Introduction
Breast cancer (BC) is a malignant tumor with high
incidence rates in female patients, which impacts both health and
quality of life (1). According to
data released by the International Agency for Research on Cancer,
part of the World Health Organization, there were 2.26 million new
cases of BC and 680,000 deaths worldwide in 2020(2). The number of new cases of BC in China
was 420,000, resulting in ~120,000 deaths. Notably, an aging
population increases the overall risk of BC (3).
Glucose metabolism in tumor cells is characterized
by increased glucose uptake and aerobic glycolysis. Since aerobic
glycolysis occurs in cancer cells, the efficiency of sugar
utilization is notably higher than that of healthy cells. Despite
sufficient oxygen, cancer cells use aerobic glycolysis to produce
energy (4). Aerobic glycolysis
plays a fundamental role in cytoskeletal remodeling and cell
motility in BC (5). In addition,
aerobic glycolysis promotes the migration and proliferation of BC
cells, and inhibits apoptosis (6).
These results indicate that aerobic glycolysis serves an important
role in BC cells.
Homeobox (HOX) genes are highly conserved at the
genomic level, and play a key role in the regulation of numerous
biological processes, including apoptosis, cell differentiation,
transfer and receptor signal transduction (7). As a member of the HOX gene family,
HOXC13 is a homologous box gene and is a key transcription factor
for the growth and development of mammals (8). A previous study demonstrated that
HOXC13 expression is higher in BC than in gastric, colon and other
types of cancer, and results of a survival analysis demonstrated
that high expression levels of HOX transcript antisense RNA and
HOXC13 are associated with a poor prognosis (9). However, the role of HOXC13 in BC is
yet to be fully elucidated. Notably, HOXC13 can promote the
proliferation of esophageal squamous cell carcinoma by inhibiting
caspase 3 transcription (10). In
addition, when compared with healthy tissue, HOXC13 mRNA expression
levels are increased in lung cancer tissue. HOXC cluster antisense
RNA 2 (HOXC-AS2) overexpression has been reported to significantly
activate HOXC13 protein and mRNA expression, and HOXC-AS2 gene
knockdown can inhibit the proliferation and migration of lung
cancer cells (11). In addition,
HOXC13 knockdown reverses the increased proliferation and migration
of lung cells (11). HOXC13 can
also promote cervical cancer cell proliferation and invasion, and
the Warburg effect via the β-catenin/c-Myc signaling pathway
(12). Thus, it was hypothesized
that HOXC13 may serve a role in the proliferation, migration and
glycolysis of BC.
DNA methyltransferase 3α (DNMT3A) is highly
expressed in a variety of malignant tumors and is a key enzyme
required for the re-methylation of mammalian genes. DNMT3A may
serve as a potential therapeutic target in the prevention of cancer
recurrence (13,14). Downregulated DNMT3A expression
decreases the Warburg effect, proliferation and invasion of ovarian
cancer cells (15). In addition,
DNMT3A promotes colorectal cancer progression by regulating
DOC-2/DAB2 interactive protein-mediated MEK/ERK activation
(16). Upregulated DNMT3A
expression promotes progression of BC (17). However, the regulatory association
between HOXC13 and DNMT3A in BC, and the regulatory effects of
DNMT3A on BC cell proliferation, invasion and glycolysis are yet to
be fully elucidated.
Thus, the present study knocked down HOXC13 and
overexpressed DNMT3A concurrently to determine the roles of HOXC13
and DNMT3A in BC, and the underlying regulatory mechanisms in BC
cell proliferation, invasion and glycolysis.
Materials and methods
The application of UALCAN and JASPAR
databases
The UALCAN database based on The Cancer Genome Atlas
was used to predict HOXC13 expression in patients with BC (18). The JASPAR database was used to
predict promoter binding sites for the transcription factors HOXC13
and DNMT3A (19).
Cell culture
The human BC cell lines MCF-7 (cat. no. TCHu 74),
BT-549 (cat. no. TCHu 93) and MDA-MB-231 (cat. no. TCHu227) were
purchased from The Cell Bank of Type Culture Collection of The
Chinese Academy of Sciences. Human mammary epithelial MCF-10A cells
(cat. no. BNCC337734) were purchased from BeNa Culture Collection
(Beijing Beina Chunglian Institute of Biotechnology). Cells were
cultured in DMEM (Gibco; Thermo Fisher Scientific, Inc.)
supplemented with 10% fetal bovine serum (FBS; Gibco; Thermo Fisher
Scientific, Inc.) at 37˚C in a humidified atmosphere containing 5%
CO2.
