Introduction
Colon cancer has high prevalence worldwide and its
rates of morbidity and mortality have maintained an upward trend
over the last 5-10 years (1-3).
Although early-stage colon cancer generally has a favorable
outcome, the majority of patients with colon cancer are diagnosed
at a late stage and have a poor prognosis due to local or even
distant metastasis (4-6).
Therefore, investigation of the molecular mechanism of metastasis
is crucial to address an unmet clinical need.
Ion channels are emerging as pivotal modulators of
different aspects of cancer cell behavior. The abnormal expression
of genes encoding ion-channel proteins, particularly potassium
(K+) channel proteins, has attracted considerable
attention (7,8). Potassium voltage-gated channel
subfamily E (KCNE) proteins are single transmembrane-segment
voltage-gated potassium (Kv) channel ancillary subunits that
associate with pore-forming subunits to form complexes with unique
characteristics (9). The KCNE
family comprises five members (KCNE1-5), which can interact with a
wide range of channels (10). It
has been documented that KCNE4 is highly expressed in multiple
types of tumors and participates in tumor vascularization, so as to
promote the proliferation, migration and invasion of cancer cells
(11,12). Notably, Wu et al (12) revealed that KCNE4 is upregulated in
samples from patients with high-risk colorectal cancer (CRC)
compared with low-risk CRC, and markedly elevated KCNE4 expression
is highly associated with poor progression-free survival in CRC.
Furthermore, The University of Alabama at Birmingham Cancer data
analysis portal (UALCAN) database shows that KCNE4 is significantly
elevated in colon cancer tissues, and KCNE4 expression is
positively associated with cancer stage and negatively associated
with patient survival in colon cancer.
The genes positively correlated with KCNE4 in colon
cancer may be identified based on analysis performed using
LinkedOmics database. Moreover, this database shows a strongly
positive correlation between KCNE4 and EGF containing fibulin
extracellular matrix protein 2 (EFEMP2) using Pearson's correlation
analysis. EFEMP2 is a member of the fibulin family of glycoproteins
(13). It is involved in the
maintenance of cell morphology, as well as the growth, adhesion and
movement of cells, and is closely associated with the development
of various types of tumors (14,15).
It has been reported that EFEMP2 is highly expressed in colon
cancer cells and may promote colon cancer cell invasion and growth
through the ERK1/2 signaling pathway (16). Additionally, EFEMP2 expression is
also upregulated in the tumor tissues of patients with CRC, even at
an early stage (17).
Therefore, the current study aimed to elucidate the
functional role of KCNE4 in the biological behaviors of colon
cancer cells and to investigate the molecular mechanism underlying
the involvement of KCNE4 and its highly correlated gene EFEMP2 in
the malignant progression of colon cancer.
Materials and methods
UALCAN analysis
KCNE4 expression in colon cancer tissues was
compared with that in normal tissues using the UALCAN database
(http://ualcan.path.uab.edu), and the
associations between KCNE4 expression and the pathological stages
of colon cancer and the survival probability of patients with colon
cancer were also investigated.
LinkedOmics analysis
The LinkedOmics database (http://www.linkedomics.org/) is a web-based platform
for the analysis of 32 The Cancer Genome Atlas cancer-associated
multi-dimensional datasets. The KCNE4-associated genes were
analyzed using Pearson's correlation coefficient and presented in a
volcano plot, heat map and scatter plot.
Cell culture
The human colon cancer cell lines HCT-116 (cat. no.
TCHu 99), SW480 (cat. no. TCHu172), SW1116 (cat. no. TCHu174),
Caco2 (cat. no. TCHu146) and LoVo (cat. no. TCHu 82) were purchased
from The Cell Bank of Type Culture Collection of The Chinese
Academy of Sciences. The NCM460 normal colonic mucosa cell line
(cat. no. CP-H040) was purchased from Procell Life Science &
Technology Co., Ltd. All cells were cultured in Dulbecco's modified
Eagle's medium (HyClone; Cytiva) supplemented with 10% fetal bovine
serum (FBS; Hyclone; Cytiva), 100 U/ml penicillin and 100 µg/ml
streptomycin in a humidified atmosphere with 5% CO2 at
37˚C.
Cell transfection
Small interfering RNA (siRNA) targeting KCNE4
(si-KCNE4; 5'-GATTGGCTGTAAGTATCTCTA-3') and scrambled negative
control (si-NC, 5'-GGATCACTAGTTACGATTGTT-3') were obtained from
Shanghai GenePharma Co., Ltd. The EFEMP2 overexpression plasmid
(Ov-EFEMP2) was established by inserting the EFEMP2 gene into a
pcDNA3.1 vector (Shanghai GenePharma Co., Ltd.), whereas an empty
vector served as the NC (Ov-NC). Transfection with si-KCNE4 (50
nM), si-NC (50 nM), Ov-EFEMP2 (0.5 µg) or Ov-NC (0.5 µg) was
performed using Lipofectamine® 2000 (Invitrogen; Thermo
Fisher Scientific, Inc.) at 37˚C for 4 h strictly according to the
manufacturer's guidelines. After another 48 h of incubation at
37˚C, the cells were collected for subsequent experiments.
Cell Counting Kit-8 (CCK-8) assay
The viability of HCT-116 cells was determined using
a CCK-8 assay. Cells (5,000 cells/well) grown in 96-well plates
were cultured for 48 h. Afterwards, 10 µl CCK-8 solution (Beyotime
Institute of Biotechnology) was added to each well and incubated at
37˚C for 2 h. The optical density at 450 nm was detected using a
microplate-reader (Bio-Rad Laboratories, Inc.).
Colony formation assay
The colony-forming ability of HCT-116 cells was
determined via colony formation assay. Cells plated at a density of
500 cells/well in 6-well plates were maintained in complete medium
for 10 days. After that, the cells were fixed with 4%
paraformaldehyde at room temperature for 15 min and stained with
0.1% crystal violet solution (Beijing Solarbio Science &
Technology Co., Ltd.) at room temperature for 10 min. Visible
colonies were photographed under a light microscope (Olympus
Corporation) and were counted using ImageJ software (Version 1.46;
National Institutes of Health). Colonies consisted of ≥50
cells.
Wound healing assay
The migratory ability of HCT-116 cells was assessed
using a wound healing assay. Cells (1x105 cells/well)
were seeded into 6-well plates and cultured up to 90% confluence.
Then, the cells were scratched with a sterile 200-µl pipette tip
and the detached cells were washed away with PBS. Afterwards, the
cells were incubated in serum-free medium at 37˚C for 24 h and the
area of the wound was photographed at 0 and 24 h under a microscope
(magnification, x100). Migration was assessed as follows: Migration
(%)=[(0 h average scratch distance-24 h average scratch distance)/0
h average scratch distance] x100.
