Introduction
Endothelial progenitor cells (EPCs) are the
precursors of vascular endothelial cells (1), which can be mobilized from the bone
marrow to peripheral blood in response to physiological or
pathological conditions for endothelial repair and
neovascularization (2,3). EPC-induced vasculogenesis has been
considered to provide a novel therapeutic approach for patients
with heart and limb ischemia. Various approaches have been explored
to enhance EPC grafting, including local EPC delivery, promotion of
EPC mobilization, EPC function enhancement, and in vitro EPC
expansion (4). However, the
biology of EPCs is complex and understanding of the precise
mechanisms that regulate EPC differentiation is limited (5).
N6-methyladenosine (m6A) is the most
common internal modification in eukaryotic RNA (6,7). It
can regulate multiple physiological processes, including stem cell
differentiation, animal growth and development, Drosophila
sex determination and DNA damage repair (8). The effectors of m6A
include ‘writers’ and ‘erasers’, which install and remove the
methyl group, respectively, and ‘readers’, which recognize
methylation (9-12).
The core writer complex, consisting of methyltransferase-like
(METTL)3 and METTL14, forms a stable heterodimer complex that
catalyzes m6A modification (9). It has been established that Wilms'
tumor 1-associated protein (WTAP) binds to this heterodimer complex
and facilitates its nuclear localization, thus promoting the
deposition of m6A (13). Accordingly, the m6A
levels are largely dependent on this methyltransferase complex.
Moreover, it is now understood that WTAP is involved in several
biological functions, including embryo development, cell cycle
progression, cell differentiation, pre-mRNA splicing, and antiviral
responses (14-16).
Notably, m6A has been studied in stem
cell differentiation. Previous studies have documented the effect
of enzymes associated with m6A regulation on stem cells.
In this respect, it has been reported that YTHDF2 is essential for
self-renewing hematopoietic stem cells (17). Notably, Chen et al (18) revealed that doxycycline-induced
fusion of dCas13a with the catalytic domain of ALKBH5 could
demethylate m6A-enriched SOX2 and control the
differentiation of human embryonic stem cells. Moreover, elevated
levels of m6A have been documented in cancer stem cells
(CSCs) (19,20). Although it has been established
that m6A can promote the expression of oncogenes in CSCs
(19), it can also promote the
tumor phenotype of CSCs and metastasis (21). While previous studies have
primarily focused on m6A in the context of CSCs,
investigations into m6A in EPCs have been relatively
scarce (22). The present study
focused on the m6A ‘writer’ WTAP in EPC differentiation.
The study aimed to investigate the dynamics of m6A
levels during the differentiation process of HPB-EPCs and identify
the key enzyme involved in regulating m6A that impacted
this process. These findings may illuminate an effective mechanism
for promoting vascular repair through m6A.
Materials and methods
Isolation of HPB-EPCs, overexpression,
and knockdown of WTAP
The present study was approved by the Medical Ethics
Committee of Fudan University Pudong Medical Center (Shanghai,
China; approval no. 2020-SZR-04) and written informed consent was
obtained from the participants. Venous blood samples (15 ml) from
three healthy male donors aged 25-28 years were aseptically
collected in the blood collection room at the hospital's
experimental center by a trained nurse. Then the peripheral blood
was diluted with PBS before being added to human lymphocyte
separation medium (cat. no. P8610; Beijing Solarbio Science and
Technology, Co., Ltd.). After centrifugation at 500 x g for 20 min
at room temperature, the mononuclear cell layer was isolated and
washed twice with an equal volume of PBS containing 2% fetal bovine
serum (FBS; Gibco; Thermo Fisher Scientific, Inc.). The
differentiation of EPCs was induced using complete endothelial cell
medium (ECM; cat. no. 1001; ScienCell Research Laboratories, Inc.)
containing 5% FBS, 5% endothelial cell growth supplement (cat. no.
1052; ScienCell Research Laboratories, Inc.), 100 U/ml penicillin
and 100 µg/ml streptomycin (cat. no. 0503; ScienCell Research
Laboratories, Inc.). The cells were cultured in an incubator
containing 5% CO2 at 37˚C, and cell growth was monitored
every other day using an inverted light microscope. The first day
of induction in vitro (1D) marks the initiation, while the
fourteenth day of induction (14D) refers to a specific time point
during the process.
To induce overexpression of WTAP, the coding
sequence of WTAP was inserted into the pLVX-Puro plasmid (cat. no.
HH-LV-048; HedgehogBio Science and Technology, Ltd.). For WTAP
knockdown, WTAP-specific short hairpin (sh)RNA #1-3 and the
scrambled (scr) sequence that was used as a negative control (NC),
were ligated to the linearized pLKO.1-Puro plasmid (cat. no.
HH-shRNA-004; HedgehogBio Science and Technology, Ltd.). The study
utilized a second-generation lentiviral packaging system comprising
of three plasmids to conduct the experiments. Table I provides a comprehensive overview
of the sequence information for shRNA #1-3 and Scr (NC). The 5 µg
recombinant plasmids, along with the psPAX2 packaging plasmid and
pMD2G envelope plasmid at a ratio of 4:3:1, were transfected into
293T cells (cat. no. FH0244; Fuheng Biotechnology, Ltd.) using
Lipofectamine™ 3000 (cat. no. L3000015; Invitrogen; Thermo Fisher
Scientific, Inc.). A total of 48 h post-transfection, the
supernatant was collected and filtered to obtain the virus for
subsequent transduction. For overexpression of WTAP, a blank vector
was used as an NC. Well-cultured HPB-EPCs at a confluence of 40%
were pre-selected and the aforementioned viral particles were added
to the supernatant of HPB-EPCs at an MOI of 200. After being
cultured for 24 h in an incubator set at 37˚C, the virus-containing
culture medium was replaced with fresh culture medium. A total of 3
days post-transduction, fluorescence expression in cells was
observed and stable cell lines were screened using 5 µg/ml
puromycin for 24 h prior to subsequent experiments, while a
maintenance dose of 2 µg/ml puromycin was used during the
experiment.
 | Table IshRNA and primer sequences used in
the present study. |
Table I
shRNA and primer sequences used in
the present study.