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA was isolated from human mammary epithelial
MCF-10A cells and human BC cells using TRIzol® reagent
(Invitrogen; Thermo Fisher Scientific, Inc.) and
reverse-transcribed into cDNA using the PrimeScript RT Reagent kit
(Takara Bio, Inc.), according to the manufacturer's protocol. qPCR
was performed using the SYBR Green kit (Takara Bio, Inc.) on the
ABI 7500 Real-Time PCR System (Applied Biosystems; Thermo Fisher
Scientific, Inc.). Thermocycling conditions were as follows:
Initial denaturation for 5 min at 95˚C, followed by 40 cycles of 15
sec at 95˚C and 34 sec at 60˚C and extension at 72˚C for 30 sec,
with a final extension step at 72˚C for 10 min. GAPDH was used as
the internal control and expression levels were calculated using
the 2-ΔΔCq method (20). The sequences of primers used for
RT-qPCR were as follows: HOXC13 forward, 5'-CCATAACCGAACCCACGGAA-3'
and reverse, 5'-AATTGGGGCCATTCGGGATT-3'; DNMT3A forward,
5'-GGCCATACGGTGGAGCC-3' and reverse, 5'-TGTTGAGCCCTCTGGTGAAC-3' and
GAPDH forward, 5'-AATGGGCAGCCGTTAGGAAA-3' and reverse,
5'-GCGCCCAATACGACCAAATC-3'.
Western blot analysis
Total cellular protein was extracted using RIPA
lysis buffer (Beyotime Institute of Biotechnology) on ice. The
total protein concentration was measured with a BCA protein assay
kit (Takara Biotechnology Co., Ltd.). A total of 30 µg/lane protein
was separated by SDS-PAGE on 10% gels and transferred onto PDVF
membranes (MilliporeSigma) that were then blocked with 5% BSA for 2
h at room temperature. The following antibodies were added and
incubated at 4˚C overnight: Anti-HOXC13 (1:1,000; cat. no.
ab168368; Abcam), anti-E-cadherin (1:1,000; cat. no. ab40772;
Abcam), anti-N-cadherin (1:1,000; cat. no. ab76011; Abcam),
anti-Vimentin (1:1,000; cat. no. ab92547; Abcam), anti-hexokinase
II (HK2; 1:1,000; cat. no. ab209847; Abcam), anti-pyruvate kinase
M2 (PKM2; 1:1,000; cat. no. ab85555; Abcam), anti-DNMT3A (1:1,000;
cat. no. ab188470; Abcam) and anti-GAPDH (1:1,000; cat. no. ab8245;
Abcam). Horseradish peroxidase-conjugated anti-rabbit IgG antibody
(1:5,000; cat. no. ab288151; Abcam) or anti-mouse IgG antibody
(1:2,000; cat. no. ab6728; Abcam) was added and incubated at 4˚C
overnight. Protein bands were visualized using an enhanced
chemiluminescence (ECL)-Plus kit (Thermo Fisher Scientific, Inc.).
Blots were analyzed using ImageJ software (1.42q; National
Institutes of Health).
Cell transfection
Cells were placed into 6-well plates until they
reached 70-80% confluence. Small interfering (si)RNAs targeting
HOXC13 (si-HOXC13#1 and si-HOXC13#2) and the corresponding negative
control (NC; si-NC) were purchased from GeneChem, Inc.; the
sequences were as follows: si-HOXC13#1 sense,
5'-UUAACAUUAAAUACUCUUCUG-3' and antisense,
5'-GAAGAGUAUUUAAUGUUAAGG-3'; si-HOXC13#2 sense,
5'-UCCUUAACAUUAAAUACUCUU-3' and antisense,
5'-GAGUAUUUAAUGUUAAGGAAA-3' and si-NC sense,
5'-UUCUCCGAACGUGUCACGUTT-3' and antisense,
5'-ACGUGACACGUUCGGAGAATT-3'. pcDNA3.1 vectors containing
full-length HOXC13 gene were used and empty pcDNA3.1 vectors
(Shanghai GenePharma, Co., Ltd.) were used as the negative control.
pcDNA3.1 containing HOXC13 (Oe-HOXC13) and DNMT3A (Oe-DNMT3A) and
the corresponding NC plasmid (Oe-NC) were synthesized by Shanghai
GenePharma, Co., Ltd. Transfection with aforementioned plasmids and
siRNAs at a final concentration of 10 nM was performed using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) at 37˚C for 48 h, according to the manufacturer's
instructions and the transfection was completed. Further
experiments were performed after 48 h.
