Introduction
With the rapid development of medical technology,
some hematological malignancies can be cured by hematopoietic stem
cell transplantation (HSCT). To effectively avoid graft-versus-host
disease (GVHD), the calcineurin inhibitors, ciclosporin or
tacrolimus, are often used as long-term preventive drugs following
transplantation (1). To destroy
the normal immunity is inevitable, inhibiting the proliferation of
T-cells, the maturation of dendritic cells and the activation of
neutrophils (2,3). Therefore, the use of
immunosuppressive agents following haploidentical HSCT (haplo-HSCT)
leads to patients becoming susceptible to bacterial and fungal
infections, which is the cause of increased non-relapse mortality
in patients (4). Invasive fungal
infection has caused serious fatal damage to patients who have
undergone HSCT, including Aspergillus spp. and
Pneumocystis (5,6). In addition to these common pathogens,
patients are also vulnerable to some uncommon microorganisms
following transplantation. Infections with rare pathogenic
microorganisms are mainly described in case reports, and there is a
lack of systematic research, including the analysis of conditions,
diagnosis, treatment and traceability (7-10).
As a result, the accurate identification of pathogens and the
optimal management of uncommon disseminated fungal infections is
warranted.
Some species of the genus Microascus are
known to be opportunistic pathogens, mainly causing superficial
tissue infections, and they represent some of the principal causes
of non-dermatophytic onychomycoses. As to Microascus, the
morphological and molecular identification of the etiological agent
has not yet been fully established. Microascus cirrosus
(M. cirrosus) species account for only 2.1% of the genus and
are rare isolates of clinical origin, whereas they induce the
majority of human infections among the Microascus genus
(11). To the best of the authors'
knowledge, M. cirrosus Curzi was reported to cause the first
disseminated infection in a pediatric bone marrow transplant
recipient in 1994(12). Currently,
there are a few species of M. cirrosus which have been
publicly reported to cause human cutaneous and pulmonary infection
(10,12-16).
The majority of these were determined by cultivation and
morphological recognition. The morphological identification of the
etiological agent has not been confirmed at the molecular level,
and the real prevalence of M. cirrosus species in clinical
infection remains unknown.
Over the past decades, the development of molecular
diagnostic technology has greatly improved the efficiency of
pathogen detection in human infections. Notably, metagenomic
next-generation sequencing (mNGS) has been widely used to diagnose
lung infections in immunocompromised adults (17-21).
In the present study, extensive conventional microbiologic testing
and mNGS failed to identify Microascus species, challenging
the accurate diagnosis and individualized therapy of pulmonary
fungal infection in the patient. In the present study, a
retrospective analysis of the detailed process assisted in the
identification and optimal management of fatal lung infection
caused by Microascus species. Complete laboratory
identification methods from morphology to gene sequencing were
established to identify the invasive fungal infection by
Microascus in immunosuppressive patients following
haplo-HSCT, including growth and morphological features, extensive
drug resistance, the first whole genome information, genetic
evolution, protein fingerprint and pathological characteristics.
This provides necessary etiological information for investigating
the real prevalence of Microascus infection in
immunosuppressive patients, as Microascus is widely
distributed worldwide (22,23),
and China has the largest HSCT population (24).
Materials and methods
Fungal isolation
Fiberoptic bronchoscopy was performed on the patient
at Dushu Lake Hospital Affiliated to Soochow University, Suzhou,
China and bronchoalveolar lavage fluid (BALF) was collected from
the infected area (left upper bronchus) for laboratory examination,
including bacterial culture, 1,3-β-D-glucan (GM) test and mNGS on
March 10, 2021. The BALF was inoculated according to the method of
bacterial culture, and cultured on a Columbia blood agar plate
(Autobio Diagnostics Co., Ltd.) and chocolate agar plate (Autobio
Diagnostics Co., Ltd.) at 35˚C and 5% CO2. Dozens of
white filamentous-like fungal colonies were formed on the plate and
one pure fungal colony was transferred to Sabouraud agar (Autobio
Diagnostics Co., Ltd.) for fungal culture at 28˚C and 35˚C.
Morphological and physiological
assessments
Due to the lack of the whole genome of M.
cirrosus in the National Center for Biotechnology Information
(NCBI) database, mNGS testing (BGI) did not include its genetic
information. Due to the lack of protein fingerprint information of
the fungi, rapid identification methods, such as matrix-assisted
laser desorption/ionization (MALDI)-time-of-flight (TOF)-mass
spectrometry (MS; IVD MALDI Biotyper System; Bruker Daltonics;
Bruker Corporation) could not identify the species. The fungus was
inoculated on a chocolate agar plate, Columbia blood plate,
nutrient agar, Sabouraud agar and Luria-Bertani (LB) broth [Sangon
Biotech (Shanghai) Co., Ltd.] respectively, to observe the growth
status of the colony every day. The conidia and septate hyphae were
observed in all visual fields under an Olympus CX33 light
microscope (Olympus Corporation) with lactophenol cotton blue (BaSO
Diagnostics Inc.) staining.
Resistance to common antifungal
drugs
Broth dilution antifungal susceptibility testing was
performed according to the Clinical and Laboratory Standards
Institute (CLSI) M38-A2(25). In
total, four types of antifungal agents were tested, including
amphotericin B (CAS: 1397-89-3; Bio Basic, Inc.), caspofungin
acetate (CAS: 179463-17-3; Beijing Jin Ming Biotechnology Co.,
Ltd.), fluconazole (CAS: 86386-73-4; Rhawn Reagent) and
fluorocytosine (CAS: 2022-85-7; Rhawn Reagent). Different reagent
grades were tested in the following concentrations according the
manufacturers' instructions: Amphotericin B 0.03-16 µg/ml,
caspofungin 0.03-16 µg/ml, fluconazole 0.12-64 µg/ml,
fluorocytosine 0.12-64 µg/ml. The susceptibility of this fungus to
each drug was determined.
Multi-site sequence analysis
To accurately identify the fungus, four nuclear DNA
regions were amplified and sequenced. Including large subunit
ribosomal RNA gene (LSU) and internal transcribed spacer (ITS)
regions of the rRNA operon, fragments of the translation elongation
factor 1α (EF-1α) and β-tubulin genes (TUB), following the criteria
described in the study by Sandoval et al (26). DNA extraction was conducted using
the Ezup Column Fungi Genomic DNA Purification kit [Sangon Biotech
(Shanghai) Co., Ltd.] following the manufacture's protocols. Next,
2X SanTaq PCR Mix [Sangon Biotech (Shanghai) Co., Ltd.], the
aforementioned primers, fungal nucleic acid and pure water were
added to the PCR reaction tube to yield a total volume of 50 µl,
which was amplified on a SLAN-96P (Shanghai Hongshi Medical). The
reactions were performed according to the following conditions: 1
cycle at 95˚C for 5 min, followed by 40 cycles at 95˚C for 5 sec,
52-58˚C for 30 sec, 72˚C for 30 sec. The amplification was carried
out with the primers, as previously described by Brasch et
al (27). The PCR product was
entrusted to Sangon Biotech (Shanghai) Co., Ltd. for Sanger
sequencing and the sequencing instrument used was the Applied
Biosystems 3730XL (Applied Biosystems; Thermo Fisher Scientific,
Inc.). Consensus sequences obtained for each locus were aligned
with sequences of Microascus species retrieved from GenBank
(http://www.ncbi.nlm.nih.gov/genbank/), using the
ClustalW algorithm under MEGA-X v10.0.4 software (28,29).
Phylogenetic reconstructions by maximum likelihood (ML) approaches
were performed using MEGA-X v10.0.4. The fungal nucleic acid was
amplified by PCR with the primers (Table I), as previously described by
Brasch et al (26,27). and four products were obtained
(LSU568bp, ITS618bp, EF-1α898bp and TUB523bp).
 | Table IPrimer list. |
Table I
Primer list.
| Gene | Primer sequence
(5'-3') |
|---|
| NL1-F |
GCATATCAATAGCGGAGGAAAAG |
| NL4-R |
GGTCCGTGTTTCAAGACGG |
| ITS5-F |
GGAAGTAAAAGTCGTAACAAGG |
| ITS4-R |
TCCTCCGCTTATTGATATGC |
| EF1T-F |
ATGGGTAAGGARGACAAGAC |
| 1567R-R |
ACHGTRCCRATACCACCSATCTT |
| Bt2a-F |
GGTAACCAAATCGGTGCTGCTTTC |
| Bt2b-R |
ACCCTCAGTGTAGTGACCCTTGGC |
Genomic DNA extraction and whole
genome sequencing
The fungus was cultured in LB broth for ~3 days,
forming a white, pom-pom-shaped fungus. The fungal mass was
collected by centrifugation (4˚C and 10,000 x g for 10 min) and
washing with saline. The cell wall was destroyed by grinding in
liquid nitrogen. The Ezup Column Fungi Genomic DNA Purification kit
[Sangon Biotech (Shanghai) Co., Ltd.] was used to extract genomic
DNA according to the manufacture's protocols for library
construction and next-generation sequencing.