| shRNA or
primer | Sequence,
5'-3' |
|---|
| Human WTAP shRNA
#1 |
GGUUCGAUUGAGUGAAACATT |
| Human WTAP shRNA
#2 |
GCUUUGGAGGGCAAGUACATT |
| Human WTAP shRNA
#3 |
GGGCAACACAACCGAAGAT |
| Scr (NC) |
CAACAAGATGAAGAGCACCAAC |
| CD133 | F:
AGTCGGAAACTGGCAGATAGC |
| | R:
GGTAGTGTTGTACTGGGCCAAT |
| KDR | F:
GGCCCAATAATCAGAGTGGCA |
| | R:
CCAGTGTCATTTCCGATCACTTT |
| vwF | F:
CCGATGCAGCCTTTTCGGA |
| | R:
TCCCCAAGATACACGGAGAGG |
| CD31 | F:
AACAGTGTTGACATGAAGAGCC |
| | R:
TGTAAAACAGCACGTCATCCTT |
| METTL3 | F:
TCTCCACGCCAGATGCTC |
| | R:
ACAGTCCCTGCTACCTCCC |
| METTL14 | F:
CCTCCCATGTACTTACAAGCC |
| | R:
TAGCAGTGATGCCAGTTTCTC |
| WTAP | F:
ATGGCGAAGTGTCGAATGC |
| | R:
CCAACTGCTGGCGTGTCTC |
| FTO | F:
ACTTGGCTCCCTTATCTGACC |
| | R:
TGTGCAGTGTGAGAAAGGCTT |
| ALKBH5 | F:
CGGCGAAGGCTACACTTACG |
| | R:
CCACCAGCTTTTGGATCACCA |
| IGF2BP1 | F:
TCCCCGATGAGCAGATAGC |
| | R:
CTGGGTCTGTTTTGTGATGTTG |
| IGF2BP2 | F:
ATGAAACAGGGACCAAGATAAC |
| | R:
GTTGAAAAGATGCCAAGTGC |
| IGF2BP3 | F:
GATTAAATCTGAACGCCTTGG |
| | R:
TGGCACCGACTGATAGAGC |
| YTHDF1 | F:
ACCTGTCCAGCTATTACCCG |
| | R:
TGGTGAGGTATGGAATCGGAG |
| YTHDF2 | F:
AGCCCCACTTCCTACCAGATG |
| | R:
TGAGAACTGTTATTTCCCCATGC |
| YTHDF3 | F:
TCAGAGTAACAGCTATCCACCA |
| | R:
GGTTGTCAGATATGGCATAGGCT |
| GAPDH | F:
GGAGCGAGATCCCTCCAAAAT |
| | R:
GGCTGTTGTCATACTTCTCATGG |
Cell proliferation assay
Exponential phase HPB-EPCs were resuspended with
complete ECM following digestion with trypsin. The cells were
counted and seeded into a 96-well plate at a density of 2,000
cells/well. After incubation for 0, 24, 48, and 72 h, the culture
medium containing 10% Cell Counting Kit 8 (CCK8; Dojindo
Laboratories, Inc.) was added to the cells and incubated for 2 h
within the CO2 incubator, all at 37˚C. Subsequently, the
absorbance of the samples was measured at a wavelength of 450 nm
using a spectrophotometer.
Cell invasion assay
The cell invasion assay was performed using
Transwell plates (24-well insert; pore size, 8 µm; BD Biosciences).
The filter membrane of the chamber was coated with 60 µl Matrigel
(1:8 dilution; BD Biosciences) for 1 h at 37˚C. The upper chamber
was seeded with 100 µl serum-free medium containing
2x104 HPB-EPCs and the lower chamber was seeded with 600
µl complete ECM. After incubation for 24 h at 37˚C, the chamber was
fixed with 4% paraformaldehyde for 30 min and stained with 0.1%
crystal violet for 30 min at room temperature. Finally, a
magnifying light microscope (Leica DMI3000B; Leica Microsystems
GmbH) was used to count the number of invaded cells at the bottom
of the chamber.
Tube formation assay
Precooled 96-well plates were seeded with 100
µl/well Matrigel (BD Biosciences) and incubated at 37˚C for 30 min.
Subsequently, the stably transfected cells were trypsinized,
resuspended in complete ECM, seeded at 5x104/well in the
aforementioned 96-well plates, and incubated for another 6 h at
37˚C. Finally, images were captured using the white light channel
of a fluorescence microscope (Nikon Corporation). The number and
length of tubes were counted and analyzed by ImageJ (version 1.8.0;
National Institutes of Health).
Liquid chromatography with tandem mass
spectrometry (LC-MS/MS) assay
TRIzol® Regent (cat. no. 15596026;
Invitrogen; Thermo Fisher Scientific, Inc.) was employed to isolate
total RNA from HPB-EPCs on 1D and 14D. Oligo dT magnetic beads
(cat. no. 19820; Yeasen Biotechnology, Ltd.) were used to purify
mRNA from total RNA. Subsequently, 200 ng purified mRNA was
incubated with nuclease P1 (0.5 U; MilliporeSigma) at 42˚C for 1 h
in a reaction system containing 10 mM NH4OAC (pH, 5.3;
25 µl). Then, NH4HCO3 (1 M; 3 µl) and alkaline
phosphatase (1 µl; 1 U/µl; MilliporeSigma) were added and incubated
at 37˚C for 2 h. After neutralization with 1 µl HCl (3 M), samples
were diluted to 50 µl and filtered through a 0.22-µM filter
(MilliporeSigma). The separation of all samples (10 µl per
injection) was achieved using reverse-phase ultra-performance LC
through an ACQUITY UPLC T3 column (Waters Technologies, Inc.). The
flow rate was 0.3 ml/min. Analysis was performed using a TripleTOF
6600 tandem mass spectrometer (SCIEX Technologies, Inc.) in
positive electrospray ionization mode. The interface heater
temperature was 550˚C. The curtain gas was set at 30 PSI, and both
Ion source gas1 and Ion source gas2 were all set at 55 PSI. All
nucleosides were quantified using retention times, and ion mass
transitions (m/z) from 268.1 to 136.1 [Adenosine (A)] and 282.1 to
150.1 (m6A). Quantification was performed using standard
curves generated within the same experimental batch. A calibration
curve was derived from these standard curves to calculate the
m6A to A ratios (23).
Immunofluorescence
The cells were fixed with 4% paraformaldehyde for 20
min at room temperature and were then incubated with 0.3% Triton
X-100 for 10 min, and the non-specific binding sites were blocked
with 5% BSA (cat. no. SW3015; Beijing Solarbio Science and
Technology, Co., Ltd.) for 30 min at room temperature.
Subsequently, the cells were incubated with a primary antibody
against m6A (cat. no. A-1801; EpiGentek, Inc.) at a
dilution of 1:100 overnight at 4˚C, followed by a 1:1,000 dilution
of Alexa Fluor® 594-conjugated goat anti-rabbit
secondary antibody (cat. no. 8889; Cell Signaling Technology, Inc.)
at room temperature for 1 h. Finally, nuclear staining was
performed with DAPI for 1 min at room temperature, and cells were
observed with EVOS™ FL Auto 2 imaging system (Invitrogen; Thermo
Fisher Scientific, Inc.).
