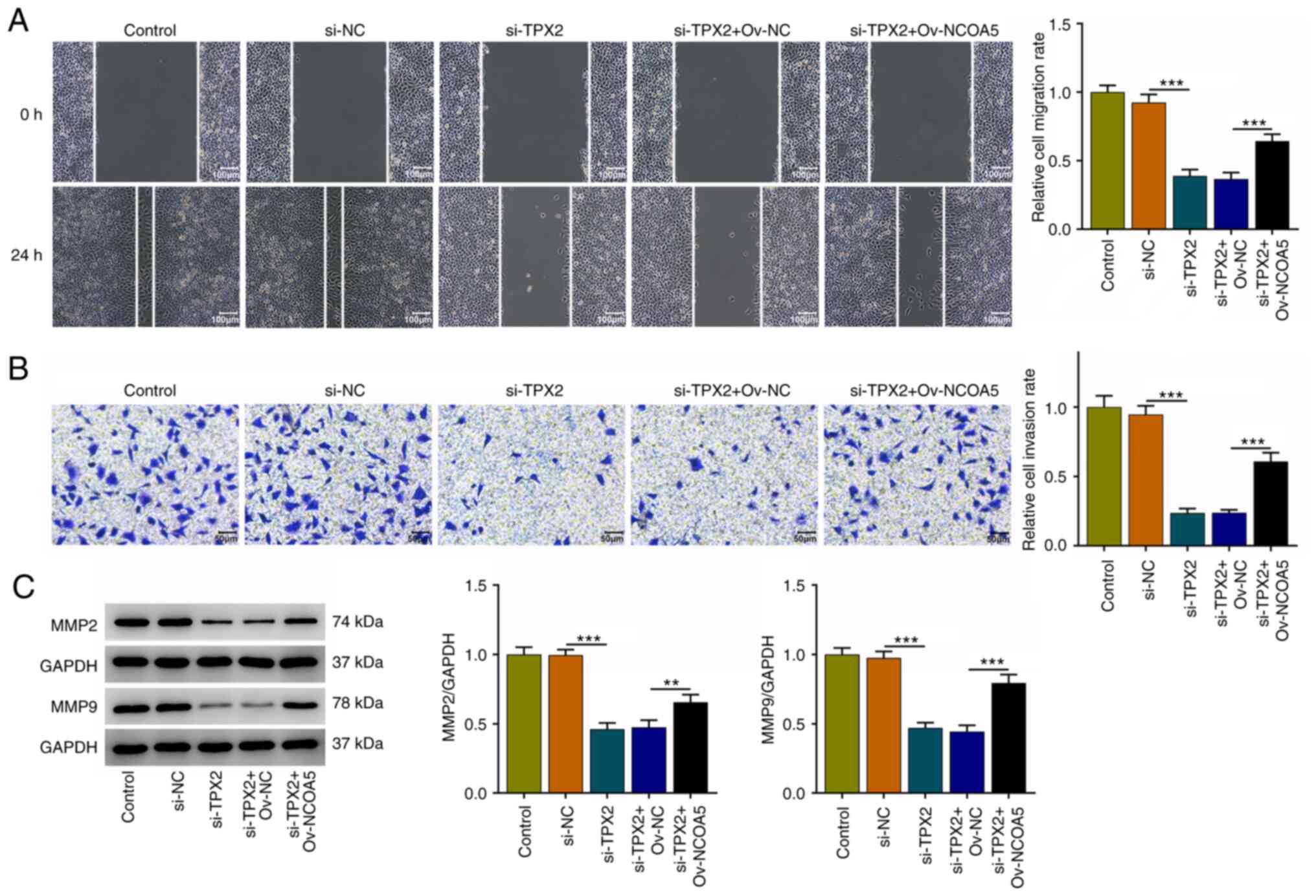
TPX2 knockdown restrains the migration
and invasion of breast cancer cells by downregulating NCOA5
Moreover, it was discovered that TPX2 knockdown
suppressed the migratory and invasive capabilities of MCF7 cells,
which were reversed by NCOA5 overexpression (Fig. 7A and B). Besides, increases in MMP2 and MMP9
expressions also suggested that upregulation of NCOA5 reversed the
suppressive effects of TPX2 knockdown on the migration and invasion
of breast cancer cells (Fig. 7C).