Transwell assay
The invasive ability of HCT-116 cells was assessed
by performing a Transwell assay. The cells were re-suspended in
serum-free medium and seeded at a density of 2x104
cells/well into the upper compartment of a Transwell chamber
(Corning, Inc.), which was precoated with 50 µl Matrigel (Beijing
Solarbio Science & Technology Co., Ltd.) at 37˚C for 30 min.
The lower chambers were filled with 600 µl FBS-containing medium.
After 24 h of incubation at 37˚C, the invaded cells were fixed with
4% paraformaldehyde and stained with 0.1% crystal violet solution
at room temperature for 20 min. Finally, the invaded cells were
photographed and counted under a light microscope (magnification,
x200).
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA was isolated from cells using
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc.). Subsequently, the RNA was reverse transcribed into cDNA
using a PrimeScript RT Kit (Takara Bio, Inc.). The RT conditions
were performed at 30˚C as the priming stage for 10 min, 42˚C as the
RT step for 30 min and 95˚C as the RT inactivation stage for 5 min.
Then, qPCR reactions were performed on an ABI PRISM 7900 Real-Time
PCR system (Thermo Fisher Scientific, Inc.) using SYBR Premix Ex
Taq kit (Takara Bio, Inc.). The thermocycling conditions were as
follows: 95˚C for 10 min, followed by 35 cycles of 95˚C for 15 sec
and 65˚C for 60 sec. The sequences of the primers were as follows:
KCNE4 forward: 5'-CACCGCTACCTGAAAACCCT-3' and reverse:
5'-TTGATCGTGGCAGAGTGAGC-3'; EFEMP2 forward:
5'-GAGTGTCTGACCATCCCTGAG-3' and reverse:
5'-GCCGTGTAGGTCGTTGATGAC-3'; GAPDH forward:
5'-CAGGAGGCATTGCTGATGAT-3' and reverse: 5'-GAAGGCTGGGGCTCATTT-3'.
GAPDH served as the internal control. Relative gene expression
levels of KCNE4 and EFEMP2 were normalized to GAPDH and calculated
via the 2-ΔΔCq method (18).
Western blotting analysis
Total protein was isolated from cells using RIPA
lysis buffer (Beyotime Institute of Biotechnology) and protein
concentrations were quantified using a BCA protein assay kit
(Beyotime Institute of Biotechnology). Equal amounts of protein
samples (30 µg/lane) were subjected to 10% sodium dodecyl sulfate
(SDS)-polyacrylamide gel electrophoresis and then transferred onto
polyvinylidene fluoride membranes (MilliporeSigma). After blocking
in 5% skimmed milk for 1 h at room temperature, the membranes were
incubated with specific antibodies against KCNE4 (ab254642;
1:1,000), proliferating cell nuclear antigen (PCNA; ab92552;
1:1,000), Ki-67 (ab92742; 1:5,000), MMP2 (ab181286; 1:1,000), MMP9
(ab137867; 1:1,000), EFEMP2 (ab125073; 1:1,000) and GAPDH
(ab181602; 1:10,000), all from Abcam, overnight at 4˚C. The next
day, the membranes were incubated with goat anti-rabbit horseradish
peroxidase-conjugated IgG secondary antibody (ab205718; 1:50,000;
Abcam) for 1.5 h at room temperature. GAPDH served as the internal
control. Signals of the immunoblots were developed using a BeyoECL
plus kit (Beyotime Institute of Biotechnology) and analyzed with
ImageJ software v1.8.0 (National Institutes of Health).
Co-immunoprecipitation (Co-IP)
The interaction between KCNE4 and EFEMP2 was
verified via Co-IP. Cells were lysed using IP lysis buffer
(Beyotime Institute of Biotechnology). The supernatant was
collected after centrifugation at 13,000 x g for 10 min at 4˚C.
Next, anti-KCNE4 (ab254642; 1:200; Abcam) or anti-IgG (ab205718;
1:1,000; Abcam) was added to 250 µl cell lysate and incubated
overnight at 4˚C. On the following day, the cell lysates were
cultured with 25 µl protein A/G agarose beads (Santa Cruz
Biotechnology, Inc.) for 4 h at 4˚C to bind the antibody complexes.
Following centrifugation at 1,000 x g for 2 min at 4˚C, the agarose
beads were washed in lysis buffer 3-4 times. Finally, a total of 15
µl 2X SDS sample buffer was added and the samples were boiled at
100˚C for 5 min, followed by western blot analysis to detect EFEMP2
and KCNE4, as aforementioned.
Statistical analysis
Data from three independent experiments are
displayed as the mean ± standard deviation (SD). One-way analysis
of variance followed by Tukey's post hoc test was employed for
analyzing the statistical significance of differences among
multiple groups. P<0.05 was considered to indicate a
statistically significant difference.