A DNA library with a 500 bp insert size was
constructed and sequenced in Illumina's HiSeq platform (Illumina,
Inc.) with a pair-end 150 bp sequencing strategy. The sequence
reads were assembled using SPAdes v3.5.0(30). The Protein Coding Genes and rRNA of
M. cirrosus SZ 2021 were then predicted by Prokka (31). The toxicity factors of M.
cirrosus SZ 2021 were analyzed by PHIB-BLAST (http://phi-blast.phi-base.org/). Genome
sequencing and analysis were performed by Sangon Biotech (Shanghai)
Co., Ltd. As there is currently no whole M. cirrosus genome
available for reference in the NCBI database, the assembled
sequence of M. cirrosus SZ 2021 was uploaded onto the NCBI
public database for research. The mNGS sequence of the patient's
sample was aligned with the genome sequence of M. cirrosus
SZ 2021 using Burrows-Wheeler Aligner software (bwa-0.7.17)
(32).
Identification by MALDI-TOF-MS
For sample preparation, briefly, after the fungi
were cultured in Yeast Malt Broth, at 35˚C for 2 days; the samples
were transferred to Eppendorf (EP) tubes containing 1 ml of
high-performance liquid chromatography (HPLC) water, washed and
pelleted following centrifugation at 25˚C and 8,000 x g for 10 min.
The pellet collected was dissolved in 300 µl HPLC water and washed
twice using HPLC water. Subsequently, 900 µl ethanol were added and
removed following centrifugation at 25˚C at 10,000 x g for 10 min
and air-drying. The sample was then transferred to a grinder for
grinding and 100 µl of 70% formic acid was injected into the
grinder for ~5 min. The homogenate was then transferred to a clean
1.5-ml EP tube, and an equal amount of acetonitrile was added
followed by centrifugation (25˚C and 10,000 x g for 10 min) again.
A total of 1 µl of the supernatant was pipetted onto the target,
overlaid with 1 µl α-cyano-4-hydroxycinnamic acid matrix and
analyzed using MALDI-TOF-MS (Bruker Daltonics; Bruker Corporation).
A MBT Compass Explorer (Bruker Daltonics; Bruker Corporation) was
used for the analysis of the results.
Murine models of pulmonary infection
caused by M. cirrosus
Male C57BL/6 mice (6 weeks old; weight 18±1 g; n=12)
were purchased from Xinchen Biotechnology Co. Ltd. The animals were
housed in controlled temperature (24±1˚C) and humidity (40-60%)
under a natural 12 h/12 h light/dark cycle, with standard chow and
water provided ad libitum. All animal experiments were
carried out in accordance with the National Institute of Health
guide for the care and use of laboratory animals (33).
The mice were randomly divided into four groups,
with three mice in each group. One group served as a healthy
control, while another group was directly infected with M.
cirrosus. The remaining two groups received 1 mg/kg tacrolimus
(FK506) intraperitoneally every day, as previously described
(34); of these groups, one was
used to establish a pulmonary infection model caused by M.
cirrosus. For infection, the mice were lightly anesthetized by
the inhalation of isoflurane (2% concentration, approximately 2-3
min) and immobilized in an upright position using rubber bands
attached to a cardboard for oropharyngeal aspiration. A blunt 24G
needle attached to a 1-ml syringe was advanced into the trachea to
deliver the indicated number of conidia (5x108) in a
volume of 0.05 ml saline. After 48 h, the mice were then painlessly
sacrificed by cervical dislocation. The absence of a heartbeat and
pupil dilation for 5 min was used to confirm mortality. The
experimental procedures were performed in accordance with the
conditions specified and approved by the Animal Experimentation
Ethics Committee of Dushu Lake Hospital affiliated to Soochow
University (Suzhou, China; approval no. 220138).
Analysis of M. cirrosus SZ
2021-challenged mice
The concentration of IL-6 in serum samples was
measured using enzyme linked immunosorbent assay (ELISA) according
to the manufacturer's instructions [cat nos. EK206/3-96;
Multisciences (Lianke) Biotech Co., Ltd.]. In colony forming unit
(CFU) assays, the half of the left lung tissue was harvested and
homogenized with glass beads on a Mini-Bead beater (KZ-II; Wuhan
Servicebio Technology Co., Ltd.), and serially diluted onto
chocolate agar plates in duplicate, and the CFU was determined
after 3 days. The remaining lung tissue was immersed in formalin
liquid for histological analysis. Paraffin-embedded sections (4 µm)
prepared from the lungs of these mice were stained with hematoxylin
and eosin (H&E) at room temperature for the evaluation of
airway inflammation. Specifically, after dewaxing using xylene, the
slices were treated with different alcohol concentrations (100,
100, 95 and 85%, for 1 min each), and then stained with hematoxylin
for 10 min after being washed with water. The slices were
differentiated with 1% hydrochloric acid alcohol for 5 sec, and
then different concentrations of alcohol (95 and 95%, for 10 sec
each; 100 and 100%, for 30 sec each) were used for dehydration
after washing with water. Following this, the slices were treated