Cell Counting Kit (CCK)-8
Transfected BC cells were seeded in 96-well plates
at the density of 5x103 cells/well. After 24, 48 and 72
h of culture, CCK-8 solution (cat. no. ab228554; Abcam) was added
to each well for 2 h at 37˚C, according to the manufacturer's
instructions. The absorbance in each well was measured using a
microplate reader at 450 nm.
5-ethynyl-2'-deoxyuridine (EdU)
staining
Cell proliferation was determined via uptake of EdU
into DNA using a Click-iT EdU Microplate Assay kit (Invitrogen;
Thermo Fisher Scientific, Inc.), according to the manufacturer's
instructions. Cells were seeded into 96-well plates
(3x103 cells/well) and 10 µl EdU solution was added to
each well for 18 h at 37˚C. Following nuclear staining with DAPI
for 30 min at room temperature, cells were visualized under a
BZ-8000 fluorescence microscope (Keyence Corporation).
Apoptosis assay
The apoptosis of cells was assessed via Annexin
V-FITC/PI staining (Invitrogen; Thermo Fisher Scientific, Inc.) and
flow cytometry, according to the manufacturer's instructions. The
cells were washed with PBS and the cell concentration was adjusted
to 7x104 cells/ml. Cells were resuspended in 500 µl
binding buffer containing 5 µl Annexin V-FITC and 5 µl PI
(Invitrogen; Thermo Fisher Scientific, Inc.). Samples were
incubated at 4˚C for 15 min in the dark and analyzed using BD
FACSAria flow cytometry (BD Biosciences) with excitation at 488 nm
and emission measured at 560 nm. Data were analyzed using CellQuest
16.0 software (BD Biosciences).
Transwell assay
The Transwell assay was conducted using a Transwell
chamber coated with Matrigel (pore size, 8 µm; BD Biosciences) for
30 min at 37˚C. A total of 2x104 cells in serum-free
medium were plated in the upper chambers, and medium containing 20%
FBS was added to the lower chambers. After 24 h at 37˚C, cells in
the upper chambers were removed, and remaining cells were fixed
with 4% methanol for 40 min at 4˚C. After staining with 0.1%
crystal violet at room temperature for 30 min, migrated cells were
visualized under a light microscope and counted using ImageJ
software (1.42q; National Institutes of Health).
Wound healing assay
Cells were plated in 6-well plates at
1x105 cells/well using serum-free DMEM at 37˚C for 24 h
until cells reached 90% confluence. Linear wounds were created
using a 200-µl pipette tip and cells were incubated overnight at
37˚C with 5% CO2. Wounds were imaged at 0 and 24 h under
a light microscope. Cell migration rate=(0 h scratch width-scratch
width after culture)/0 h scratch width x100.
Extracellular acidification rate
(ECAR) assay
Seahorse XF96 Extracellular Flux Analyzer (Seahorse
Bioscience) was used for measuring ECAR. Cells (density,
1x105) were plated in XF96 cell culture plates (Seahorse
Bioscience) and incubated at 37˚C for 8 h. Following the addition
of glucose, oligomycin (oxidative phosphorylation inhibitor) and
2-DG (glycolytic inhibitor), cells were equilibrated with
bicarbonate-free buffered DMEM (Gibco; Thermo Fisher Scientific,
Inc.) for 1 h at 37˚C without CO2, immediately before
the extracellular flux assay.
3-bromopyruvate (3BP; Sigma-Aldrich; Merck KGaA)
solution in assay medium (final concentration, 10X; Gibco; Thermo
Fisher Scientific, Inc.) was loaded into the sensor cartridge
(final concentration, 20 or 100 µM). The instrument was set to
acquire consecutive measurements for 1 h with mixing. 3BP was
injected after acquiring one measurement as the baseline ECAR.
Lactic acid measurement
A lactic acid assay kit (cat.no. A019-2-1; Nanjing
Jiancheng Bioengineering Institute) was used to measure lactic acid
production, according to the manufacturer's instructions. Cells
were counted directly using ImageJ software (1.42q; National
Institutes of Health) for data normalization and to determine the
final amount of lactate production.