Results
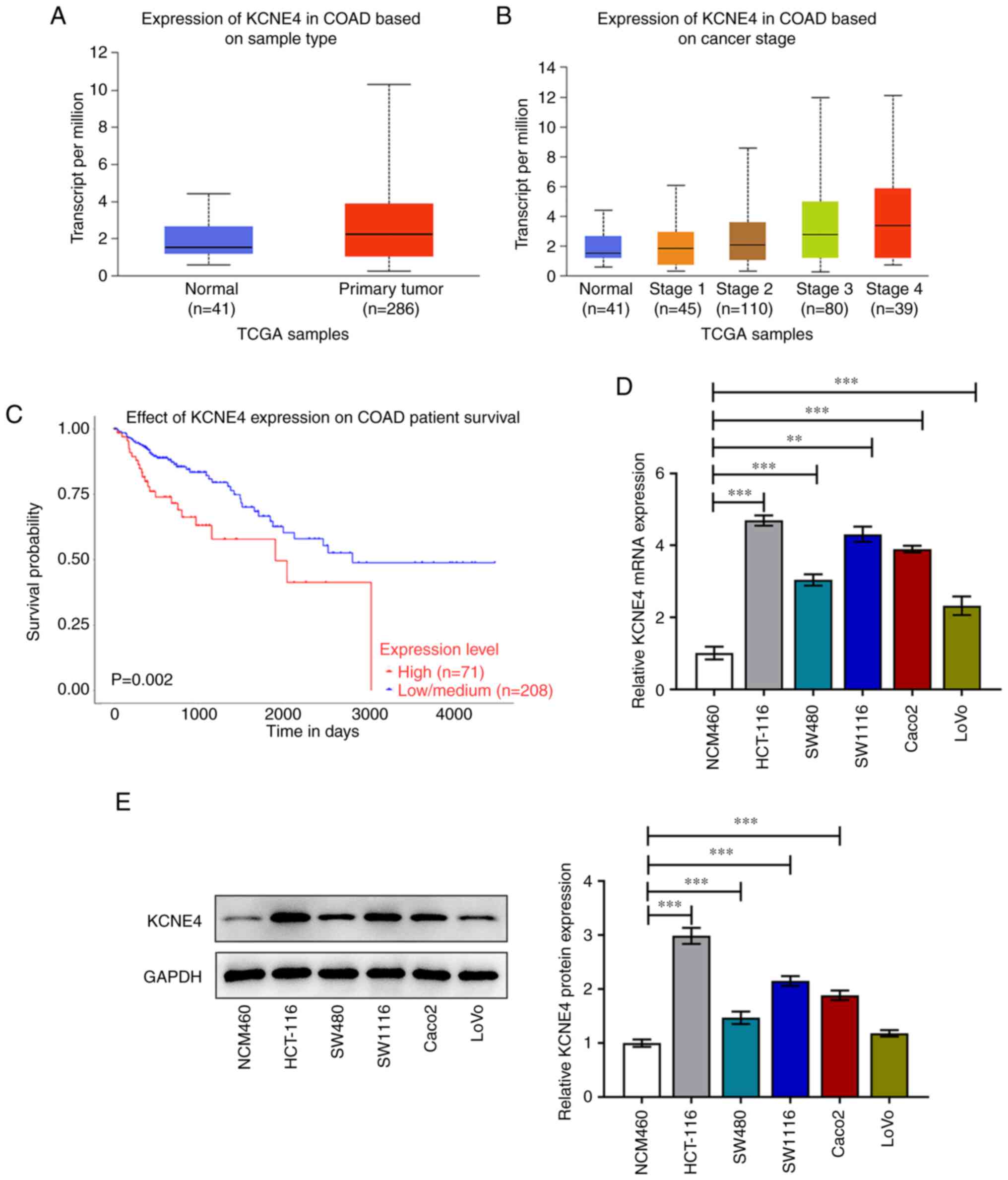
KCNE4 is highly expressed in colon
cancer
KCNE4 expression in colon cancer tissues and normal
tissues, as well as the relationships of KCNE4 expression with the
pathological stage of colon cancer and the survival probability of
patients with colon cancer were explored using the UALCAN database.
The UALCAN data indicated that KCNE4 was upregulated in colon
cancer tissues compared with normal tissues (Fig. 1A). In addition, the expression
level of KCNE4 in colon cancer tissues was positively associated
with the cancer stage; KCNE4 expression in late-stage colon cancer
was higher than that in early-stage colon cancer (Fig. 1B). Furthermore, patients with colon
cancer whose tissues had high KCNE4 expression had a shorter
duration of survival than those with low KCNE4 expression (Fig. 1C). In the present study,
differences in the expression levels of KCNE4 in the HCT-116,
SW480, SW1116, Caco2 and LoVo human colon cancer cell lines and the
NCM460 normal colonic mucosa cell line were assessed via RT-qPCR
and western blot assays. The KCNE4 mRNA (Fig. 1D) and protein (Fig. 1E) expression levels in the colon
cancer cells were markedly increased compared with those in NCM460
cells, and the expression levels in HCT-116 cells were particularly
high. Therefore, HCT-116 cells were selected for subsequent
experiments.
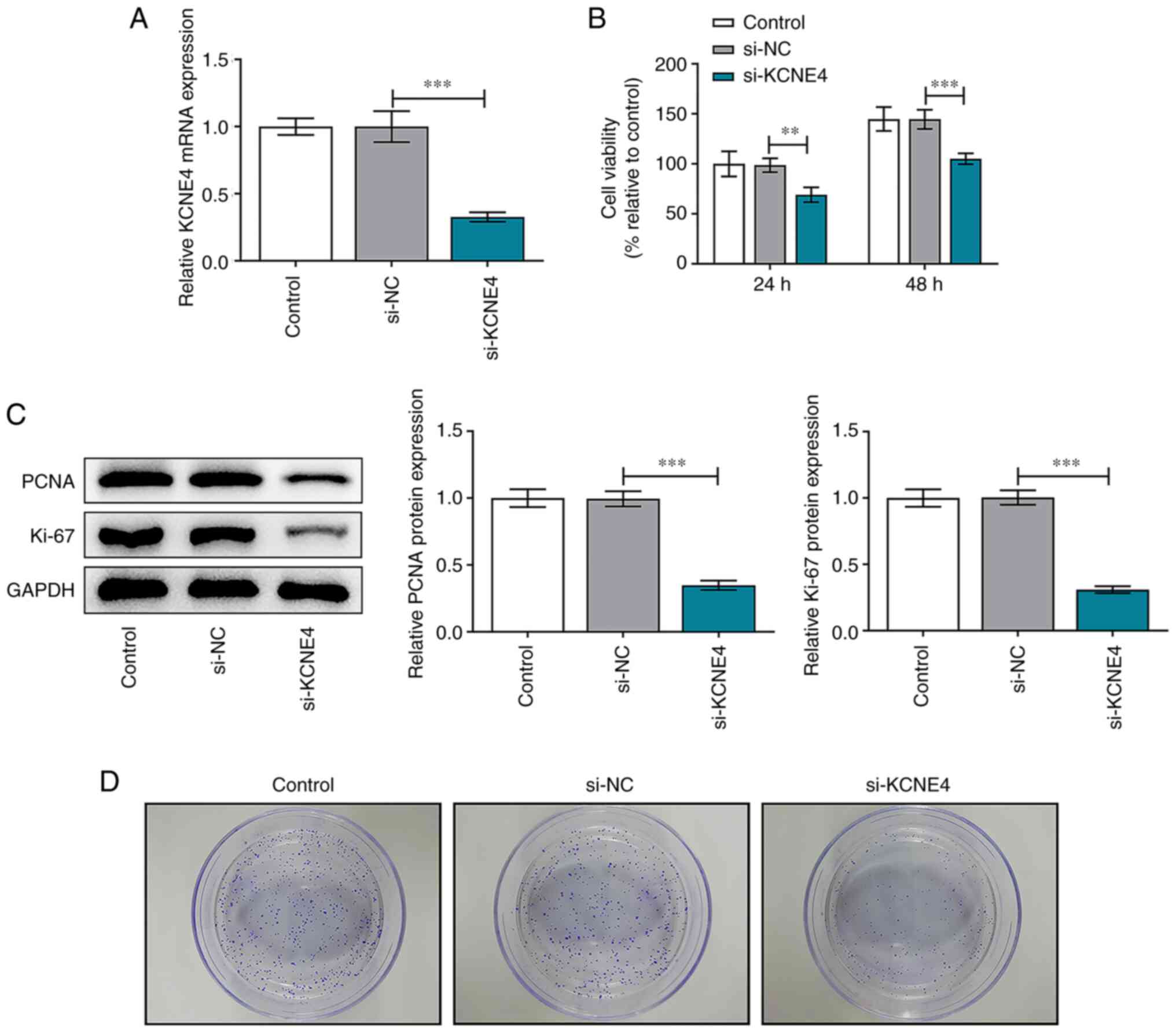
KCNE4 knockdown suppresses the
proliferative and colony-forming abilities of colon cancer
cells
HCT-116 cells were transfected with si-KCNE4 or
si-NC and transfection efficiency was validated via RT-qPCR.