with xylene for transparency and sealed with neutral gum. Olympus
BX43 light microscope (Olympus Corporation) was used to capture
images from all visual fields.
Statistical analysis
Data are expressed as the mean ± standard error of
the mean (SEM). The Student's t-test assuming equal variances was
performed to examine the differences between groups and
associations were investigated using regression analysis. P<0.05
was considered to indicate a statistically significant difference.
Data were analyzed using GraphPad prism 8 (Dotmatics).
Results
Isolation of M. cirrosus from the
patient
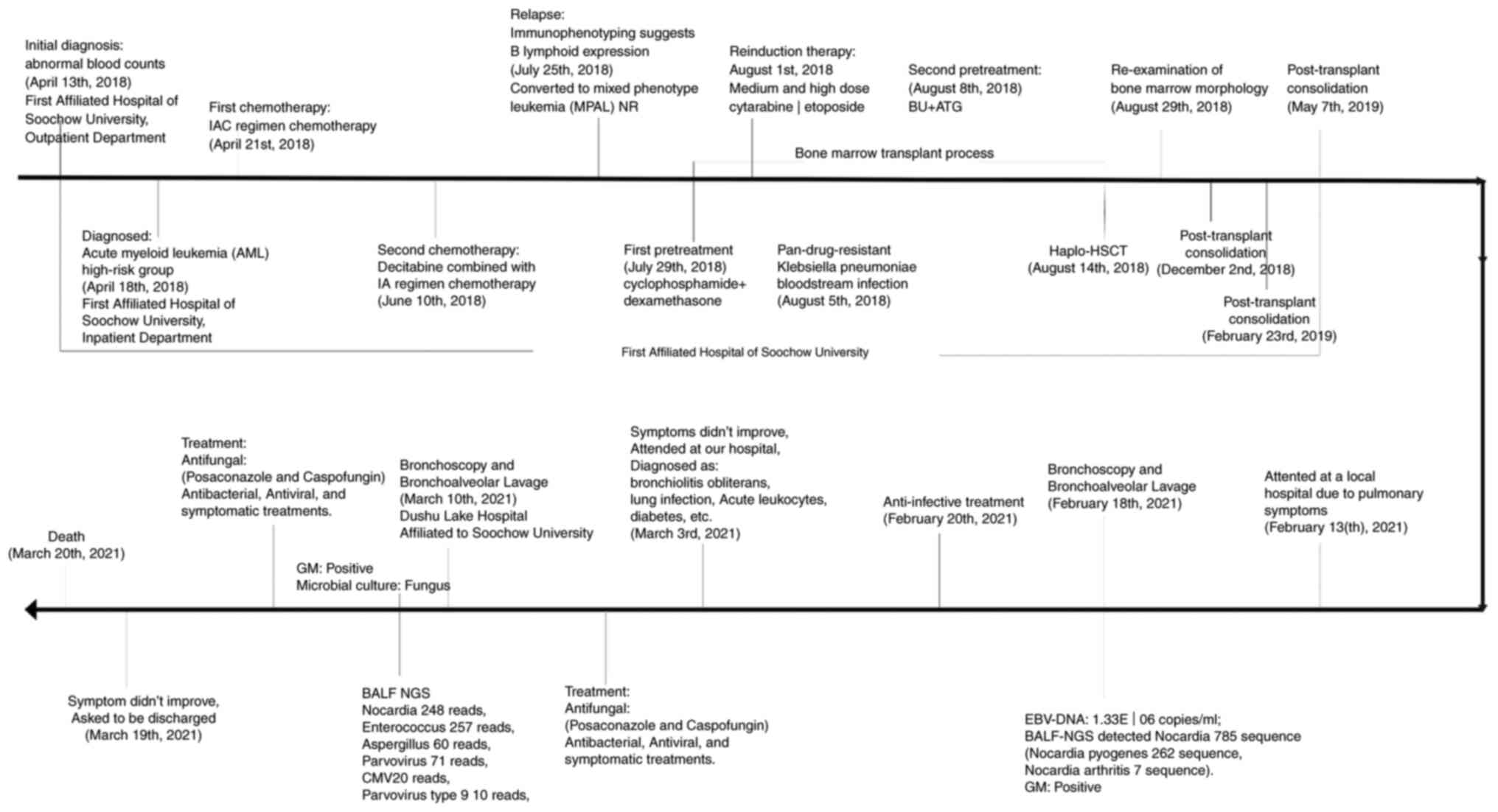
A 40-year-old male was diagnosed with acute myeloid
leukemia in a high-risk group on April 18, 2018 and underwent two
rounds of chemotherapy. On August 14, 2018, the patient received
haplo-HSCT, and the transplant was successfully reconstructed on
August 29, 2018. During this time, he continued immunosuppressive
therapy with anti-rejection agent (cyclosporine, 50 mg, twice a
day) for 3 years in order to prevent GVHD. The patient did not
relapse with any hematonosis. Therefore, he was admitted to a local
hospital due to aggravated pulmonary symptoms on February 13, 2021,
with self-reported ‘chest tightness and asthma over the past 6
months, which worsened half a month ago’. He was transferred to
Dushu Lake Hospital Affiliated to Soochow University on March 3,
2021 as his pulmonary infection symptoms did not improve following
anti-infection treatment at the local hospital. Following
admission, the doctor systematically inquired about the medical
history and performed relevant examinations. A chest computed
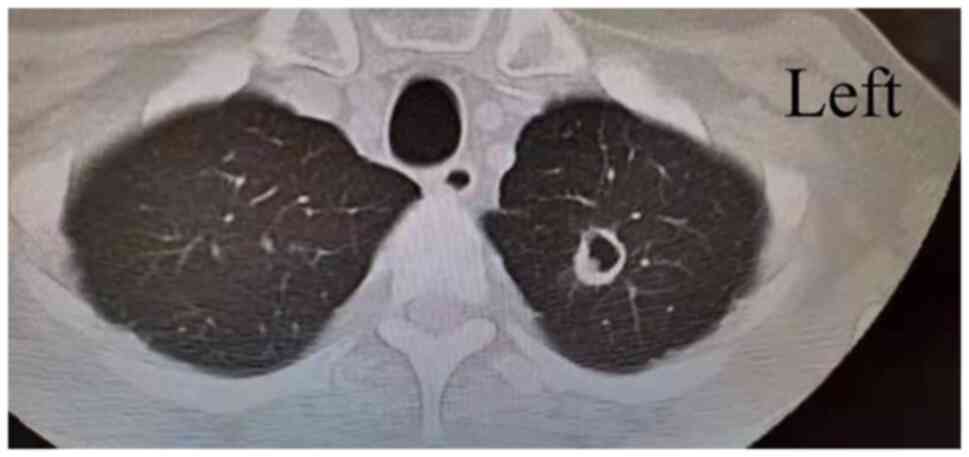
tomography (CT) examination presented one infectious cavity in the
upper lobe of the left lung (Fig.
1). In addition, the scattered small dots and patches in both
lungs were new foci. A number of routine laboratory examinations
also suggested infection. To identify the pathogens, a fiberoptic
bronchoscopy was performed to collect BALF from left upper bronchus
for culture, GM and mNGS on March 10, 2021. Given these findings,
posaconazole and caspofungin were used for antifungal therapy,
meropenem and co-sulfamethoxazole for antibacterial, ganciclovir
and ribavirin for antiviral. Antifungal drugs had been used for ~2
weeks, as clinical symptoms, CT examination and laboratory
examinations suggested fungal infection in the lung. Finally, all
these therapies failed to control this fatal pulmonary infection,
and the patient succumbed due to respiratory failure on March 20,
2021. The patient's disease progression is presented in Fig. 2. In combination with the patient's
medical history, symptoms and laboratory tests, the patient was
finally diagnosed with the following: i) Bronchiolitis obliterans;
ii) lung infection; iii) respiratory failure; iv) acute leukocytes;
v) status of hematopoietic stem cell transplantation; vi) chronic
GFHD; vii) diabetes; viii) electrolyte disorder; and ix)
hypoalbuminemia. The treatment of this patient was based on the
study by Williams (35). The
5-year survival rate of patients with bronchiolitis obliterans is
not high (36), and the mortality
of this patient may be a result of disease progression. M.
cirrosus, as an infectious factor, is likely to be one of the
critical factors that promoted the progression and deterioration of
the disease in this patient.
Results of laboratory tests for the
patient
Blood cell analysis suggested mild anemia
(hemoglobin, 106 g/l; reference range, 130-175 g/l) with a normal
white blood cell count (4.82x109/l; reference range,
3.5-9.5x109/l; neutrophils, 4.42x109/l;
reference range, 1.8-6.3x109/l; lymphocytes,
0.27x109/l; reference range, 1.1-3.2x109/l).
As to inflammatory markers, the level of procalcitonin was 0.18
ng/ml (reference range, <0.05 ng/ml) and that of hypersensitive
C-reactive protein (hs-CRP) was 59.33 mg/l (reference range, 0-6
mg/l); significantly increased. No significant abnormalities were
found in blood coagulation functions. Liver and kidney functions
exhibited mild abnormality, 9.03 mmol/l uric acid (reference range,
3.1-8 mmol/l) and 82 U/l alanine transaminase (reference range,
9-50 U/l). Sputum was repeatedly subjected to bacterial culture and
no pathogenic microorganisms were found. BALF was collected on
February 18, 2021. GM tests were positive (5.65; reference range,
<0.5) and mNGS presented Nocardia nova with 785 reads,
cytomegalovirus with 109 reads, Epstein-Barr virus with 26 reads in
this BALF. As symptoms worsened, BALF from the left upper bronchus
was collected on March 10, 2021 for mNGS. Nocardia nova (248
reads), Enterococcus (257 reads), Aspergillus (60
reads), parvovirus (71 reads) and cytomegalovirus (20 reads) were
detected by mNGS in the BALF. Some white velvety colonies grew in
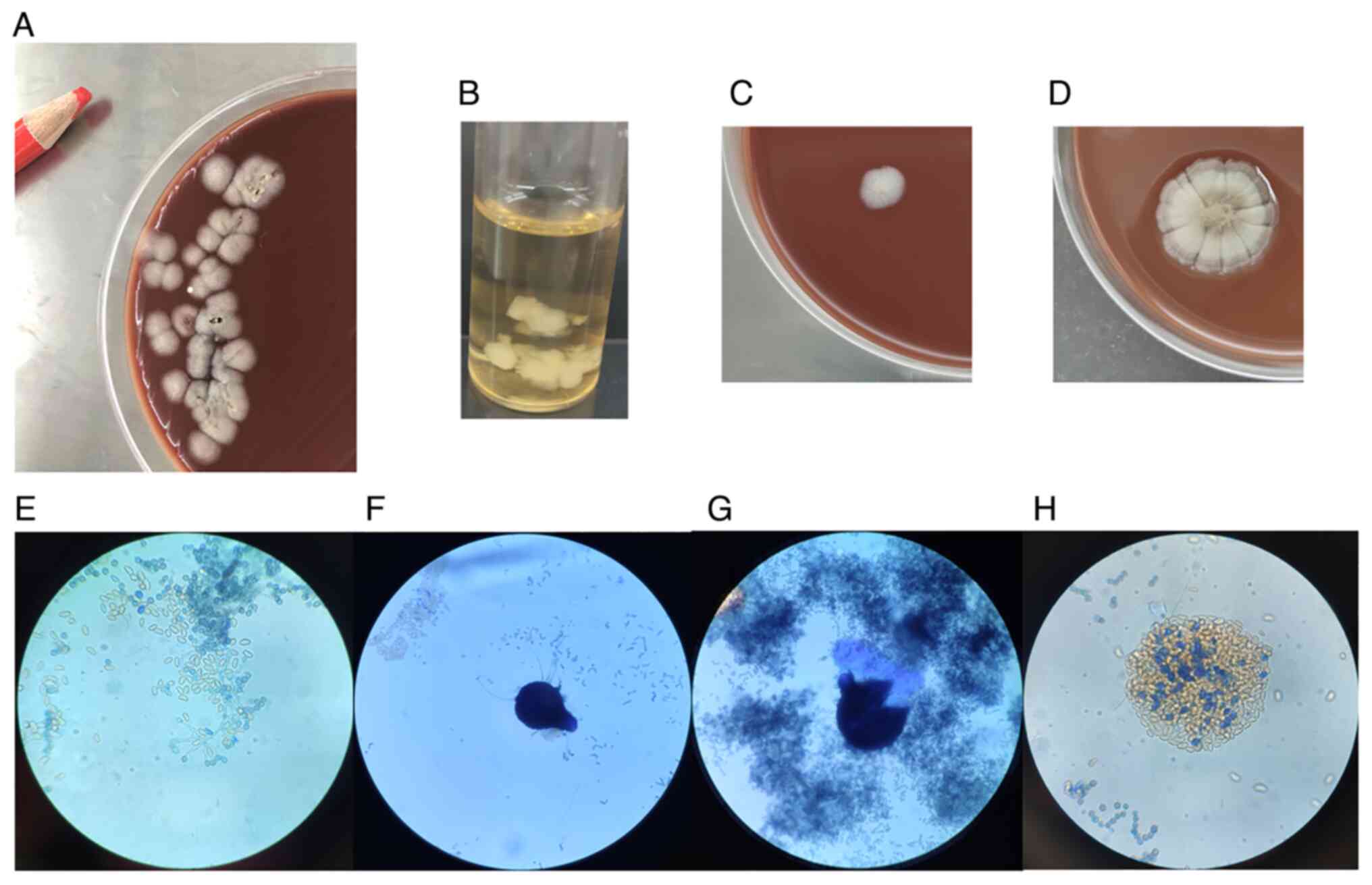
the BALF following 3 days of incubation (Fig. 3), with no evidence of
mycobacterial, or additional fungal pathogens. As the accurate
morphology of this fungus had not been established and M.