Western blotting
Protein samples were extracted from the experimental
cells using protein lysis buffer, which contained RIPA lysis buffer
(cat. no. PC101; Epizyme Biomedical Technology, Ltd.) and 1X
protease inhibitor cocktail (cat. no. GRF101; Epizyme Biomedical
Technology, Ltd.). The extracted proteins were then quantified
using the BCA method, denatured using sample buffer, separated by
10% SDS-PAGE with loading of 20 µg protein per well and
electrophoretically transferred to PVDF membranes. The membranes
were then exposed to a blocking solution (cat. no. PS108P; Epizyme
Biomedical Technology, Ltd.) for 10 min at room temperature and
incubated overnight with the primary antibodies at 4˚C. The
membranes were then washed three times with TBST (containing 0.1%
Tween20) and incubated with an HRP-conjugated secondary antibody
for 1 h at room temperature. Signals were detected and captured
using a filesystem (GBOX; Syngene) with a luminescence solution
(cat. no. SQ201L; Epizyme Biomedical Technology, Ltd.) (liquids A
and B; 1:1 ratio). The primary antibodies utilized in this
investigation were WTAP (cat. no. 41934; Cell Signaling Technology,
Inc.) and β-actin (cat. no. 4970; Cell Signaling Technology, Inc.),
both at a dilution of 1:1,000. The secondary antibody employed was
HRP-linked anti-rabbit antibody (cat. no. 7074; Cell Signaling
Technology, Inc.) at a dilution of 1:3,000.
Reverse transcription-quantitative PCR
(RT-qPCR)
RNA samples were extracted from cells using
TRIzol® Regent (cat. no. 15596026; Invitrogen; Thermo
Fisher Scientific, Inc.) and reverse transcribed (cat. no. RR037A;
Takara Bio, Inc.) according to the manufacturer's instructions.
cDNA was diluted in nuclease-free water and RT-qPCR was performed
using 50 ng diluted cDNA, the TB Green® Premix Ex Taq™
kit (cat. no. RR420A; Takara Bio, Inc.), and the ABI 7500 Real-Time
PCR system (Applied Biosystem; Thermo Fisher Scientific, Inc.)
according to the manufacturer's instructions. The thermocycling
protocol was established based on the manufacturer's instructions
and the specifications of the RT-PCR instrument utilized, as
follows: Denaturation for 1 cycle at 95˚C for 30 sec, followed by
PCR for 40 cycles at 95˚C for 5 sec and 60˚C for 34 sec, and then
melting for 1 cycle at 95˚C for 15 sec, 60˚C for 1 min and 95˚C for
15 sec. GAPDH was employed as the housekeeping gene in this
experiment. All primers used in the present study were sourced from
Thermo Fisher Scientific, Inc., and are listed in Table I. The final results were analyzed
using the 2-ΔΔCq method (17).
Statistical analysis
Statistical analysis was conducted using Prism 9
software (GraphPad; Dotmatics). Data are presented as the mean ±
standard error of the mean. The significance level (α) was set at
0.05. P<0.05 was used to indicate a statistically significant
difference. Comparisons between two groups were evaluated using
unpaired two-tailed Student's t-test for data exhibiting normal
distribution based on the Shapiro-Wilk normality test. For multiple
comparisons, one-way ANOVA and LSD post hoc test was performed.