Transfection with si-KCNE4 significantly downregulated KCNE4
expression compared with transfection with si-NC (Fig. 2A). CCK-8 assay results indicated
that KCNE4 knockdown significantly reduced the viability of HCT-116
cells (Fig. 2B). Furthermore,
reduced expression levels of PCNA and Ki-67 in the HCT-116 cells
transfected with si-KCNE4 were observed, which also suggested that
the viability of the colon cancer cells was repressed by the
downregulation of KCNE4 (Fig. 2C).
In addition, the colony formation assay revealed that the
downregulation of KCNE4 suppressed the colony formation ability of
the HCT-116 cells (Fig. 2D).
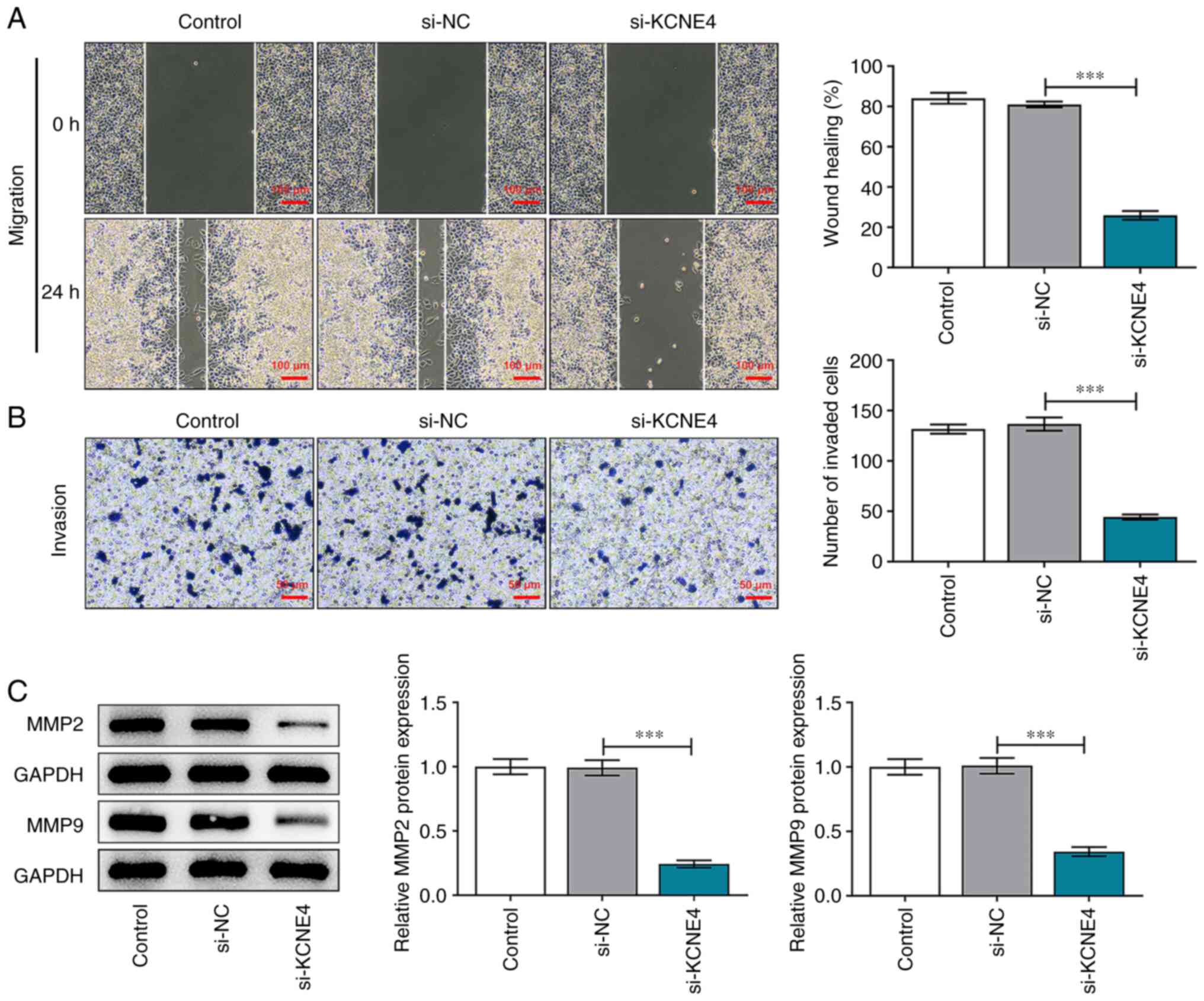
KCNE4 knockdown weakens the migratory
and invasive capacities of colon cancer cells
Wound healing and Transwell assays demonstrated that
the migratory and invasive abilities of HCT-116 cells were
significantly suppressed by KCNE4 knockdown (Fig. 3A and B). MMP2 and MMP9 are gelatinases that are
able to degrade and remodel the extracellular matrix (19). Therefore, MMP2 and MMP9 play
important roles in promoting the metastasis of cells. Decreased
expression levels of MMP2 and MMP9 in the HCT-116 cells transfected
with si-KCNE4 compared with those transfected with si-NC also
indicated that the downregulation of KCNE4 restrained the migration
and invasion of the colon cancer cells (Fig. 3C).