cirrosus was not listed in the MALDI-TOF-MS library, this
fungus was not correctly identified as a Microascus species
until 1 month later. Numerous results presented some fungal
infection in the lungs, including GM, mNGS and pure fungal
colonies.
Morphological and physiological
characteristics of the fungus
The fungus was detected from the BALF of the patient
by growing in both a Columbia blood plate and chocolate agar plate
for 2 days. The primary colonies formed on chocolate agar plate
were white and 3-4 mm in diameter (Fig. 3A). The fungus was then incubated on
a number of media to observe growing at different temperatures. The
fungus could grow on a Columbia blood plate, chocolate agar plate,
nutrient agar, Sabouraud agar and LB broth (Fig. 3B), and grew well at 35 and 28˚C. It
could grow at a more rapid rate and into a larger size on a
chocolate agar plate compared with Sabouraud agar. With prolonged
cultivation, it presented different conies. Overall, white colonies
3-4 mm in diameter could be observed on the chocolate agar plate
for 3 days (Fig. 3C). Colonies
>12 mm in diameter with folds around the periphery and a dark
pigment in the center could be observed on the culture medium for
~1 week (Fig. 3D). The fungus
could also grow at the bottom of a liquid medium. After ~1 week of
cultivation on chocolate agar, the ball-shaped, smooth conidia and
septate hyphae could be observed under the microscope by staining
with lactophenol cotton blue (Fig.
3E). For ~4 weeks, in the dark and brown part of the fungal
colony, ascoma and ascospores were observed as the forms of sexual
reproduction of the fungus. Microscopically, the ascoma were large
and spherical-shaped, containing ascospores which were half-moon
shaped (Fig. 3F-H). Based on the
presence of branched conidiophores bearing cylindrical anniellids
in brush-like groups and on the development of small black ascomata
(cleistothecia) after 4 weeks, it was morphologically identified as
Microascus spp. Compared with the morphological description
of M. cirrosus from seven clinical cases (Table II), the M. cirrosus
presented typical colonies and a microscopic morphology. Compared
with other filamentous fungi, such as Aspergillus,
Microascus species usually only have short chains of conidia
without apical cysts.
 | Table IISummary of patients with
Microascus cirrosus. |
Table II
Summary of patients with
Microascus cirrosus.
| First author/s,
year | Microascus
species | Infection Site | Type of transplant
or other factors | Age/Sex and
area | Antifungal agent or
therapeutic methods | Outcome | (Ref.) |
|---|
| de Vroey, 1992 | Microascus
cirrosus | Toenail | No | 56/F (Belgium
) | Imidazole
griseofulvin ketoconazole | Not cured | (13) |
| de Vroey, 1992 | Microascus
cirrosus | Toenail | No | 63/F (Italy) | Griseofulvin
miconazole | Not cured | (13) |
| Krisher, 1995 | Microascus
cirrosus | Lung | AML, Auto-BM | 12/M (America
) | AMB | No recurrence | (12) |
| Ustun, 2006 | Microascus
cirrosus | Lung | AML, BMT | 49/M (America
) | VOR, AMB and TER;
Surgery | Succumbed due to
AML relapse during infection | (14) |
| Miossec, 2011 | Microascus
cirrosus | Multiple
organs | SOT for cystic
fifibrosis | 36/M (France) | VOR and CAS | Succumbed (9
days) | (10) |
| Taton, 2018 | Microascus
cirrosus | Lung | Bilateral lung
transplant | 60/F (Belgium
) | VOR, CAS, TER and
AMB | Cure | (15) |
| Gao, 2018 | Microascus
cirrosus | Left ankle
skin | Systemic
corticosteroids for two months | 17/F (China) | itraconazole | Cure | (16) |
| Present case | Microascus
cirrosus SZ 2021 | Lung | MPAL,
haplo-HSCT | 40/M (China) | POS and CAS | Succumbed | Present case |
Fungal drug sensitivity
The minimum inhibitory concentrations of this fungus
against amphotericin B, caspofungin, fluconazole and fluorocytosine
were as follows: Amphotericin B ≥16 µg/ml, caspofungin ≥16 µg/ml,
fluconazole ≥64 µg/ml, fluorocytosine ≥64 µg/ml, according to
previously published criteria (25). The fungus are insensitive to the
antifungal drugs, suggesting that the treatment with common
antifungal drugs may be ineffective. This is generally consistent
with the results of in vitro drug susceptibility studies on
Microascus spp. by Sandoval-Denis et al (11) and Gao et al (16).
Multi-site sequence analysis
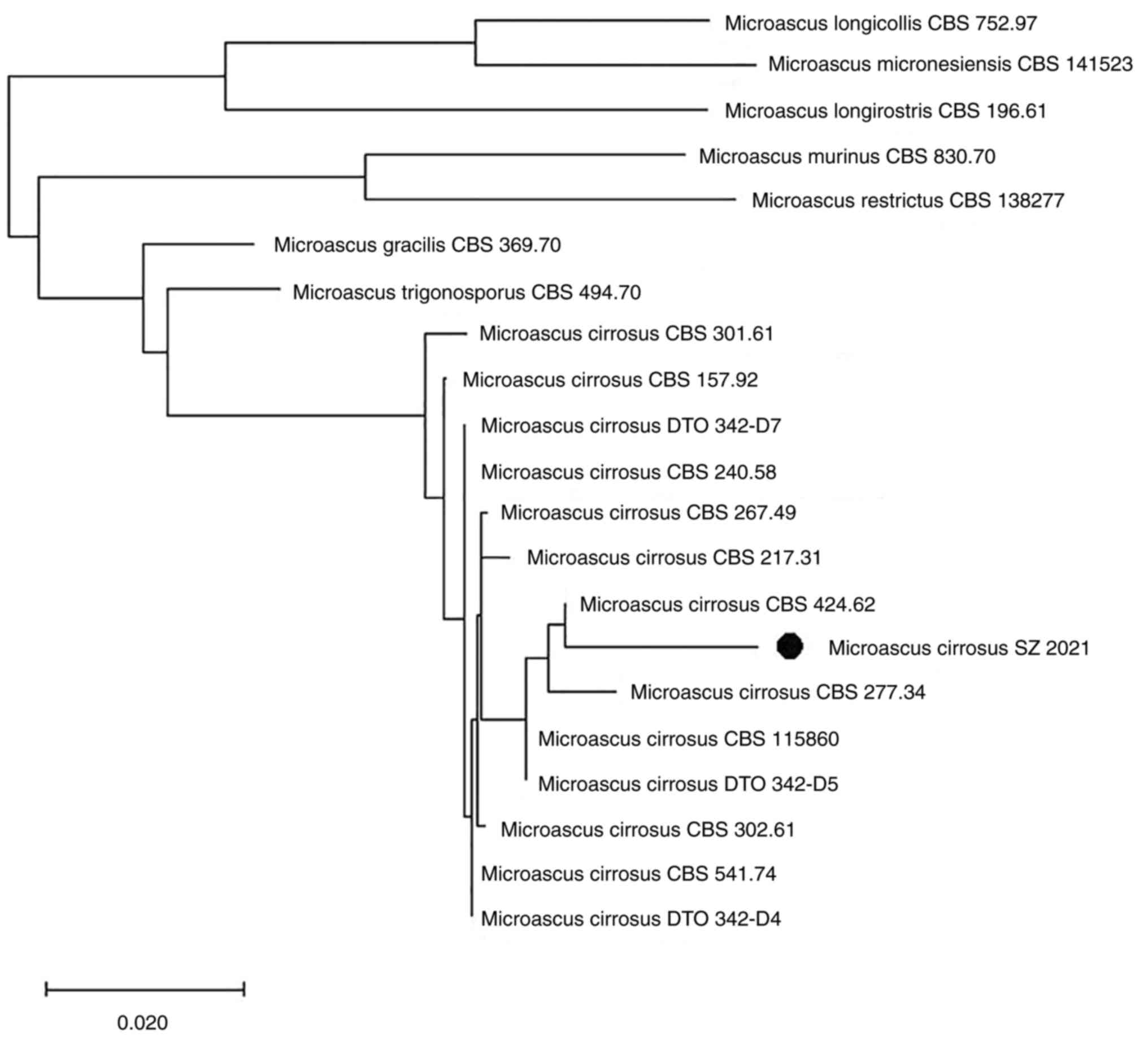
The analysis revealed that this fungus represented a
new genotype in the genus closely related to M. cirrosus
(Fig. 4). Based on its genetic
characteristics and on its distinct morphological features, it was
proposed as a novel genotype of M. cirrosus, termed M.