Immunofluorescence, western blotting, and qPCR were performed with
at least three independent biological replicates. The sample size
was not predetermined using a statistical method, but a minimum of
three samples were included in each experimental group and
condition.
Results
M6A levels are increased
with the differentiation of EPCs
It has been established that the differentiation of
EPCs can be identified by detecting specific cell markers (24,25).
To evaluate the differentiation of HPB-EPCs, the expression levels
of cell surface markers were detected at different time points (1D
and 14D) using RT-qPCR. The expression levels of the cell lineage
marker CD133 were significantly downregulated with increased
culture duration (Fig. 1A). By
contrast, markers associated with endothelialization, including
KDR, von Willebrand factor (vWF), and CD31, were significantly
upregulated on 14D compared with on 1D (Fig. 1A). These findings suggested that
EPCs could differentiate into endothelial cells during prolonged
culture in vitro. Furthermore, m6A modification
levels were investigated at different time points during EPC
culture using LC-MS/MS and immunofluorescence techniques. Table SI provides information regarding
the raw and normalized peaks detected by LC-MS/MS. The results
revealed that the differentiated EPCs exhibited higher levels of
m6A modification than undifferentiated cells (Fig. 1B and C).
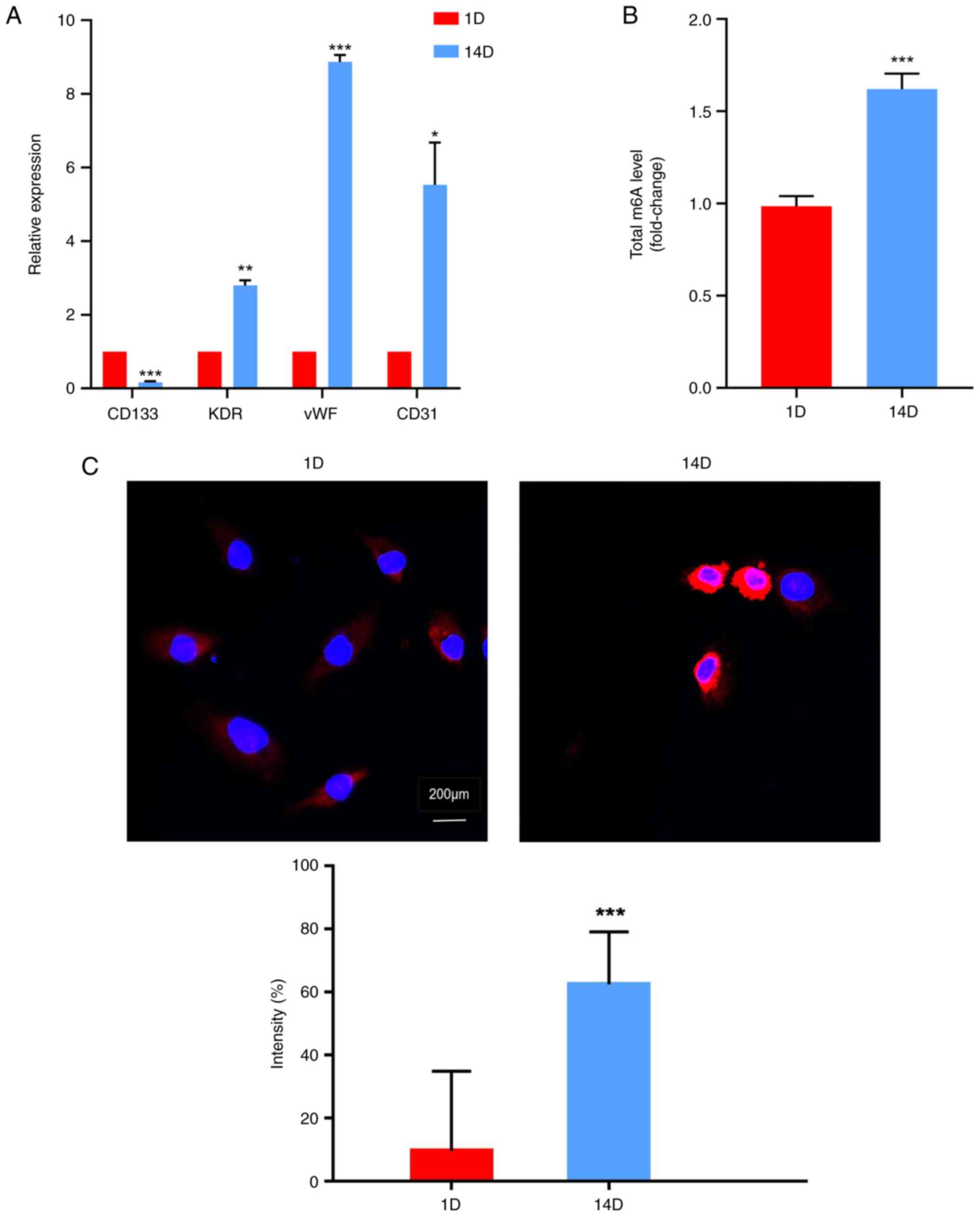 | Figure 1M6A levels are increased
with the differentiation of EPCs. (A) Relative expression levels of
cell surface markers of human peripheral blood-derived EPCs at
different time points (1D and 14D) were detected by quantitative
PCR. Expression levels of cell lineage marker CD133 and
reendothelialization markers KDR, vWF, and CD31 were assessed at
different time points during EPC culture. CD133 expression
decreased over time, whereas KDR, vWF and CD31 expression
increased. m6A levels were quantified in differentiated
EPCs by (B) LC-MS/MS and (C) immunofluorescence assays.
Differentiated EPCs exhibited higher m6A levels.
*P<0.05, **P<0.01,
***P<0.001 vs. 1D. D, day; EPC, endothelial
progenitor cell; m6A, N6-methyladenosine; vWF, von
Willebrand factor. |
WTAP contributes to increased
m6A levels in EPCs
The mRNA expression levels of multiple
m6A-related enzymes were detected during EPCs
differentiation by RT-qPCR. Compared with pre-differentiation EPCs,
the mRNA expression levels of a variety of methyltransferases
(‘writers’, such as METTL14 and WTAP), demethylases (‘erasers’,
such as FTO and ALKBH5), and methylation-recognition enzymes
(‘readers’, for example, IGF2BP1-3 and YTHDF1-2) were significantly
increased (except for METTL3 and YTHDF3) after differentiation
(Fig. 2A), suggesting that
m6A was increased after HPB-EPCs differentiation. Among
all enzymes, the mRNA expression levels of WTAP were increased the
most after the differentiation of HPB-EPCs. Western blotting
further verified that the protein expression levels of WTAP were