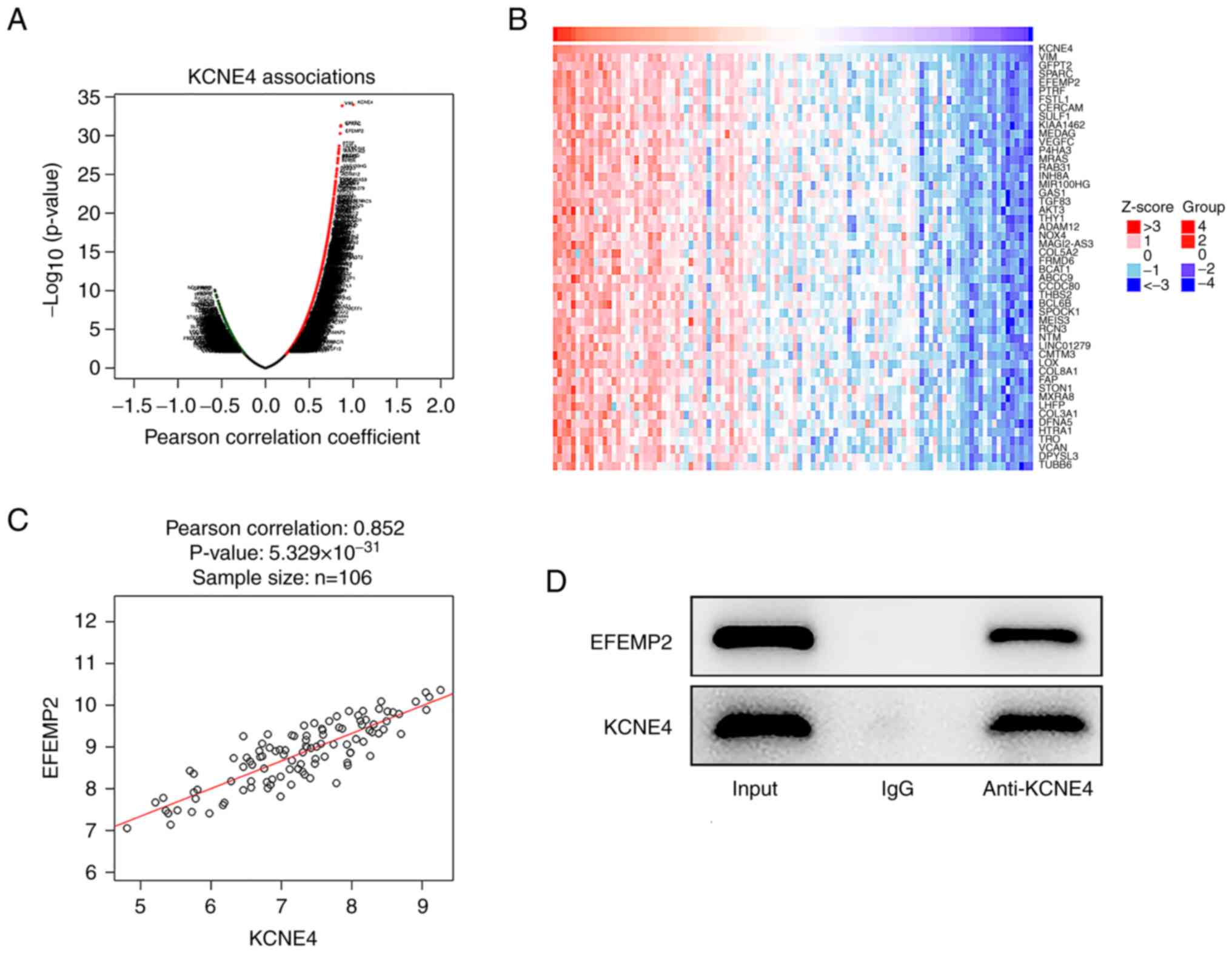
Positive correlation between KCNE4 and
EFEMP2
Genes associated with the key gene KCNE4 were
identified using the LinkedOmics database. The genes indicated to
be highly associated with KCNE4 in colon cancer are displayed in a
volcano plot (Fig. 4A). The top 50
genes significantly positively correlated with KCNE4 in colon
cancer are shown in a heat map (Fig.
4B). Using Pearson's correlation analysis, it was revealed that
KCNE4 exhibited a strongly positive correlation with EFEMP2
(Fig. 4C). Moreover, the
interaction between KCNE4 and EFEMP2 was validated via Co-IP
analysis. EFEMP2 protein was detected in the anti-KCNE4 group,
indicating that KCNE4 interacted with and bound to EFEMP2 (Fig. 4D).
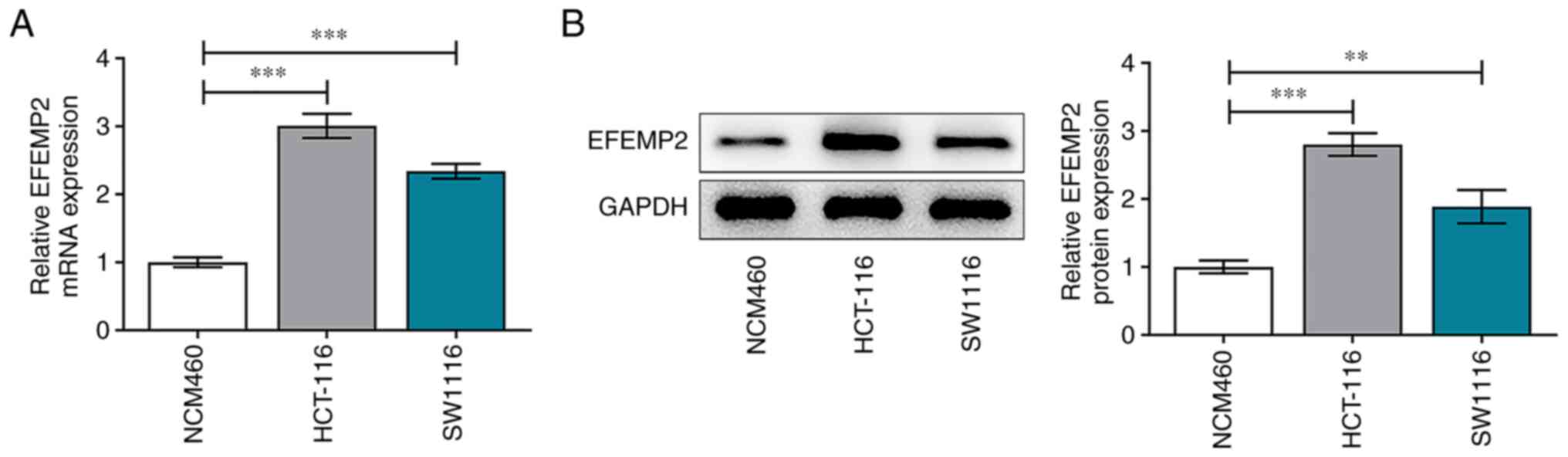
EFEMP2 is highly expressed in colon
cancer
Expression differences of EFEMP2 in the HCT-116 and
SW1116 human colon cancer cell lines and the NCM460 normal colonic
mucosa cell line were assessed via RT-qPCR and western blot assay.
In comparison with those in NCM460 cells, EFEMP2 mRNA (Fig. 5A) and protein (Fig. 5B) expression levels in both colon
cancer cell lines were significantly upregulated.