cirrosus SZ 2021.
Whole genome sequence of M. cirrosus
SZ 2021 by second-generation sequencing
By performing de novo sequencing on the case
fungus, its whole nucleic acid information was obtained. The total
number of bases after quality control were ~32.61 Mb and the GC
Bases Ratio was 53.98%. This second-generation sequencing project
was deposited in the NCBI under BioProject: PRJNA835605, BioSample:
SAMN28105390 and GenBank: JAMBUN000000000. The version described in
the present study is version JAMBUN010000000.
According to gene prediction, M. cirrosus SZ
2021 has a total of 9,939 coding genes, including the virulence
gene, FKS1. Through PHIB-BLAST, it was found that FKS1 of M.
cirrosus SZ 2021 was partially consistent with that of
Aspergillus fumigatus.
Re-analysis of sequences from BALF
detected by mNGS
As no whole genomic information of Microascus
species were available in public databases, no sequences of this
fungus had been reported by mNGS. The results of mNGS from BALF had
always recommended Nocardia and Aspergillus for
dozens of reads. The initial mNGS sequence from BALF was reanalyzed
by comparing the whole genome sequence of M. cirrosus SZ
2021. No Aspergillus gene sequence was reported in the
former BALF on February 18, 2021 in the local hospital, whereas
four reads from M. cirrosus could be detected by
retrospective bioinformatics analysis. In the BALF sample where the
M. cirrosus colonies were cultured (March 10, 2021, in Dushu
Lake Hospital Affiliated to Soochow University), more than a
thousand sequences (1,000 reads) could be aligned, including 660
no-human reads and unclassified reads. The initial BALF mNGS
sequence from the patients who underwent HSCT and developed
pulmonary infection, but failed to be cured in Dushu Lake Hospital
Affiliated to Soochow University were also reanalyzed. Different
numbers of sequences of M. cirrosus could be detected in
another two cases (a 23-year-old male was diagnosed with acute
myeloid leukemia and another 31-year-old male was diagnosed with
acute lymphoblastic leukemia; both had received HSCT), namely three
reads and 13 reads, respectively, with no verification of cultural
colonies, but favored by re-analysis of the mNGS sequences. In
conclusion, the sequences from BALF detected by mNGS demonstrated
that M. cirrosus could be detected correctly and rapidly by
mNGS, as there is correct genomic information in the database.
Distinguished features of M. cirrosus
detected using MALDI-TOF-MS
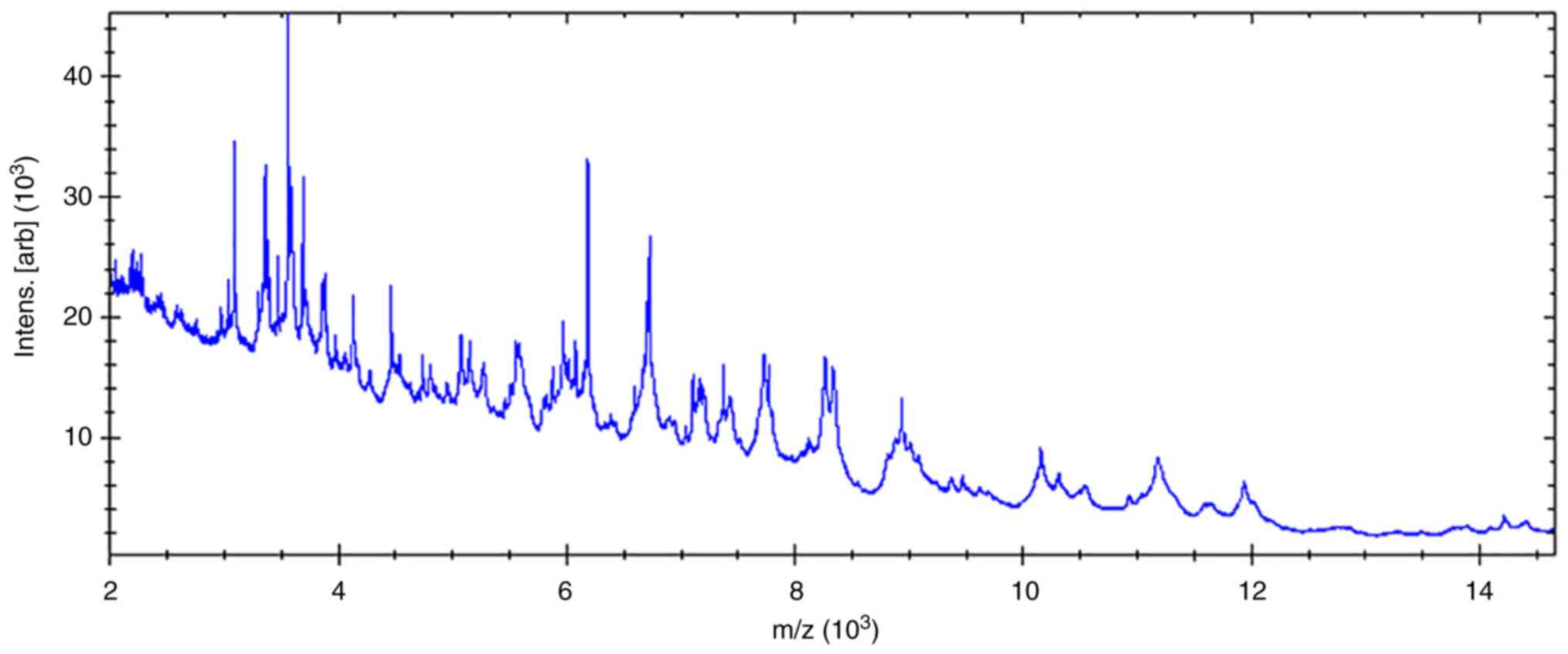
The protein fingerprint of M. cirrosus SZ
2021 was obtained by MALDI-TOF-MS (Fig. 5); however, the strain name was not
identified, probably as the MS identification database did not
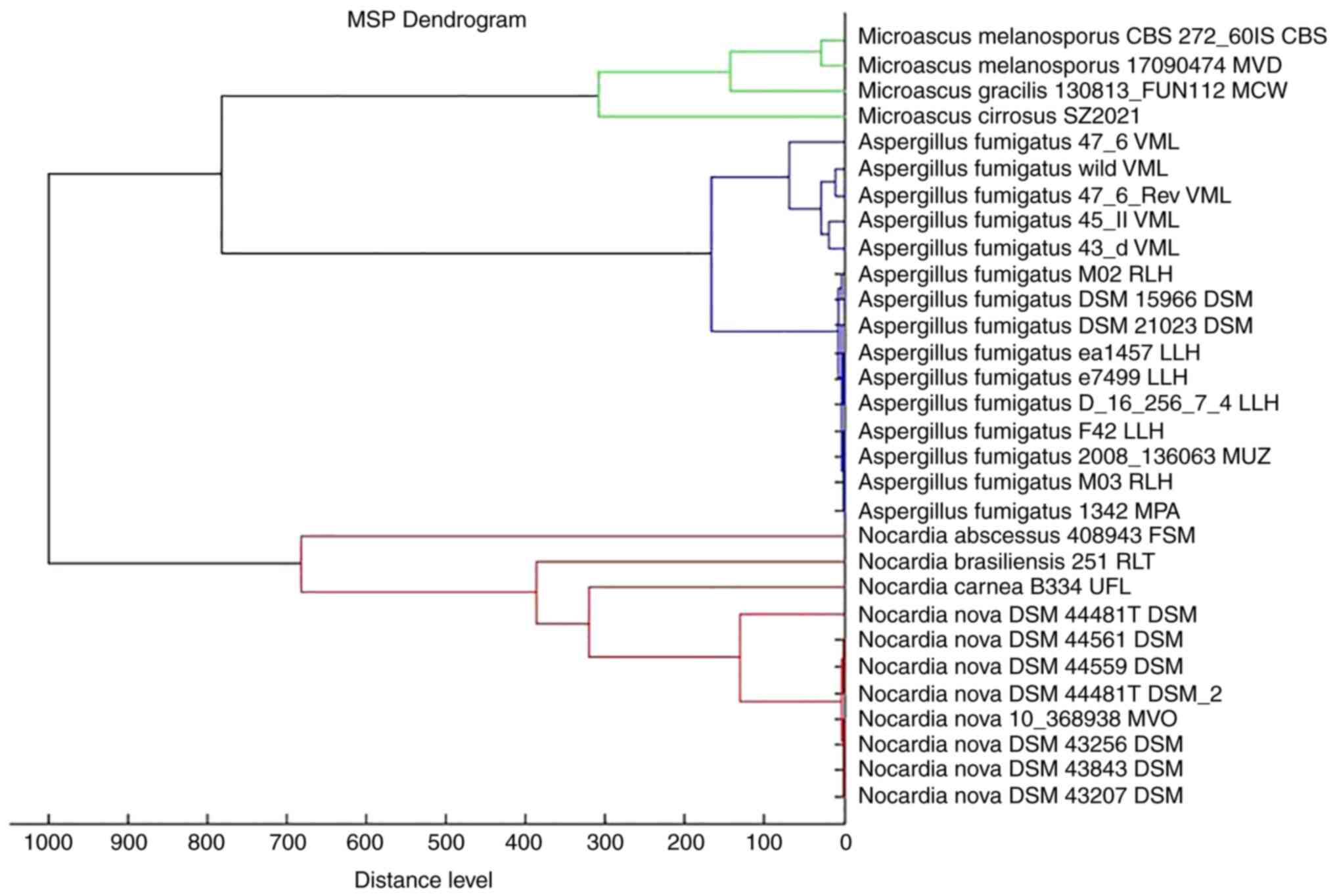