significantly increased after differentiation (Fig. 2B), suggesting that WTAP may play a
major role in EPCs differentiation. To investigate the effect of
WTAP on EPCs differentiation, overexpression and knockdown of WTAP
were successfully induced in HPB-EPCs and were verified using
western blotting. Compared to the control group, the overexpression
group exhibited an increase in WTAP protein expression. While
EPC-shWTAP-1 did not markedly alter the expression level, the
EPC-shWTAP-2 and EPC-shWTAP-3 groups, which were selected for
subsequent experiments, exhibited a significant reduction in
protein expression compared with the negative control group
(EPC-scr; Fig. 2C). Subsequently,
an immunofluorescence assay was employed to detect alterations in
m6A levels resulting from overexpression and knockdown
of WTAP in HPB-EPCs. Overexpression of WTAP in HPB-EPCs led to an
increase in m6A level, whereas knockdown of WTAP
resulted in a downregulation of m6A level (Fig. 2D). These findings suggested that
WTAP may have a crucial role in regulating m6A during
EPC differentiation. Subsequently, the present study evaluated the
effects of changes in WTAP expression on EPC proliferation,
invasion, and tube formation.
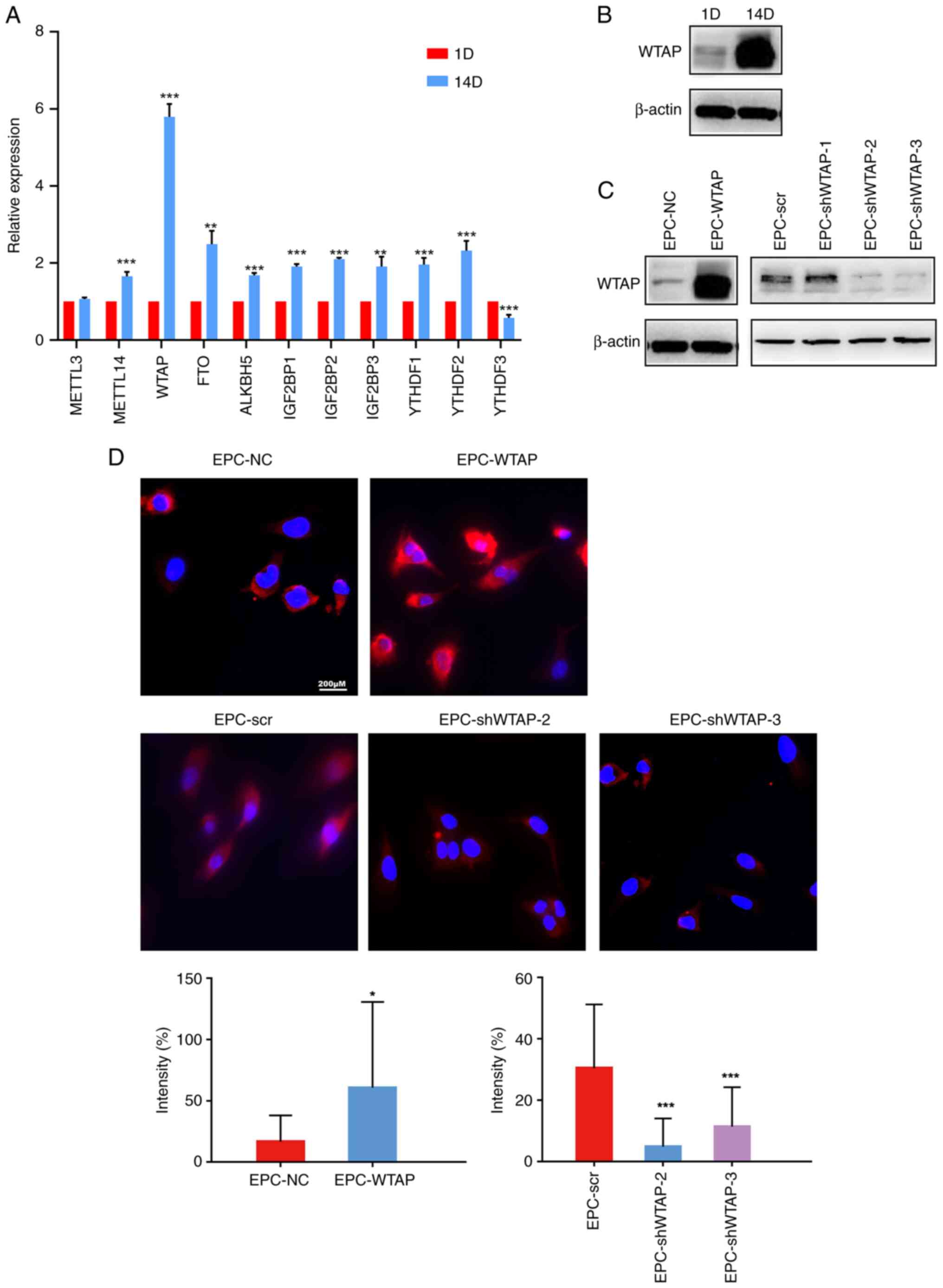 | Figure 2WTAP contributes to increased
m6A levels in EPCs. (A) mRNA expression levels of
m6A-related enzymes during EPC differentiation, as
detected by quantitative PCR. After differentiation, the expression
levels of METTL14, WTAP, FTO, ALKBH5, IGF2BP1-3 and YTHDF1-2 were
significantly higher than in EPCs before differentiation.
**P<0.01, ***P<0.001 vs. 1D. (B)
Western blotting was used to verify the protein expression levels
of WTAP, which were also increased after cell differentiation. (C)
WTAP expression was validated by western blot analysis following
overexpression and knockdown of WTAP. Compared with the control
group, the overexpression group exhibited an increase in WTAP
protein expression, whereas the knockdown group showed a decrease
in WTAP protein expression (with the exception of EPC-shWTAP-1).
(D) Immunofluorescence assays were performed to validate
alterations in m6A following the successful
overexpression and knockdown of WTAP in HPB-EPCs. Overexpression of
WTAP in HPB-EPCs increased m6A levels, whereas knockdown
of WTAP resulted in a decrease in m6A levels.
*P<0.05 vs. EPC-NC; ***P<0.001 vs.
EPC-scr. D, day; EPC, endothelial progenitor cell; m6A,
N6-methyladenosine; NC, negative control; scr, scrambled; sh, short
hairpin; WTAP, Wilms' tumor 1-associated protein. |
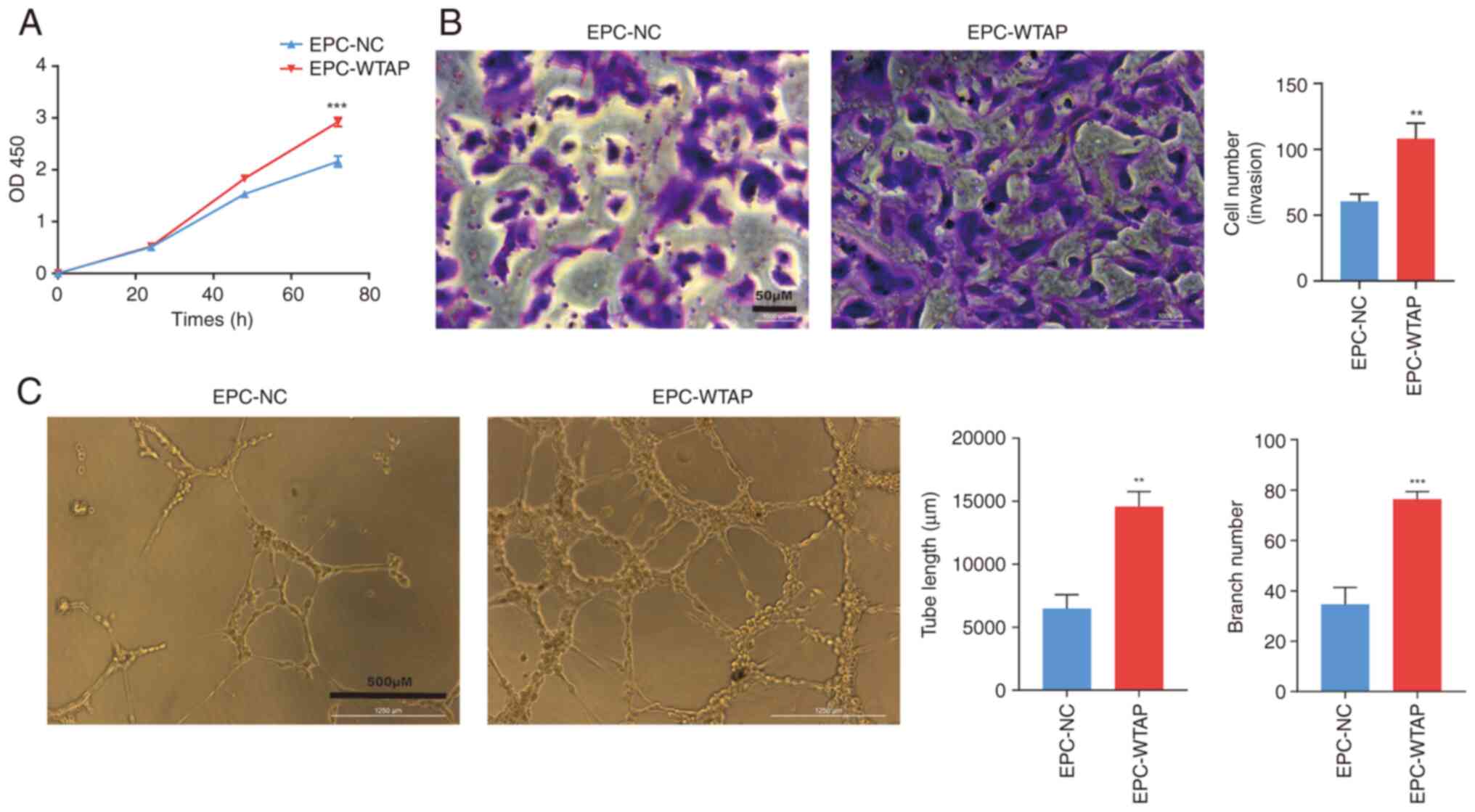
Overexpression of WTAP promotes the
differentiation of EPCs
Compared with the EPC-NC group, the proliferation of
EPCs was increased after WTAP overexpression in a time-dependent
manner and was significant after 72 h of culture (Fig. 3A). During the Transwell assay,
HPB-EPCs with WTAP overexpression exhibited a higher number of
cells crossing the filter membrane compared with that in the EPC-NC
group, suggesting that the overexpression of WTAP enhanced the
invasive ability of the cells (Fig.