KCNE4 knockdown suppresses the
proliferative and colony formation abilities of colon cancer cells
by downregulating EFEMP2
Rescue experiments were carried out to explore
whether KCNE4 mediates colon cancer progression via the regulation
of EFEMP2 expression. The transfection efficiency of Ov-EFEMP2 was
validated via RT-qPCR, and this vector markedly upregulated EFEMP2
expression (Fig. 6A). The
downregulation of KCNE4 reduced the viability of the HCT-116 cells,
and this reduction was attenuated by EFEMP2 overexpression
(Fig. 6B). Increased PCNA and
Ki-67 expression levels in the HCT-116 cells co-transfected with
si-KCNE4 and Ov-EFEMP2 compared with those transfected with
si-KCNE4 and Ov-NC also indicated that the upregulation of EFEMP2
abolished the suppressive effects of KCNE4 knockdown on the
viability of colon cancer cells (Fig.
6C). In addition, the decreased colony formation of HCT-116
cells transfected with si-KCNE4 was attenuated by EFEMP2
overexpression (Fig. 6D). In
summary, these results indicate that KCNE4 knockdown suppressed the
proliferative and colony formation abilities of colon cancer cells
via the downregulation of EFEMP2 expression.
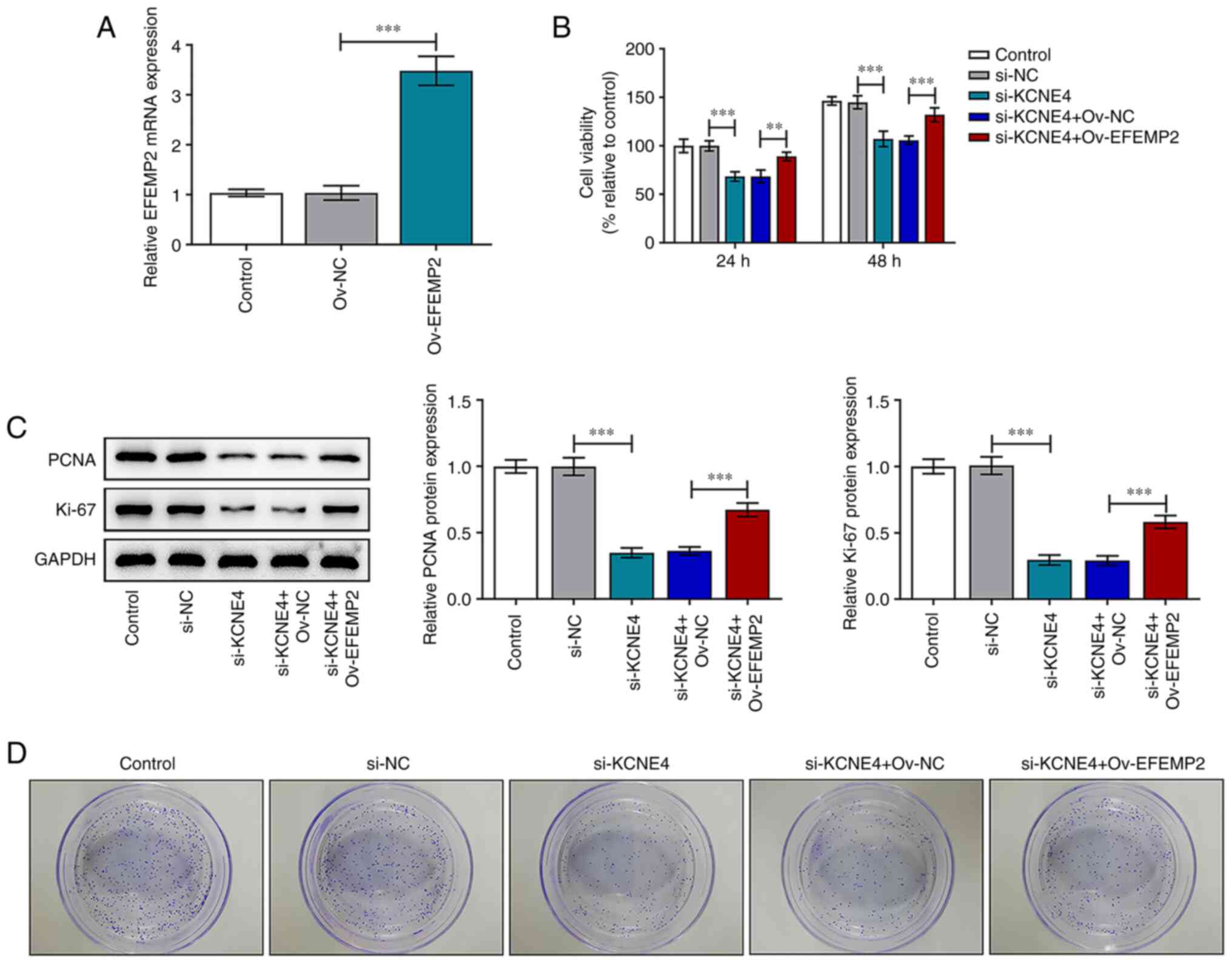 | Figure 6KCNE4 knockdown suppresses the
proliferative and colony-forming abilities of colon cancer cells
via the downregulation of EFEMP2. (A) HCT-116 cells were
transfected with Ov-EFEMP2 or Ov-NC and the transfection efficiency
was validated via the detection of the EFEMP2 mRNA level in the
HCT-116 cells using a reverse transcription-quantitative PCR assay.
HCT-116 cells were transfected with si-KCNE4 or co-transfected with
si-KCNE4 and Ov-EFEMP2 and (B) cell viability was detected using a
Cell Counting Kit-8 assay, (C) PCNA and Ki-67 expression levels
were detected via western blot analysis and (D) colony formation
was evaluated. **P<0.01, ***P<0.001.