contain strain information. Subsequently, a main spectrum profile
dendrogram was constructed by combining M. cirrosus SZ 2021
with other Microascus spp., Aspergillus fumigatus and
Nocardia nova. It was found that M. cirrosus SZ 2021
was relatively close to Microascus gracilis, classified as a
different cluster from Aspergillus fumigatus and Nocardia
nova (Fig. 6).
Results of animal models of pulmonary
infection by M. cirrosus SZ 2021
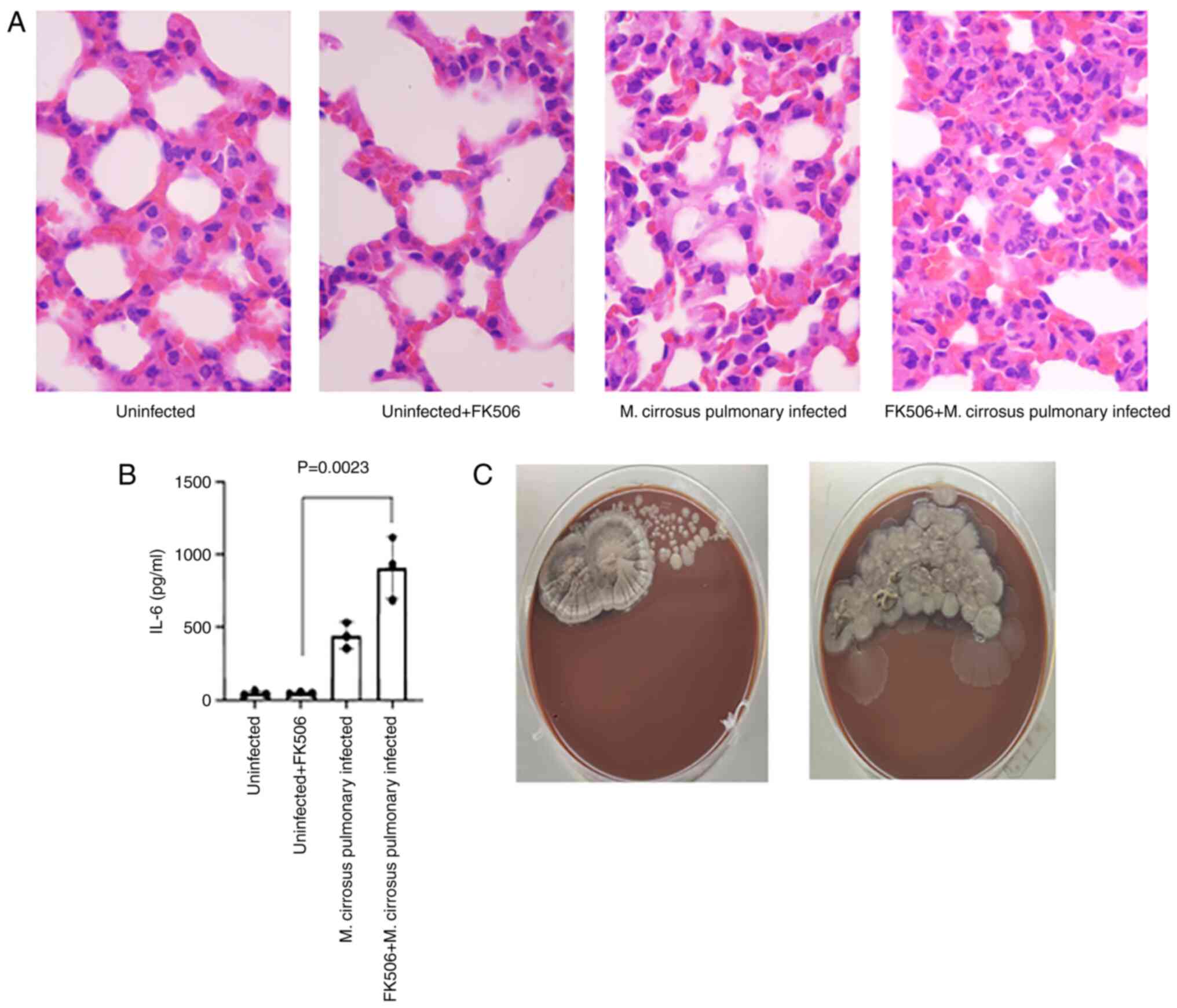
Prior to sacrifice, one mouse died following
infection by M. cirrosus SZ 2021 in the immunosuppressed
group pre-treated with FK506. The H&E-stained sections of the
lung tissue also revealed a greater amount of inflammatory cell
infiltration in M. cirrosus-infected mice from the
immunosuppressed group (Fig. 7A).
The levels of IL-6, as a prognostic biomarker, which can detect an
immediate response to infection compared with hs-CRP and
procalcitonin (37), were also
examined. Following infection with M. cirrosus SZ 2021, it
was observed that the IL-6 level was significantly increase in
mouse serum (P<0.05), particularly in mice pre-treated with
FK506 (Fig. 7B). The CFU assays
revealed no M. cirrosus growth, with the exception of two
mice from the immunosuppressed group. The mouse with the highest
M. cirrosus colony count was the one that died prior to
sacrifice (Fig. 7C).
Discussion
In the present study, the discovery of the real
pathogenic fungus of M. cirrosus posed a challenge, due to
the limited knowledge available on the Microascus genus. No
Microascus species were contained in the clinical database,
although mNGS is used extensively in China. Only one of the
repeated culture tests of sputum and BALF presented some pure
fungal colonies and, furthermore, they were not correctly
identified as M. cirrosus due to limited information. Thus,
their extensive drug resistance was not detected and recognized in
time and effective therapy was not administered; the patient
succumbed due to fatal lung infection, even though antifungal drugs
had been used continuously according to the repeated hints of
fungal etiology by GM tests. Indeed, through retrospective
reanalysis of the original sequencing of the first BALF sample by
mNGS, DNA sequence fragments of M. cirrosus were discovered
with 13 reads, which indicated that the patient likely carried this
fungus from the beginning. Following treatment with anti-fungal
drugs for half a month, M. cirrosus spread and replicated
proficiently within the lungs due to its extensive drug resistance,
conditions which favored its growth. It therefore appears that at
times of poor efficacy by routine anti-fungal infection treatment,
uncommon fungal pathogens may be misdiagnosed by a single test.