To conclude, TPX2 knockdown could repress the migratory and
invasive capacities of breast cancer cells by downregulating NCOA5
expression.
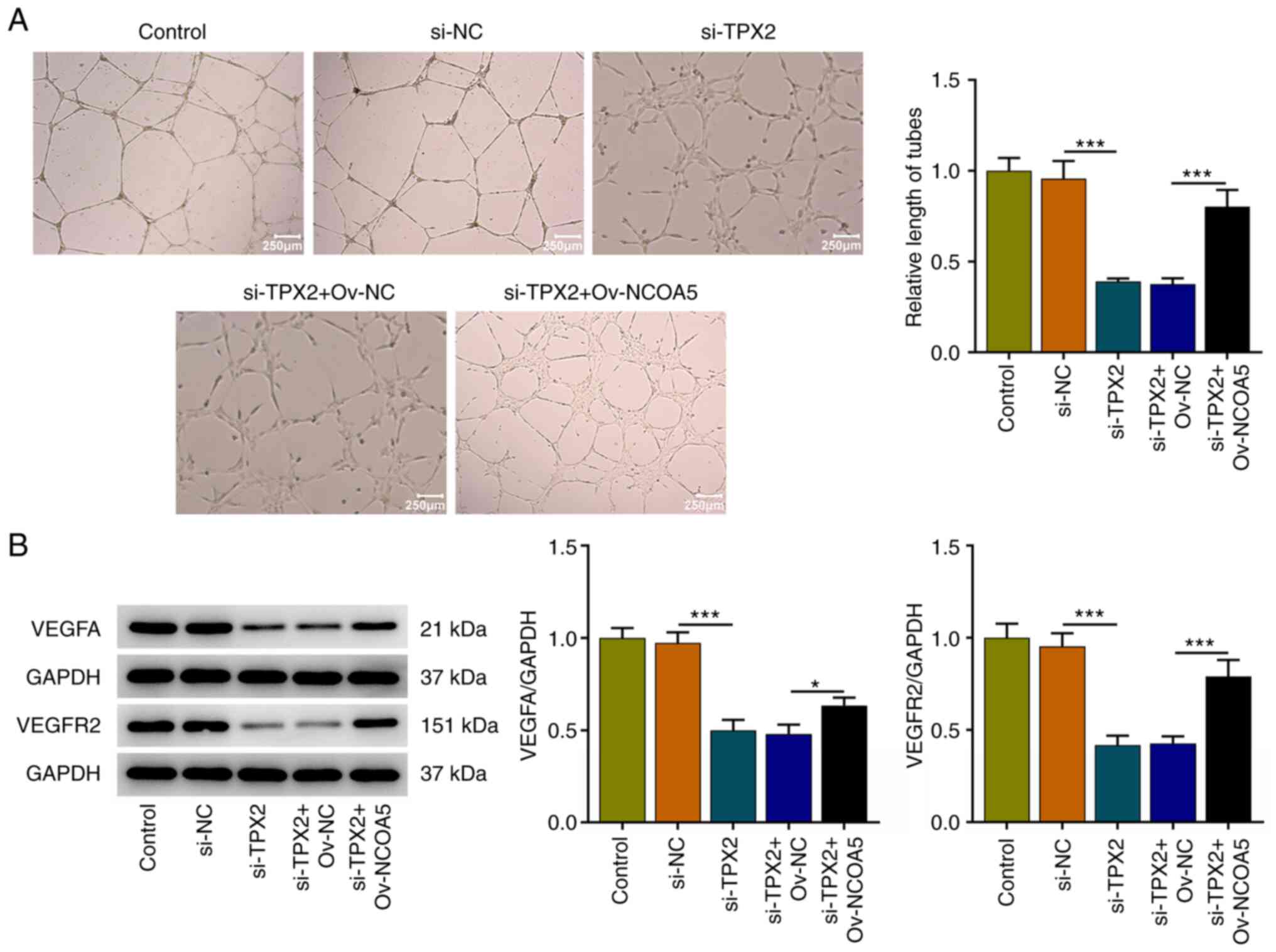
TPX2 knockdown induces weaker in vitro
angiogenesis by downregulating NCOA5
The tube formation assay of HUVECs revealed that
TPX2 knockdown induced a weaker in vitro angiogenesis, which
were reversed by NCOA5 overexpression (Fig. 8A). Meanwhile, increased expressions
of VEGFA and VEGFR2 in HUVECs together demonstrated that
upregulation of NCOA5 reversed the suppressive effect of TPX2
knockdown on in vitro angiogenesis (Fig. 8B). Overall, TPX2 knockdown could
arrest in vitro angiogenesis by downregulating NCOA5
expression.
Discussion
Breast cancer is one of the most common malignant
tumors in women worldwide. Although systemic therapies have
improved the prognosis of breast cancer patients, recurrence and
metastasis are barriers to the successful treatment of patients
with breast cancer (17).
Meanwhile, understanding of the pathogenesis and mechanisms of
breast cancer remains greatly limited. Recently, early diagnosis
and molecular targeted therapy for breast cancer patients have
become research hotspots (18).
Therefore, it is in urgent need to identify new genes involved in
breast cancer, aiming to aid the development of faster and safer
diagnostic methods and to improve breast cancer prognosis and
treatment.
Evidences suggest that NCOA5 is highly expressed in
colorectal cancer (6),
hepatocellular carcinoma (7) and
breast cancer (8) tissues or cell
lines. Interestingly, Tan et al (19) have reported that NCOA5 is
upregulated in breast cancer tissues and cell lines, and NCOA5
knockdown could inhibit the viability, migration and
epithelial-mesenchymal transition (EMT) of breast cancer cell