KCNE4, potassium voltage-gated channel subfamily E member 4;
EFEMP2, EGF containing fibulin extracellular matrix protein 2;
Control, untransfected cells; si-KCNE4, small interfering RNA
targeting KCNE4; si-NC, small interfering RNA negative control;
Ov-EFEMP2, vector overexpressing EFEMP2; Ov-NC, empty vector
negative control; PCNA, proliferating cell nuclear antigen. |
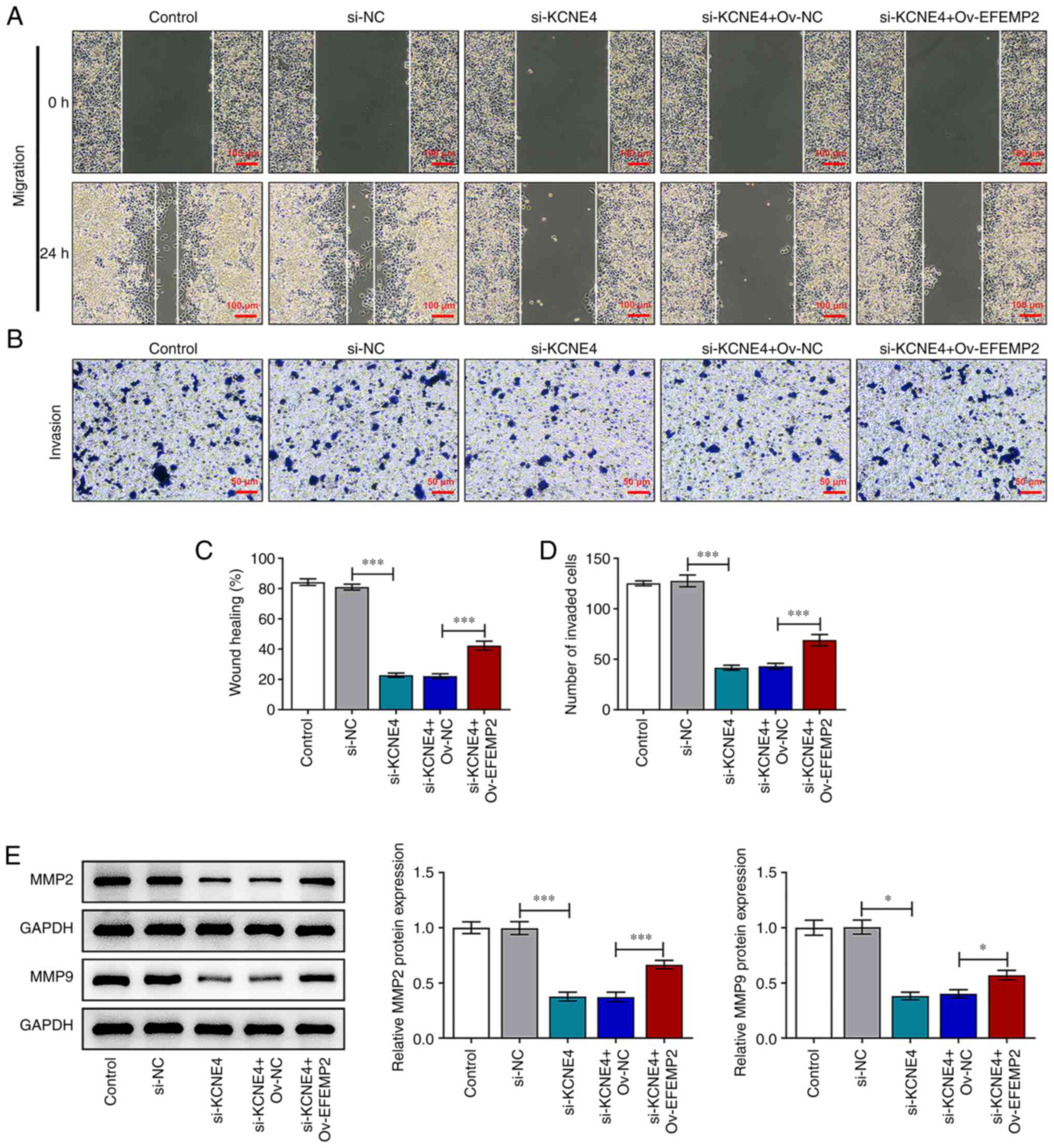
KCNE4 knockdown weakens the migratory
and invasive capacities of colon cancer cells by downregulating
EFEMP2
The downregulation of KCNE4 repressed the migratory
and invasive abilities of colon cancer cells, and these changes
were reversed by EFEMP2 overexpression (Fig. 7A-D). Furthermore, increased MMP2
and MMP9 expression levels in the HCT-116 cells transfected with
si-KCNE4 and Ov-EFEMP2 compared with those transfected with
si-KCNE4 and Ov-NC also indicated that upregulation of EFEMP2
abolished the suppressive effects of KCNE4 knockdown on the
migration and invasion of colon cancer cells (Fig. 7E). In conclusion, these results
suggest that KCNE4 knockdown weakens the migratory and invasive
capacities of colon cancer cells via the downregulation of EFEMP2
expression.
Discussion
Colon cancer is a malignant tumor of the
gastrointestinal tract, which mostly occurs in the rectum and colon
(1,2). Tumor metastasis results in the poor
outcome of patients in the advanced stages of colon cancer
(4). Although several biomarkers
associated with the development of colon cancer have been
documented, further animal experiments and even long-term clinical
trials are required for the application of conventional biomarkers
in the diagnosis and treatment of colon cancer. Therefore, it is
urgently important to develop more effective biomarkers that are
highly associated with colon cancer metastasis, to expand the
screening range of clinical gene therapy for colon cancer.
Ion channels allow the passage of ions from one side
of the membrane to the other (20). Ion channels play critical roles in
the generation and facilitation of the transmission of information
within various tissues and cells (21). There is evidence to suggest that
the abnormal modulation of ion channels and ancillary proteins
could lead to serious diseases, including multiple cancers
(22,23). Pivotal roles have been indicated
for a variety of ion channels, including KCNE, in the development
of several tumors. KCNE4 is the largest of the KCNE subunits and
can regulate a variety of Kv channels (24). Furthermore, Wu et al
(12) reported that the high
expression of KCNE4 in CRC predicts a poor prognosis. In the
current research, it was demonstrated that KCNE4 was highly
expressed in colon cancer cells, and the knockdown of KCNE4
suppressed the proliferative, colony-forming, migratory and
invasive capacities of colon cancer cells.
The positive correlation of the gene EFEMP2 with
KCNE4 in colon cancer was identified using the LinkedOmics
database. In a previous study, Zuo et al (25) discovered that EFEMP2 enhanced the
invasive ability of breast cancer cells. In addition, Wang et
al (26) confirmed that EFEMP2
expression is elevated in gliomas, and the silencing of EFEMP2
inhibits glioma cell proliferation and invasion. Furthermore, Yuen
et al (27) reported that
EFEMP2 is closely associated with colon cancer progression.
Importantly, EFEMP2 has been exposed to function as a modulator in
the biological behaviors of colon cancer cells (16). In the present study, the Co-IP
assay verified that KCNE4 interacted with and bound to EFEMP2, and
the suppressive effects of KCNE4 knockdown on the proliferative,
colony-forming, migratory and invasive abilities of colon cancer
cells were abolished by EFEMP2 overexpression.
In conclusion, the downregulation of KCNE4 inhibited
the proliferation, colony formation, migration and invasion of
colon cancer cells via the suppression of EFEMP2 expression. These
results indicated that KCNE4 and EFEMP2 serve as oncogenes,
contributing to the malignant progression of colon cancer. These
findings are beneficial to the development of a promising approach
for the treatment of colon cancer. However, in vivo analysis
should be conducted to support the findings of this study. In
addition, the in-depth mechanism underlying the effects of
KCNE4/EFEMP2 in tumor promotion requires further investigation to
elucidate the predictive values of KCNE4/EFEMP2 in the treatment of
colon cancer.