Glucose consumption
Glucose consumption was determined using a Glucose
Assay kit (Sigma-Aldrich; Merck KGaA), according to the
manufacturer's instructions. Cells were seeded in a 96-well plate
(density, 1x104) in serum-free DMEM (Gibco; Thermo
Fisher Scientific, Inc.) and 10 µl/well 2-deoxy-d-glucose was added
at 37˚C for 30 min. A total of 50 µl/well 2-deoxy-d-glucose uptake
assay working solution was added and cells were incubated at room
temperature for 90 min. The optical density ratio at 570/610 nm was
measured using a spectrophotometer (Thermo Fisher Scientific,
Inc.).
Luciferase reporter assay
DNA fragments containing wild-type (WT) or mutant
sequences for HOXC13 binding were synthesized and cloned into
luciferase reporter vectors (pGL3-Basic; Addgene, Inc.). DNMT3A
wild-type or mutant plasmids and Oe-HOXC13 or Oe-NC were
co-transfected into cells using Lipofectamine 3000 (Invitrogen;
Thermo Fisher Scientific, Inc.) for 48 h, according to the
manufacturer's protocol. Transfected cells (1x105 per
well) were seeded into 96-well plates and luciferase activity was
determined 48 h after transfection using
Dual-Luciferase® Reporter (DLR™) Assay System (Promega
Corporation), according to the manufacturer's instructions. Firefly
luciferase activity was normalized to Renilla luciferase
activity.
Chromatin immunoprecipitation
(ChIP)
A ChIP assay was performed using a EZ-Magna ChIP A/G
kit (cat. no. 17-10086; Merck KGaA). DNA and protein were
cross-linked in 1% formaldehyde for 10 min at 37˚C, extracted using
300 µl SDS lysis buffer (EMD Millipore) and lysed using sonication
(20 kHz; 4 pulses of 12 sec each, followed by 30 sec rest on ice
between each pulse). Anti-HOXC13 antibody (1:100; cat. no.
ab168368; Abcam) and IgG antibody (1:100; cat. no. ab90285; Abcam)
were used for incubation with the supernatants. Following
purification of the precipitated DNA, RT-qPCR was performed as
aforementioned and the same primer pairs were used as those used
for qPCR.
Statistical analysis
All data were analyzed using SPSS software (version
26.0; IBM Corp.) and are presented as the mean ± standard
deviation. All experiments were performed in triplicate. Unpaired
Student's t-test was used for comparison between two groups and one
way-ANOVA followed by Tukey's post hoc test was used for comparison
between multiple groups. Survival analysis was performed using the
Kaplan-Meier method. P<0.05 was considered to indicate a
statistically significant difference.
Results
HOXC13 is highly expressed in BC
The UALCAN database demonstrated that HOXC13
expression was significantly increased in the tissues of patients
with BC compared with the normal tissues from healthy individuals
(Fig. 1A) and high HOXC13
expression in BC based on the criteria (Cutoff-High, median 50% and
Cutoff-Low, median 50%) was associated with a poor prognosis
(Fig. 1B). In addition, HOXC13
expression in BC was associated with tumor stage (Fig. 1C) and regional lymph node
involvement (Fig. 1D). RT-qPCR and
western blot analysis demonstrated that HOXC13 expression in BC
cell lines was significantly increased compared with that in
MCF-10A cells (Fig. 1E). HOXC13
expression was highest in MDA-MB-231 cells; thus, these were
selected for use in subsequent experiments. In summary, HOXC13
displayed increased expression in BC tissues and cells.