3B). In addition, the EPC-WTAP group showed increased formation
of tubes and branching, suggesting that EPCs overexpressing WTAP
exhibited enhanced tube formation ability (Fig. 3C).
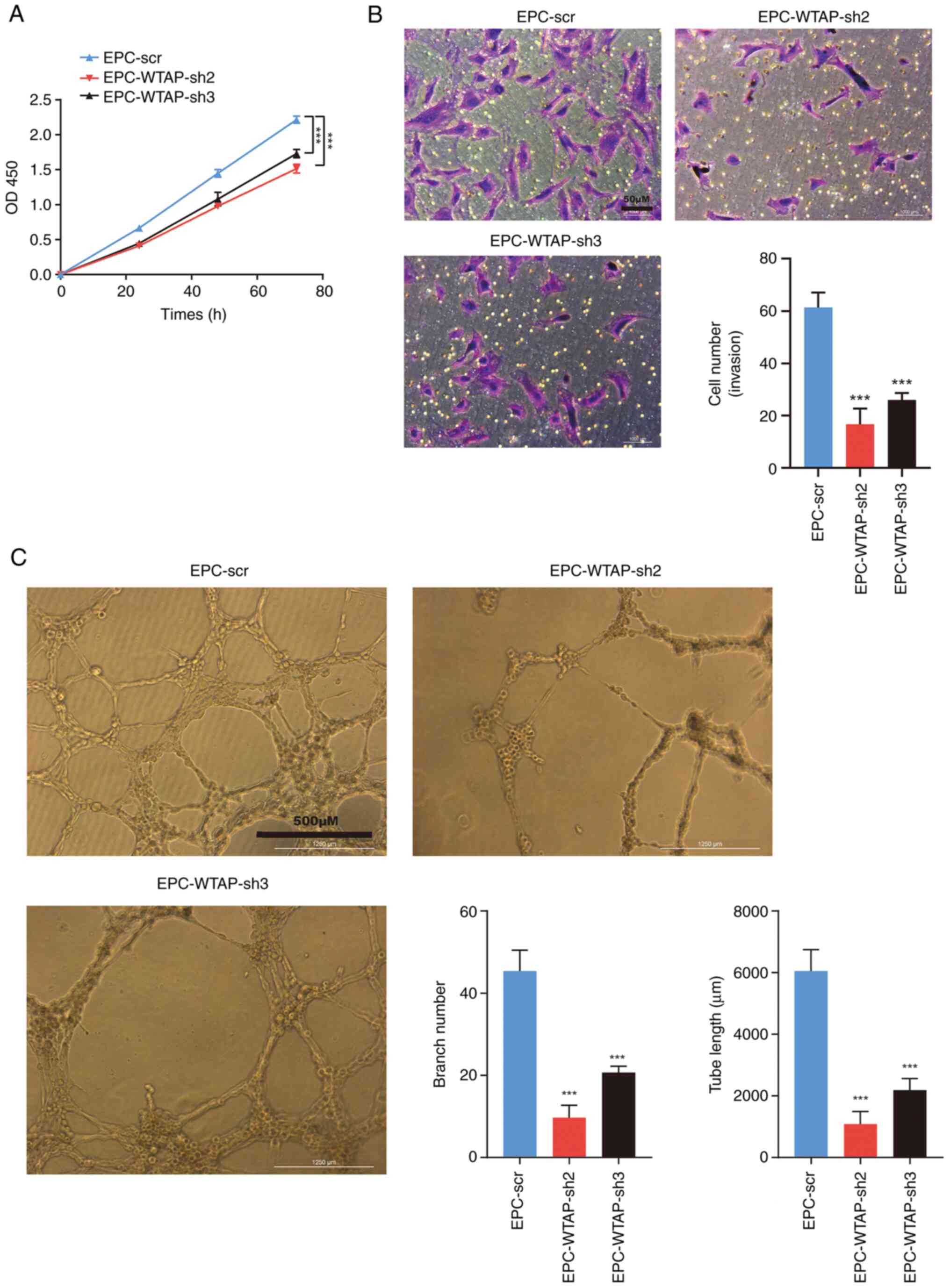
Knockdown of WTAP inhibits the
differentiation of EPCs
The present study demonstrated that the EPC-WTAP
group exhibited increased proliferation, invasion, and tube
formation. Subsequently, the present study explored the effects of
knocking down WTAP on the aforementioned functions in EPCs. The
CCK8 assay revealed that the proliferation of EPCs was decreased by
WTAP knockdown compared with in the NC group after 72 h of culture
(Fig. 4A). In contrast to the
EPC-scr group, cell invasion and tube formation were decreased
following WTAP knockdown (Fig. 4B
and C).
Discussion
Current evidence (26,27)
suggests that EPCs have similar differentiation capabilities to
stem cells, and their surface markers undergo changes at different
stages during the differentiation process. During the
differentiation of EPCs, CD133 expression is gradually
downregulated on the cell surface, whereas the immunophenotype of
differentiated EPCs is characterized by the presence of CD31,
VE-cadherin, vWF, CD146 and VEGFR2/KDR, and the lack of CD45 and
CD14 expression (24,25). The present study revealed that EPCs
exhibited differentiation ability in vitro. Notably, the
levels of m6A were increased with the differentiation of
EPCs; however, the role of m6A in EPC differentiation
remains unclear, warranting further exploration.
Little is currently known about the role of
m6A in stem cell regulation, since most studies have
focused on the role of m6A in CSCs (18-21,28).