lines. Likewise, our current research also demonstrated that NCOA5
expression was significantly elevated in breast cancer and
downregulation of NCOA5 suppressed the proliferative, migratory and
invasive capabilities of breast cancer cells. Tumor
neovascularization is one of the main characteristics of tumors
(20). It plays an important role
in the rapid proliferation of tumor cells and metastasis to distant
places (21). In breast, cancer
angiogenesis has been evidenced to be a promising therapeutic
target. For example, Zhang et al (22) demonstrated that
angiotensin-converting enzyme 2 (ACE2) could restrain the
progression of breast cancer through inhibiting angiogenesis; Cao
et al (23) discovered
decylubiquinone to repress breast cancer growth and metastasis via
restricting angiogenesis. In current research, it was confirmed
that tube formation capacity of HUVECs was arrested by NCOA5
knockdown, suggesting that downregulation of NCOA5 could inhibit
in vitro angiogenesis. Therefore, NCOA5 knockdown may exert
tumor-suppressive effect via inhibiting cell proliferation,
migration, invasion and angiogenesis in breast cancer.
TPX2 was identified as a high-confidence NCOA5
interactor based on BioPlex network data sets. Furthermore, our
work validated the interaction between TPX2 and NCOA5 by employing
Co-IP assay. TPX2 interacted with NCOA5 and there was a positive
correlation between TPX2 and NCOA5 expression. Abnormal cell cycle
mitosis is an important factor leading to tumorigenesis and growth
(11). TPX2 participates in the
assembly and stability of the spindle, which is a vital key to the
regulation of mitosis (10).
Nowadays, abundant studies have focused on the aberrant TPX2
expression in tumors and its targeted inhibition (12,13).