Apart from studying the characteristics of M. cirrosus in
detail, the timely incorporation of M. cirrosus into the
rapid mass spectrometry identification and gene information
database of fungi may be beneficial to improve the correct
detection of M. cirrosus. As this fungus is mostly resistant
to common antifungal drugs, it is difficult to treat. To date, the
majority of patients who underwent transplantation and were
infected with this fungus were not successfully cured. Perhaps it
would be beneficial to examine the use of some uncommon therapies,
including the combined use of several antifungal drugs, the
surgical resection of localized lesions, and immune regulation
therapy (Table II).
Infection with M. cirrosus has been reported
in the United States, Belgium, France, Italy and China. From the
geographical distribution point of view, M. cirrosus has
been reported as a plant-infecting pathogen with no regional
limitation (38). The
characteristics of infection are mainly lung and local skin
infections, among which lung infections are basically observed in
immunocompromised patients, and local skin infections can appear in
non-immunocompromised patients. Of note, the majority of fatal lung
infections by M. cirrosus are difficult to combat successful
in the transplant population (Table
II). The present study described a case of M. cirrosus
pulmonary infection, which is known to be the first reported
patient undergoing haplo-HSCT in China. To date, to the best of the
authors' knowledge, only one case of skin infection of M.
cirrosus has been reported in China (16). M. cirrosus infection mostly
occurs in post-transplant population (10,12,14,15),
and China has a large transplant population. Based on the whole
genome sequence of M. cirrosus, the present study
retrospectively reanalyzed the initial BALF mNGS sequence from
patients who underwent HSCT and suffered from fatal pulmonary
infection, but failed to be cured at Dushu Lake Hospital Affiliated
to Soochow University. Different numbers of sequences of M.
cirrosus could be detected in another two cases, three and 13
reads respectively, favoring the fungal etiological diagnosis by GM
tests. The data from animal experiments demonstrated that M.
cirrosus in immunosuppressed mice can cause severe pneumonia,
even leading to mortality. Therefore, the correct and rapid
diagnosis of the Microascus genus in transplant populations
is required to fully understand the epidemiology of this fungal
infection.
In the present study, the GM test and mNGS suggested
that the patient had a pulmonary fungal infection; however, this
clinical isolate was misdiagnosed. Finally, the fungus was
identified by culture, although this was time-consuming and
treatment was delayed. The development of mNGS and its wide
application in the diagnosis of infectious diseases have broken the
existing limitations of conventional diagnostic approaches
(39). Since mNGS does not require
live pathogens or cultures, its positive rate can be much higher
than conventional diagnostic approaches, and the types of pathogens
that can be detected are wider than those of conventional methods
(40). Thus, mNGS is more suitable
for the diagnosis of opportunistic pathogens and mixed infections
in immunosuppressed patients. However, metagenomic sequencing
technology also has disadvantages, such as higher costs, more
complex procedures, the background of human-derived nucleic acids,
and the need for continuous updating of databases (41,42).
The present study reported a case of M. cirrosus, which was
not regarded as a pathogen in bioinformatics analysis using mNGS.
The reasons for this may be as follows: i) M. cirrosus is
considered to be a Biological Safety Level one grade environmental
microorganism, such as M. cirrosus CBS217.31 (https://wi.knaw.nl/details/80/9762), thus, its
partial sequence information is not included in the database for
mNGS analysis; ii) there is currently no genome information of
M. cirrosus in the NCBI database. This may lead to the wrong
detection or undetected infection in immunosuppressed groups.
For this case, the assembled sequence of M.
cirrosus was provided and it was uploaded onto the NCBI public
database for research. In addition, a number of sequences of this
fungus could be detected by retrospective analysis of the mNGS
results of BALF. Thus, these data facilitate the rapid detection of
M. cirrosus by mNGS for the effective diagnosis of M.
cirrosus infection.
Compared with other filamentous fungi, such as
Aspergillus, M. cirrosus was identified with only
short chains of conidia without apical cysts in the present study.
Due to the limited information available on this fungus, M.
cirrosus may not be correctly recognized by morphological
identification even if it has been cultivated. MALDI-TOF-MS is a
common method for the rapid identification of microbial strains.
Cultured M. cirrosus can be accurately identified
MALDI-TOF-MS by a close the protein fingerprint of M.
gracilis, presenting a very different cluster from
Aspergillus fumigatus and Nocardia nova. Thus, M.
cirrosus can be rapidly detected by MALDI-TOF-MS.
In conclusion, there is an urgent need for the
extensive investigation of the frequency and pathogenicity of M.
cirrosus in immunosuppressed patients, as systemic
Microascus infections are difficult to treat and, hence, are
frequently fatal. For diagnosis, the establishment of standard
morphological and growth criteria is required. The rapid diagnosis
of M. cirrosus can be reliably achieved by mNGS, if the
vital genetic information has been uploaded onto public databases.
Due to its frequent and extensive resistance to drugs, attention
should be paid to refractory pneumonia due to M. cirrosus
when routine antifungal therapy is ineffective. In addition, it is
also necessary to explore effective treatment methods, as for
instance, the surgical removal of infection lesions combined with
the use of multiple drugs.
Acknowledgements
Not applicable.
Funding
Funding: The present study was financially supported by the
Suzhou Science and Technology Planning Project (grant nos.
SLT201921, SZM2021011, SZM2021018 and SKY2022091) and the Jiangsu
Provincial Key Research and Development Program (grant no.
BE2019656).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request. The second-generation sequencing project of Microascus
cirrosus SZ 2021 was deposited in NCBI under BioProject:
PRJNA835605 (https://www.ncbi.nlm.nih.gov/bioproject/835605),
BioSample: SAMN28105390 (https://dataview.ncbi.nlm.nih.gov/object/SAMN28105390)
and GenBank: JAMBUN000000000. The version described herein is
version JAMBUN010000000.
Authors' contributions
QH and JC conceived and designed the study. JC and
TZ completed the isolation, culture and morphological observation
of the strain. DZ provided the cases and data. JC, LZ, XH, TZ, JH,
DZ and PZ completed the molecular identification and the data
analysis. JC, QH and LW participated in the analysis or
interpretation of the data. JC participated in the drafting of the
manuscript. LW, JH and QH contributed to the revision of article.
QH and LW confirm the authenticity of all the raw data. All authors
have read and approved the final version of the manuscript.
Ethics approval and consent to
participate
The present study was conducted in accordance with
the ethical standards of the Declaration of Helsinki, and was
approved by the Institutional Review Board of Dushu Lake Hospital
Affiliated to Soochow University (Suzhou, China; approval no.
220138). Informed consent was waived due to the retrospective
nature of the study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Penack O, Marchetti M, Ruutu T, Aljurf M,
Bacigalupo A, Bonifazi F, Ciceri F, Cornelissen J, Malladi R,
Duarte RF, et al: Prophylaxis and management of graft versus host
disease after stem-cell transplantation for haematological
malignancies: Updated consensus recommendations of the European
society for blood and marrow transplantation. Lancet Haematol.
7:e157–e167. 2020.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Kang HG, Zhang D, Degauque N, Mariat C,
Alexopoulos S and Zheng XX: Effects of cyclosporine on transplant
tolerance: the role of IL-2. Am J Transplant. 7:1907–1916.
2007.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Liddicoat AM and Lavelle EC: Modulation of
innate immunity by cyclosporine A. Biochem Pharmacol. 163:472–480.
2019.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Esquirol A, Pascual MJ, Kwon M, Pérez A,
Parody R, Ferra C, Garcia Cadenas I, Herruzo B, Dorado N, Hernani
R, et al: Severe infections and infection-related mortality in a
large series of haploidentical hematopoietic stem cell
transplantation with post-transplant cyclophosphamide. Bone Marrow
Transplant. 56:2432–2444. 2021.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Roberts MB and Fishman JA:
Immunosuppressive agents and infectious risk in transplantation:
managing the ‘net state of immunosuppression’. Clin Infect Dis.
73:e1302–e1317. 2021.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Ji C, Yang Z, Zhong X and Xia J: The role
and mechanism of CARD9 gene polymorphism in diseases. Biomed J.
44:560–566. 2021.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Caira M, Trecarichi EM, Mancinelli M,
Leone G and Pagano L: Uncommon mold infections in hematological
patients: epidemiology, diagnosis and treatment. Expert Rev Anti
Infect Ther. 9:881–892. 2011.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Mohammedi I, Piens MA, Audigier-Valette C,
Gantier JC, Argaud L, Martin O and Robert D: Fatal microascus
trigonosporus (anamorph Scopulariopsis) pneumonia in a bone marrow
transplant recipient. Eur J Clin Microbiol Infect Dis. 23:215–217.