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
YX, DW and GZ searched the literature, designed the
study and performed the experiments. YX and DW collected the data,
interpreted the results and wrote the manuscript. GZ critically
revised the manuscript. YX and GZ confirm the authenticity of all
the raw data. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Martin MJ: Current stage-specific
chemotherapeutic options in colon cancer. Expert Rev Anticancer
Ther. 5:695–704. 2005.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Utomo B, Alvarez C and Baldonedo RF:
Management of colorectal cancer patients undergoing a colonic
stenting: A multidisciplinary team approach. Gastroenterol Nurs.
40:342–349. 2017.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global cancer statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249.
2021.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Willaert W, Mareel M, Van De Putte D, Van
Nieuwenhove Y, Pattyn P and Ceelen W: Lymphatic spread, nodal count
and the extent of lymphadenectomy in cancer of the colon. Cancer
Treat Rev. 40:405–413. 2014.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Zhang Z, Jia H, Wang Y, Du B and Zhong J:
Association of MACC1 expression with lymphatic metastasis in
colorectal cancer: A nested case-control study. PLoS One.
16(e0255489)2021.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Ye X, Brabletz T, Kang Y, Longmore GD,
Nieto MA, Stanger BZ, Yang J and Weinberg RA: Upholding a role for
EMT in breast cancer metastasis. Nature. 547:E1–E3. 2017.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Iorio J, Duranti C, Lottini T, Lastraioli
E, Bagni G, Becchetti A and Arcangeli A: KV11.1
potassium channel and the Na+/H+ antiporter
NHE1 K pH in colorectal cancer cells. Front Pharmacol.
11(848)2020.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Hou X, Tang L, Li X, Xiong F, Mo Y, Jiang
X, Deng X, Peng M, Wu P, Zhao M, et al: Potassium channel protein
KCNK6 promotes breast cancer cell proliferation, invasion, and
migration. Front Cell Dev Biol. 9(616784)2021.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Abbott GW: KCNE4 and KCNE5: K(+) channel
regulation and cardiac arrhythmogenesis. Gene. 593:249–260.
2016.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Yu H, Lin Z, Mattmann ME, Zou B,
Terrenoire C, Zhang H, Wu M, McManus OB, Kass RS, Lindsley CW, et
al: Dynamic subunit stoichiometry confers a progressive continuum
of pharmacological sensitivity by KCNQ potassium channels. Proc
Natl Acad Sci USA. 110:8732–8737. 2013.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Biasiotta A, D'Arcangelo D, Passarelli F,
Nicodemi EM and Facchiano A: Ion channels expression and function
are strongly modified in solid tumors and vascular malformations. J
Transl Med. 14(285)2016.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Wu B, Tao L, Yang D, Li W, Xu H and He Q:
Development of an immune infiltration-related eight-gene prognostic
signature in colorectal cancer microenvironment. Biomed Res Int.
2020(2719739)2020.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Song L, Li XX, Liu XY, Wang Z, Yu Y, Shi
M, Jiang B and He XP: EFEMP2 suppresses the invasion of lung cancer
cells by inhibiting epithelial-mesenchymal transition (EMT) and
down-regulating MMPs. Onco Targets Ther. 13:1375–1396.
2020.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Zhang D, Wang S, Chen J, Liu H, Lu J,
Jiang H, Huang A and Chen Y: Fibulin-4 promotes osteosarcoma
invasion and metastasis by inducing epithelial to mesenchymal
transition via the PI3K/Akt/mTOR pathway. Int J Oncol.
50:1513–1530. 2017.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Chen J, Liu Z, Fang S, Fang R, Liu X, Zhao
Y, Li X, Huang L and Zhang J: Fibulin-4 is associated with tumor
progression and a poor prognosis in ovarian carcinomas. BMC Cancer.
15(91)2015.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Zhao J, Xu J, Zhao J and Zhang R: EFEMP2
promotes colon cancer cell invasion and growth through the ERK1/2
signaling pathway. Int J Clin Exp Pathol. 12:851–856.
2019.PubMed/NCBI
|
|
17
|
Yao L, Lao W, Zhang Y, Tang X, Hu X, He C,
Hu X and Xu LX: Identification of EFEMP2 as a serum biomarker for
the early detection of colorectal cancer with lectin affinity
capture assisted secretome analysis of cultured fresh tissues. J
Proteome Res. 11:3281–3294. 2012.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Phillips PA, McCarroll JA, Park S, Wu MJ,
Pirola R, Korsten M, Wilson JS and Apte MV: Rat pancreatic stellate
cells secrete matrix metalloproteinases: implications for
extracellular matrix turnover. Gut. 52:275–282. 2003.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Siwy Z, Heins E, Harrell CC, Kohli P and
Martin CR: Conical-nanotube ion-current rectifiers: the role of
surface charge. J Am Chem Soc. 126:10850–10851. 2004.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Shepherd VA, Beilby MJ and Shimmen T:
Mechanosensory ion channels in charophyte cells: the response to
touch and salinity stress. Eur Biophys J. 31:341–355.
2002.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Van den Eynde C, De Clercq K and Vriens J:
Transient receptor potential channels in the
epithelial-to-mesenchymal transition. Int J Mol Sci.
22(8188)2021.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Kärki T and Tojkander S: TRPV Protein
family-from mechanosensing to cancer invasion. Biomolecules.
11(1019)2021.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Hu Z, Wei W, Zhou L, Chen M and Abbott GW:
Kcne4 deletion sex-specifically predisposes to cardiac arrhythmia
via testosterone-dependent impairment of RISK/SAFE pathway
induction in aged mice. Sci Rep. 8(8258)2018.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Zuo T, Shan J, Liu Y, Xie R, Yu X and Wu
C: EFEMP2 mediates GALNT14-dependent breast cancer cell invasion.
Transl Oncol. 11:346–352. 2018.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Wang L, Chen Q, Chen Z, Tian D, Xu H, Cai
Q, Liu B and Deng G: EFEMP2 is upregulated in gliomas and promotes
glioma cell proliferation and invasion. Int J Clin Exp Pathol.
8:10385–10393. 2015.PubMed/NCBI
|
|
27
|
Yuen HF, McCrudden CM, Huang YH, Tham JM,
Zhang X, Zeng Q, Zhang SD and Hong W: TAZ expression as a
prognostic indicator in colorectal cancer. PLoS One.
8(e54211)2013.PubMed/NCBI View Article : Google Scholar
|