Zhou et al (24) have
proposed that TPX2 overexpression could promote migration,
invasion, EMT and activities of MMPs of non-small cell lung cancer
cells. In addition, TPX2 has been proved to function as a tumor
promoter in breast cancer (14,15).
Consistently, in our study, it was also confirmed that TPX2 was
highly expressed in breast cancer, and the following experiments
revealed that TPX2 knockdown suppressed the proliferative,
migratory and invasive capabilities of breast cancer cells as well
as inhibited in vitro angiogenesis, suggesting that TPX2
knockdown was beneficial to restricting breast cancer development,
and TPX2 might be an alternative target for treatment strategies.
Furthermore, the rescue experiments revealed that upregulation of
NCOA5 reversed the suppressive effects of TPX2 knockdown on
proliferation, migration, invasion of breast cancer cells and in
vitro angiogenesis, further emphasizing the TPX2/NCOA5 axis in
regulating the development of breast cancer.
However, there were some limitations in this study.
First, all data were obtained from one single cell line, and more
cell lines might be beneficial to verify our conclusion; Secondly,
although the aberrant expression level of NCOA5 and TPX2 has been
reported in the human breast cancer tumor samples in previous
documents (14,19), the clinical validation in this
study is still helpful to improve the manuscript quality, which is
now planned in our future research.
To sum up, downregulation of TPX2 repressed
proliferation, migration and invasion of breast cancer cells as
well as restrained in vitro angiogenesis via suppressing NCOA5
expression. Findings may prompt that NCOA5 is a downstream target
of TPX2 in enhancing cell proliferation, migration, invasion and
angiogenesis of breast cancer.
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
TW, FZ and PZ searched the literature, designed the
study, performed the experiments, analyzed and interpreted the
data, and wrote the manuscript. TW and PZ confirm the authenticity
of all the raw data. All authors have read and approved the final
manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global cancer statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249.
2021.PubMed/NCBI View Article : Google Scholar
|
|
2
|
DeSantis CE, Ma J, Gaudet MM, Newman LA,
Miller KD, Goding Sauer A, Jemal A and Siegel RL: Breast cancer
statistics, 2019. CA Cancer J Clin. 69:438–451. 2019.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Henriques B, Mendes F and Martins D:
Immunotherapy in breast cancer: When, how, and what challenges.
Biomedicines. 9(1687)2021.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Liang Y, Zhang H, Song X and Yang Q:
Metastatic heterogeneity of breast cancer: Molecular mechanism and
potential therapeutic targets. Semin Cancer Biol. 60:14–27.
2020.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Chen GQ, Tian H, Yue WM, Li L, Li SH, Qi
L, Gao C, Si LB and Lu M: NCOA5 low expression correlates with
survival in esophageal squamous cell carcinoma. Med Oncol.
31(376)2014.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Sun K, Wang S, He J, Xie Y, He Y, Wang Z
and Qin L: NCOA5 promotes proliferation, migration and invasion of
colorectal cancer cells via activation of PI3K/AKT pathway.
Oncotarget. 8:107932–107946. 2017.PubMed/NCBI View Article : Google Scholar
|
|
7
|
He J, Zhang W, Li A, Chen F and Luo R:
Knockout of NCOA5 impairs proliferation and migration of
hepatocellular carcinoma cells by suppressing
epithelial-to-mesenchymal transition. Biochem Biophys Res Commun.
500:177–183. 2018.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Ye XH, Huang DP and Luo RC: NCOA5 is
correlated with progression and prognosis in luminal breast cancer.
Biochem Biophys Res Commun. 482:253–256. 2017.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Xia E, Hu W, Bhandari A, Sindan N and
Huang D: Nuclear receptor coactivator 5 is correlated with
progression in breast carcinoma. Anticancer Agents Med Chem.