It has been established that the levels of m6A are
upregulated in CSCs, and that the key enzymes involved in
m6A regulation may influence the phenotype of CSCs,
promote tumor metastasis, and influence tumor prognosis (18-21,28).
m6A can affect endothelial function and vascular
permeability, and can participate in the regulation of
atherosclerosis. Endothelial inflammation has been shown to be made
worse by METTL14(29), and
oxidative low-density lipoprotein can make human umbilical vein
endothelial cells less likely to divide and move (11). A previous study revealed that CPEB2
in glioma microvascular endothelial cells enhances SRSF5 stability
and promotes the expression of ZO-1, occludin, and claudin-5 to
protect vascular integrity through m6A modification
(upregulation of METTL3 and methylation-recognition enzyme IGF2BP3)
(30). However, studies
investigating the role of m6A in EPCs are limited. A
previous study has revealed that knockdown of METTL3 in EPCs could
result in impaired angiogenic potential; by contrast,
overexpression of METTL3 in EPCs led to enhanced tube formation
with increased tubule branching and increased angiogenesis in the
chorioallantoic membrane of chicken embryos (22). Similarly, the present study showed
that WTAP, another m6A methyl transferase, enhanced the
tubulogenic capacity of EPCs in vitro.
It is well known that METTL3, METTL14, and WTAP form
the m6A methyl transferase complex (MTC) (12). WTAP, an essential regulatory
subunit in methyltransferases, recruits the MTC to the target mRNA
(31). Current evidence suggests
that WTAP has various biological functions, including embryonic
development, cell cycle progression and differentiation, precursor
mRNA splicing, and alternative splicing (12). WTAP is also crucial in several
pathological processes, such as worsening myocardial
ischemia-reperfusion injury by increasing endoplasmic reticulum
stress (32), inducing malignant
tumor growth (12), and possibly
increasing resistance of tumors to drugs (33). The present study demonstrated that
WTAP could promote the proliferation, invasion, and angiogenesis of
EPCs, suggesting that WTAP may promote the differentiation of
EPCs.
The present study observed that the expression trend
of YTHDF3 differed from that of other m6A enzymes in
response to EPC differentiation. The other enzymes involved in
m6A modification showed an increasing trend during EPC
differentiation, whereas YTHDF3 showed a decreasing trend. The
exact role of YTHDF proteins in pluripotent stem cells remains
uncertain. After conducting phenotypic and transcriptomic analysis,
Wang et al (34) discovered
that the absence of YTHDF1 in embryonic stem cells can result in a
significant hindrance to cardiomyocyte (CM) differentiation,
whereas YTHDF3 knockdown can facilitate CM-specific gene expression
and thus promote CM differentiation. Based on these findings, it
was hypothesized that YTHDF3 may exhibit a downward trend during
EPCs differentiation. However, there is limited research on the
role of YTHDF3 in EPCs differentiation and further investigations
are needed to provide conclusive evidence supporting this
hypothesis.
To the best of our knowledge, the present study is
the first to provide evidence of the involvement of the
m6A methyltransferase WTAP in regulating EPC
differentiation; however, there are limitations. First, the present
study only included in vitro experiments, and future in
vivo vascular experiments are warranted to further evaluate the
effect of WTAP on the differentiation of EPCs. Second, due to study
limitations, RT-qPCR was used instead of flow cytometry to detect
cell surface markers and to evaluate EPCs differentiation. Third,
the present study primarily focused on observing the phenomenon
without exploring the underlying mechanisms. To elucidate the
mechanism, techniques such as gene chip analysis should be employed
to identify potential target genes and pathways involved in
WTAP-mediated EPC differentiation.
In conclusion, the present study revealed that
m6A is involved in regulating EPC differentiation, and
WTAP, one of its methyltransferases, may promote the proliferation,
invasion, and tube formation of EPCs, thus indicating that WTAP may
promote the differentiation of EPCs.
Supplementary Material
Raw and normalized peak information
(LC-MS of modified nucleosides).
Acknowledgements
Not applicable.
Funding
Funding: This research is supported by the Shanghai Pudong
Hospital (grant no. YJRCJJ201801), the Natural Science Foundation
of Shanghai (grant no. 20ZR1450100), and the Natural Science
Foundation of China (grant no. 82070587).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
JX, ZY, LN, LW and XL conceived and designed the
study. JX, LW, LN, ZY, QX and XL conducted the experiments. LN, ZY
and LW analyzed the data. LW and ZY wrote the manuscript. ZY, JX
and XL confirm the authenticity of all the raw data. JX and XL
revised the manuscript. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
Since the present study involved collecting blood
samples from participants, the contents of the experiment were
reviewed and approved by the Medical Ethics Committee of Fudan
University Pudong Medical Center, and all of the participants
provided written informed consent. In the consent form, the
experimental procedures were detailed, including the possibly
associated risks and the benefits to the participants, and it
informed them in writing that their specimens would be used only
for scientific research and there would be no commercial use.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Asahara T, Murohara T, Sullivan A, Silver
M, van der Zee R, Li T, Witzenbichler B, Schatteman G and Isner JM:
Isolation of putative progenitor endothelial cells for
angiogenesis. Science. 275:964–967. 1997.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Psaltis PJ and Simari RD: Vascular wall
progenitor cells in health and disease. Circ Res. 116:1392–1412.
2015.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Bonder CS, Sun WY, Matthews T, Cassano C,
Li X, Ramshaw HS, Pitson SM, Lopez AF, Coates PT, Proia RL, et al:
Sphingosine kinase regulates the rate of endothelial progenitor
cell differentiation. Blood. 113:2108–2117. 2009.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Tasev D, Koolwijk P and van Hinsbergh VW:
Therapeutic potential of human-derived endothelial colony-forming
cells in animal models. Tissue Eng Part B Rev. 22:371–382.
2016.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Kim J, Kim M, Jeong Y, Lee WB, Park H,
Kwon JY, Kim YM, Hwang D and Kwon YG: BMP9 induces cord
blood-derived endothelial progenitor cell differentiation and
ischemic neovascularization via ALK1. Arterioscler Thromb Vasc
Biol. 35:2020–2031. 2015.PubMed/NCBI View Article : Google Scholar
|
|
6
|
He L, Li H, Wu A, Peng Y, Shu G and Yin G:
Functions of N6-methyladenosine and its role in cancer. Mol Cancer.
18(176)2019.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Boulias K and Greer EL: Biological roles
of adenine methylation in RNA. Nat Rev Genet. 24:143–160.
2023.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Frye M, Harada BT, Behm M and He C: RNA
modifications modulate gene expression during development. Science.
361:1346–1349. 2018.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Shi H, Wei J and He C: Where, when, and
how: Context-Dependent functions of RNA methylation writers,
readers, and erasers. Mol Cell. 74:640–650. 2019.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Wang T, Kong S, Tao M and Ju S: The
potential role of RNA N6-methyladenosine in cancer progression. Mol
Cancer. 19(88)2020.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Rong J, Jie Y and Zhao H: m6A ‘writer’
KIAA1429 regulates the proliferation and migration of endothelial
cells in atherosclerosis. Mol Biotechnol. 65:1198–1206.