2004.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Baddley JW, Moser SA, Sutton DA and Pappas
PG: Microascus cinereus (Anamorph scopulariopsis) brain abscess in
a bone marrow transplant recipient. J Clin Microbiol. 38:395–397.
2000.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Miossec C, Morio F, Lepoivre T, Le Pape P,
Garcia-Hermoso D, Gay-Andrieu F, Haloun A, Treilhaud M, Leclair F
and Miegeville M: Fatal invasive infection with fungemia due to
Microascus cirrosus after heart and lung transplantation in a
patient with cystic fibrosis. J Clin Microbiol. 49:2743–7.
2011.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Sandoval-Denis M, Sutton DA, Fothergill
AW, Cano-Lira J, Gené J, Decock CA, de Hoog GS and Guarro J:
Scopulariopsis, a poorly known opportunistic fungus: Spectrum of
species in clinical samples and in vitro responses to antifungal
drugs. J Clin Microbiol. 51:3937–43. 2013.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Krisher KK, Holdridge NB, Mustafa MM,
Rinaldi MG and McGough DA: Disseminated Microascus cirrosus
infection in pediatric bone marrow transplant recipient. J Clin
Microbiol. 33:735–737. 1995.PubMed/NCBI View Article : Google Scholar
|
|
13
|
de Vroey C, Lasagni A, Tosi E, Schroeder F
and Song M: Onychomycoses due to Microascus cirrosus (syn. M.
desmosporus). Mycoses. 35:193–196. 1992.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Ustun C, Huls G, Stewart M and Marr KA:
Resistant Microascus cirrosus pneumonia can be treated with a
combination of surgery, multiple anti-fungal agents and a growth
factor. Mycopathologia. 162:299–302. 2006.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Taton O, Bernier B, Etienne I, Bondue B,
Lecomte S, Knoop C, Jacob F and Montesinos I: Necrotizing
Microascus tracheobronchitis in a bilateral lung transplant
recipient. Transpl Infect Dis. 20:2018.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Gao L, Chen J, Gao D and Li M: Primary
cutaneous infection due to Microascus cirrosus: A case report: BMC
Infect. Dis. 18(604)2018.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Chiu CY and Miller SA: Clinical
metagenomics. Nat Rev Genet. 20:341–355. 2019.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Wilson MR, Sample HA, Zorn KC, Arevalo S,
Yu G, Neuhaus J, Federman S, Stryke D, Briggs B, Langelier C, et
al: Clinical metagenomic sequencing for diagnosis of meningitis and
encephalitis. N Engl J Med. 380:2327–2340. 2019.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Gu W, Deng X, Lee M, Sucu YD, Arevalo S,
Stryke D, Federman S, Gopez A, Reyes K, Zorn K, et al: Rapid
pathogen detection by metagenomic next-generation sequencing of
infected body fluids. Nat Med. 27:115–124. 2021.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Tang W, Zhang Y, Luo C, Zhou L, Zhang Z,
Tang X, Zhao X and An Y: Clinical application of metagenomic
next-generation sequencing for suspected infections in patients
with primary immunodeficiency disease. Front Immunol.
12(696403)2021.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Azar MM, Schlaberg R, Malinis MF, Bermejo
S, Schwarz T, Xie H and Dela Cruz CS: Added diagnostic utility of
clinical metagenomics for the diagnosis of pneumonia in
immunocompromised adults. Chest. 159:1356–1371. 2021.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Issakainen J, Salonen JH, Anttila VJ,
Koukila-Kähkölä P, Castrén M, Liimatainen O, Vuento R, Ojanen T,
Koivula I, Koskela M and Meurman O: Deep, respiratory tract and ear
infections caused by Pseudallescheria (Scedosporium) and Microascus
(Scopulariopsis) in Finland. A 10-year retrospective multi-center
study. Med Mycol. 48:458–65. 2010.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Huang L, Chen W, Guo L, Zhao L, Cao B, Liu
Y, Lu B, Li B, Chen J and Wang C: Scopulariopsis/Microascus
isolation in lung transplant recipients: A report of three cases
and a review of the literature. Mycoses. 62:883–892.
2019.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Chang YJ, Pei XY and Huang XJ:
Haematopoietic stem-cell transplantation in China in the era of
targeted therapies: current advances, challenges, and future
directions. Lancet Haematol. 9:e919–e929. 2022.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Clinical and Laboratory Standards
Institute. 2008. Reference method for broth dilution antifungal
susceptibility testing of filamentous fungi: approved standard, 2nd
ed. CLSI document M38-A2. Clinical and Laboratory Standards
Institute, Wayne, PA.
|
|
26
|
Sandoval-Denis M, Gené J, Sutton DA,
Cano-Lira JF, deHoog GS, Decock CA, Wiederhold NP and Guarro J:
Redefining Microascus, Scopulariopsis and allied genera. Persoonia.
36:1–36. 2016.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Brasch J, Beck-Jendroschek V,
Iturrieta-González I, Voss K and Gené J: A human subcutaneous
infection by Microascus ennothomasiorum sp. nov. Mycoses.
62:157–164. 2019.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Thompson JD, Higgins DG and Gibson TJ:
CLUSTAL W: Improving the sensitivity of progressive multiple
sequence alignment through sequence weighting, position-specific
gap penalties and weight matrix choice. Nucleic Acids Res.
22:4673–4680. 1994.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Kumar S, Stecher G, Li M, Knyaz C and
Tamura K: MEGA X: Molecular evolutionary genetics analysis across
computing platforms. Mol Biol Evol. 35:1547–1549. 2018.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Bankevich A, Nurk S, Antipov D, Gurevich
AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S,
Prjibelski AD, et al: SPAdes: A new genome assembly algorithm and
its applications to single-cell sequencing. J Comput Biol.
19:455–77. 2012.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Seemann T: Prokka: Rapid prokaryotic
genome annotation. Bioinformatics. 30:2068–2069. 2014.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Li H and Durbin R: Fast and accurate short
read alignment with Burrows-Wheeler transform. Bioinformatics.
25:1754–1760. 2009.PubMed/NCBI View Article : Google Scholar
|
|
33
|
National Institutes of Health (1996) Guide
for the care and use of laboratory animals. 7th Edition, National
Academy Press, Washington DC.
|
|
34
|
Herbst S, Shah A, Carby M, Chusney G,
Kikkeri N, Dorling A, Bignell E, Shaunak S and Armstrong-James D: A
new and clinically relevant murine model of solid-organ transplant
aspergillosis. Dis Model Mech. 6:643–651. 2013.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Williams KM: How I treat bronchiolitis
obliterans syndrome after hematopoietic stem cell transplantation.
Blood. 129:448–455. 2017.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Ditschkowski M, Elmaagacli AH, Koldehoff
M, Gromke T, Trenschel R and Beelen DW: Bronchiolitis obliterans
after allogeneic hematopoietic SCT: Further insight-new
perspectives? Bone Marrow Transplant. 48:1224–1229. 2013.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Karakioulaki M and Stolz D: Biomarkers in
pneumonia-beyond procalcitonin. Int J Mol Sci.
20(2004)2019.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Mirzaee MR, Asgari B, Zare R and Mohammadi
M: Association of Microascus cirrosus (microascaceae, ascomycetes)
with brown leaf spot of pistachio in Iran. Plant Dis.
94(642)2010.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Jin X, Li J, Shao M, Lv X, Ji N, Zhu Y,
Huang M, Yu F, Zhang C, Xie L, et al: Improving suspected pulmonary
infection diagnosis by bronchoalveolar lavage fluid metagenomic
next-generation sequencing: A multicenter retrospective study.
Microbiol Spectr. 10(e0247321)2022.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Miao Q, Ma Y, Wang Q, Pan J, Zhang Y, Jin
W, Yao Y, Su Y, Huang Y, Wang M, et al: Microbiological diagnostic
performance of metagenomic next-generation sequencing when applied
to clinical practice. Clin Infect Dis. 67 (Suppl 2):S231–S240.
2018.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Gu W, Miller S and Chiu CY: Clinical
metagenomic next-generation sequencing for pathogen detection. Annu
Rev Pathol. 14:319–338. 2019.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Filkins LM, Bryson AL, Miller SA and
Mitchell SL: Navigating clinical utilization of
direct-from-specimen metagenomic pathogen detection: Clinical
applications, limitations, and testing recommendations. Clin Chem.
66:1381–1395. 2020.PubMed/NCBI View Article : Google Scholar
|