21:2520–2524. 2021.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Safari MS, King MR, Brangwynne CP and
Petry S: Interaction of spindle assembly factor TPX2 with
importins-α/β inhibits protein phase separation. J Biol Chem.
297(100998)2021.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Gomes-Filho SM, Dos Santos EO, Bertoldi
ERM, Scalabrini LC, Heidrich V, Dazzani B, Levantini E, Reis EM and
Bassères DS: Aurora A kinase and its activator TPX2 are potential
therapeutic targets in KRAS-induced pancreatic cancer. Cell Oncol
(Dordr). 43:445–460. 2020.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Zhang B, Zhang M, Li Q, Yang Y, Shang Z
and Luo J: TPX2 mediates prostate cancer epithelial-mesenchymal
transition through CDK1 regulated phosphorylation of
ERK/GSK3β/SNAIL pathway. Biochem Biophys Res Commun. 546:1–6.
2021.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Zhu H, Liu J, Feng J, Zhang Q, Bian T, Li
X, Sun H, Zhang J and Liu Y: Overexpression of TPX2 predicts poor
clinical outcome and is associated with immune infiltration in
hepatic cell cancer. Medicine (Baltimore).
99(e23554)2020.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Yang Y, Li DP, Shen N, Yu XC, Li JB, Song
Q and Zhang JH: TPX2 promotes migration and invasion of human
breast cancer cells. Asian Pac J Trop Med. 8:1064–1070.
2015.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Jiang Y, Liu Y, Tan X, Yu S and Luo J:
TPX2 as a novel prognostic indicator and promising therapeutic
target in triple-negative breast cancer. Clin Breast Cancer.
19:450–455. 2019.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Kumar S, Srivastav RK, Wilkes DW, Ross T,
Kim S, Kowalski J, Chatla S, Zhang Q, Nayak A, Guha M, et al:
Estrogen-dependent DLL1-mediated notch signaling promotes luminal
breast cancer. Oncogene. 38:2092–2107. 2019.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Zhang Y, Shu C, Maimaiti Y, Wang S, Lu C
and Zhou J: LRP6 as a biomarker of poor prognosis of breast cancer.
Gland Surg. 10:2414–2427. 2021.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Tan Y, Liu F and Xu P: Knockdown of NCOA5
suppresses viability, migration and epithelial-mesenchymal
transition, and induces adhesion of breast cancer cells. Oncol
Lett. 22(694)2021.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Chen WZ, Jiang JX, Yu XY, Xia WJ, Yu PX,
Wang K, Zhao ZY and Chen ZG: Endothelial cells in colorectal
cancer. World J Gastrointest Oncol. 11:946–956. 2019.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Tu J, Fang Y, Han D, Tan X, Jiang H, Gong
X, Wang X, Hong W and Wei W: Activation of nuclear factor-κB in the
angiogenesis of glioma: Insights into the associated molecular
mechanisms and targeted therapies. Cell Prolif.
54(e12929)2021.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Zhang Q, Lu S, Li T, Yu L, Zhang Y, Zeng
H, Qian X, Bi J and Lin Y: ACE2 inhibits breast cancer angiogenesis
via suppressing the VEGFa/VEGFR2/ERK pathway. J Exp Clin Cancer
Res. 38(173)2019.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Cao J, Liu X, Yang Y, Wei B, Li Q, Mao G,
He Y, Li Y, Zheng L, Zhang Q, et al: Decylubiquinone suppresses
breast cancer growth and metastasis by inhibiting angiogenesis via
the ROS/p53/BAI1 signaling pathway. Angiogenesis. 23:325–338.
2020.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Zhou F, Wang M, Aibaidula M, Zhang Z,
Aihemaiti A, Aili R, Chen H, Dong S, Wei W and Maimaitiaili A: TPX2
promotes metastasis and serves as a marker of poor prognosis in
non-small cell lung cancer. Med Sci Monit.
26(e925147)2020.PubMed/NCBI View Article : Google Scholar
|