2023.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Huang Q, Mo J, Liao Z, Chen X and Zhang B:
The RNA m6A writer WTAP in diseases: Structure, roles,
and mechanisms. Cell Death Dis. 13(852)2022.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Wang LJ, Xue Y, Li H, Huo R, Yan Z, Wang
J, Xu H, Wang J, Cao Y and Zhao JZ: Wilms' tumour 1-associating
protein inhibits endothelial cell angiogenesis by m6A-dependent
epigenetic silencing of desmoplakin in brain arteriovenous
malformation. J Cell Mol Med. 24:4981–4991. 2020.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Wang Z, Qi Y, Feng Y, Xu H, Wang J, Zhang
L, Zhang J, Hou X, Feng G and Shang W: The N6-methyladenosine
writer WTAP contributes to the induction of immune tolerance post
kidney transplantation by targeting regulatory T cells. Lab Invest.
102:1268–1279. 2022.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Horiuchi K, Kawamura T, Iwanari H, Ohashi
R, Naito M, Kodama T and Hamakubo T: Identification of Wilms' tumor
1-associating protein complex and its role in alternative splicing
and the cell cycle. J Biol Chem. 288:33292–33302. 2013.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Sacco MT, Bland KM and Horner SM: WTAP
Targets the METTL3 m6A-methyltransferase complex to
cytoplasmic hepatitis C Virus RNA to regulate infection. J Virol.
96(e0099722)2022.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Yang Z, Wang T, Wu D, Min Z, Tan J and Yu
B: RNA N6-methyladenosine reader IGF2BP3 regulates cell cycle and
angiogenesis in colon cancer. J Exp Clin Cancer Res.
39(203)2020.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Chen X, Zhao Q, Zhao YL, Chai GS, Cheng W,
Zhao Z, Wang J, Luo GZ and Cao N: Targeted RNA N6
-Methyladenosine demethylation controls cell fate transition in
human pluripotent stem cells. Adv Sci (Weinh).
8(e2003902)2021.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Dixit D, Prager BC, Gimple RC, Poh HX,
Wang Y, Wu Q, Qiu Z, Kidwell RL, Kim LJY, Xie Q, et al: The RNA m6A
Reader YTHDF2 maintains oncogene expression and is a targetable
dependency in glioblastoma stem cells. Cancer Discov. 11:480–499.
2021.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Paris J, Morgan M, Campos J, Spencer GJ,
Shmakova A, Ivanova I, Mapperley C, Lawson H, Wotherspoon DA,
Sepulveda C, et al: Targeting the RNA m6A Reader YTHDF2
selectively compromises cancer stem cells in acute myeloid
leukemia. Cell Stem Cell. 25:137–148.e6. 2019.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Zhang C, Huang S, Zhuang H, Ruan S, Zhou
Z, Huang K, Ji F, Ma Z, Hou B and He X: YTHDF2 promotes the liver
cancer stem cell phenotype and cancer metastasis by regulating OCT4
expression via m6A RNA methylation. Oncogene. 39:4507–4518.
2020.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Jiang W, Zhu P, Huang F, Zhao Z, Zhang T,
An X, Liao F, Guo L, Liu Y, Zhou N and Huang X: The RNA
methyltransferase METTL3 promotes endothelial progenitor cell
angiogenesis in mandibular distraction osteogenesis via the
PI3K/AKT Pathway. Front Cell Dev Biol. 9(720925)2021.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Lin X, Chai G, Wu Y, Li J, Chen F, Liu J,
Luo G, Tauler J, Du J, Lin S, et al: RNA m6A methylation
regulates the epithelial mesenchymal transition of cancer cells and
translation of Snail. Nat Commun. 10(2065)2019.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Medina RJ, Barber CL, Sabatier F,
Dignat-George F, Melero-Martin JM, Khosrotehrani K, Ohneda O, Randi
AM, Chan JKY, Yamaguchi T, et al: Endothelial Progenitors: A
Consensus Statement on Nomenclature. Stem Cells Transl Med.
6:1316–1320. 2017.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Avci-Adali M, Nolte A, Simon P, Ziemer G
and Wendel HP: Porcine EPCs downregulate stem cell markers and
upregulate endothelial maturation markers during in vitro
cultivation. J Tissue Eng Regen Med. 3:512–520. 2009.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Khakoo AY and Finkel T: Endothelial
progenitor cells. Annu Rev Med. 56:79–101. 2005.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Zwaginga JJ and Doevendans P: Stem
cell-derived angiogenic/vasculogenic cells: Possible therapies for
tissue repair and tissue engineering. Clin Exp Pharmacol Physiol.
30:900–908. 2003.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Li Z, Qian P, Shao W, Shi H, He XC, Gogol
M, Yu Z, Wang Y, Qi M, Zhu Y, et al: Suppression of m6A
reader Ythdf2 promotes hematopoietic stem cell expansion. Cell Res.
28:904–917. 2018.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Jian D, Wang Y, Jian L, Tang H, Rao L,
Chen K, Jia Z, Zhang W, Liu Y, Chen X, et al: METTL14 aggravates
endothelial inflammation and atherosclerosis by increasing FOXO1
N6-methyladeosine modifications. Theranostics. 10:8939–8956.
2020.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Zhang M, Yang C, Ruan X, Liu X, Wang D,
Liu L, Shao L, Wang P, Dong W and Xue Y: CPEB2 m6A methylation
regulates blood-tumor barrier permeability by regulating splicing
factor SRSF5 stability. Commun Biol. 5(908)2022.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Ping XL, Sun BF, Wang L, Xiao W, Yang X,
Wang WJ, Adhikari S, Shi Y, Lv Y, Chen YS, et al: Mammalian WTAP is
a regulatory subunit of the RNA N6-methyladenosine
methyltransferase. Cell Res. 24:177–189. 2014.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Wang J, Zhang J, Ma Y, Zeng Y, Lu C, Yang
F, Jiang N, Zhang X, Wang Y, Xu Y, et al: WTAP promotes myocardial
ischemia/reperfusion injury by increasing endoplasmic reticulum
stress via regulating m6A modification of ATF4 mRNA.
Aging (Albany NY). 13:11135–11149. 2021.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Wei W, Sun J, Zhang H, Xiao X, Huang C,
Wang L, Zhong H, Jiang Y, Zhang X and Jiang G: Circ0008399
Interaction with WTAP promotes assembly and activity of the
m6A methyltransferase complex and promotes cisplatin
resistance in bladder cancer. Cancer Res. 81:6142–6156.
2021.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Wang S, Zhang J, Wu X, Lin X, Liu XM and
Zhou J: Differential roles of YTHDF1 and YTHDF3 in embryonic stem
cell-derived cardiomyocyte differentiation. RNA Biol. 18:1354–1363.
2021.PubMed/NCBI View Article : Google Scholar
|