HOXC13 knockdown inhibits the
viability, proliferation and induces apoptosis of BC cells
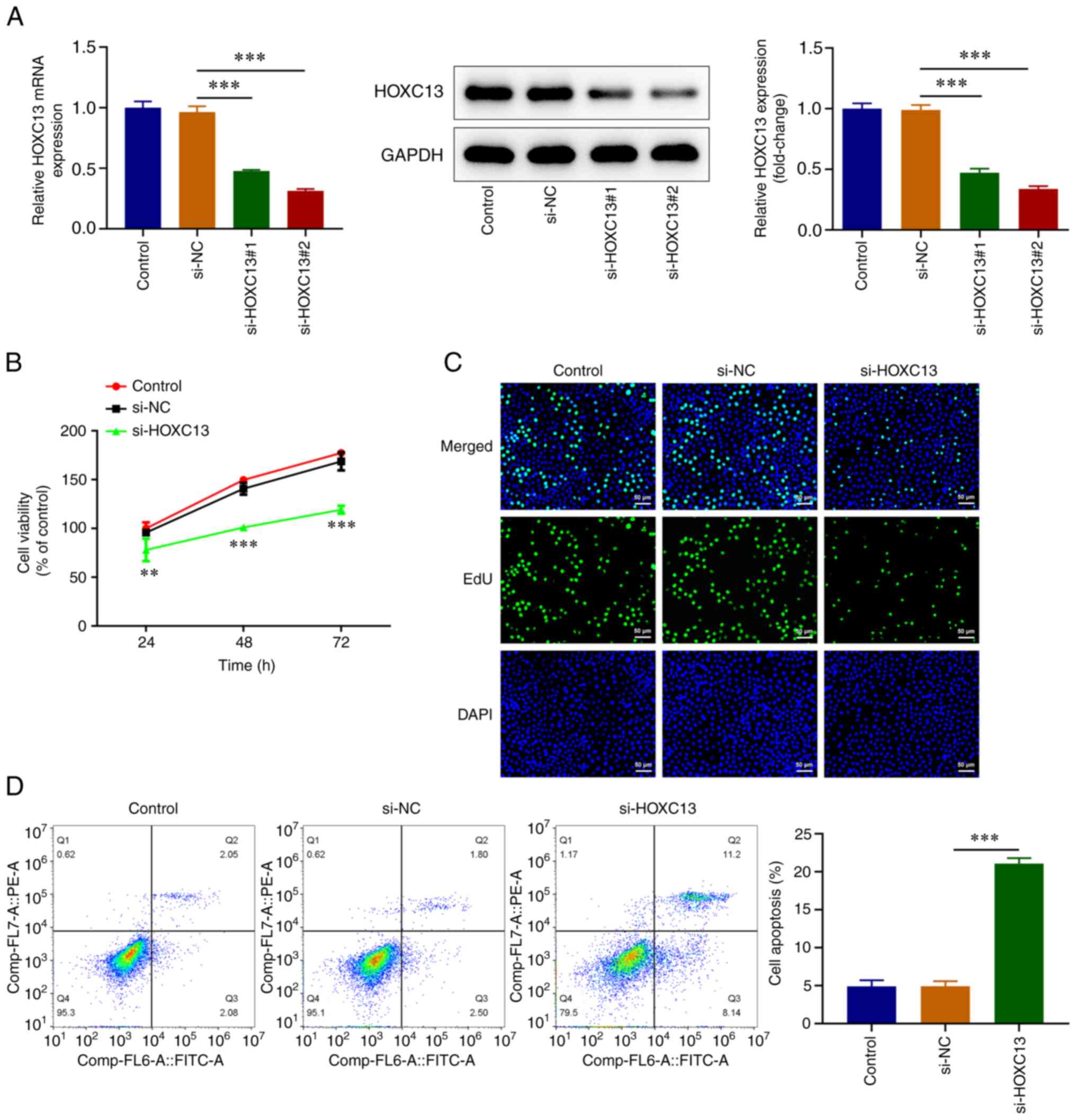
HOXC13 knockdown was performed via transfection, and
efficiency was determined using RT-qPCR and western blot analysis.
Transfection with si-HOXC13#1 and si-HOXC13#2 led to HOXC13
knockdown and transfection with si-HOXC13#2 led to the highest
reductions in HOXC13 expression. Thus, si-HOXC13#2 was selected for
subsequent experiments (Fig. 2A).
The CCK-8 assay demonstrated that cell viability was significantly
decreased in the si-HOXC13 group compared with that in the si-NC
group (Fig. 2B). In addition, EdU
staining revealed that cell proliferation was markedly decreased
following HOXC3 knockdown (Fig.
2C). Flow cytometry demonstrated that apoptosis was
significantly increased following HOXC13 knockdown (Fig. 2D). Overall, HOXC13 depletion
obstructed the viability and proliferation while it stimulated the
apoptosis of BC cells.
HOXC13 knockdown inhibits the
migration, invasion and EMT of BC cells
Cell migration and invasion were measured using
wound healing and Transwell assays. The results of the present
study demonstrated that migration and invasion were significantly
decreased in the si-HOXC13 group compared with that in the si-NC
group (Fig. 3A and B). The expression levels of
epithelial-mesenchymal transition (EMT)-associated proteins, namely
E-cadherin, N-cadherin and Vimentin, were detected using western
blot analysis. The expression levels of E-cadherin were
significantly increased in the si-HOXC13 group compared with those
in the si-NC group, whereas the expression levels of N-cadherin and
Vimentin were significantly decreased (Fig. 3C). To sum up, HOXC13 interference
impeded the migration, invasion and EMT of BC cells.
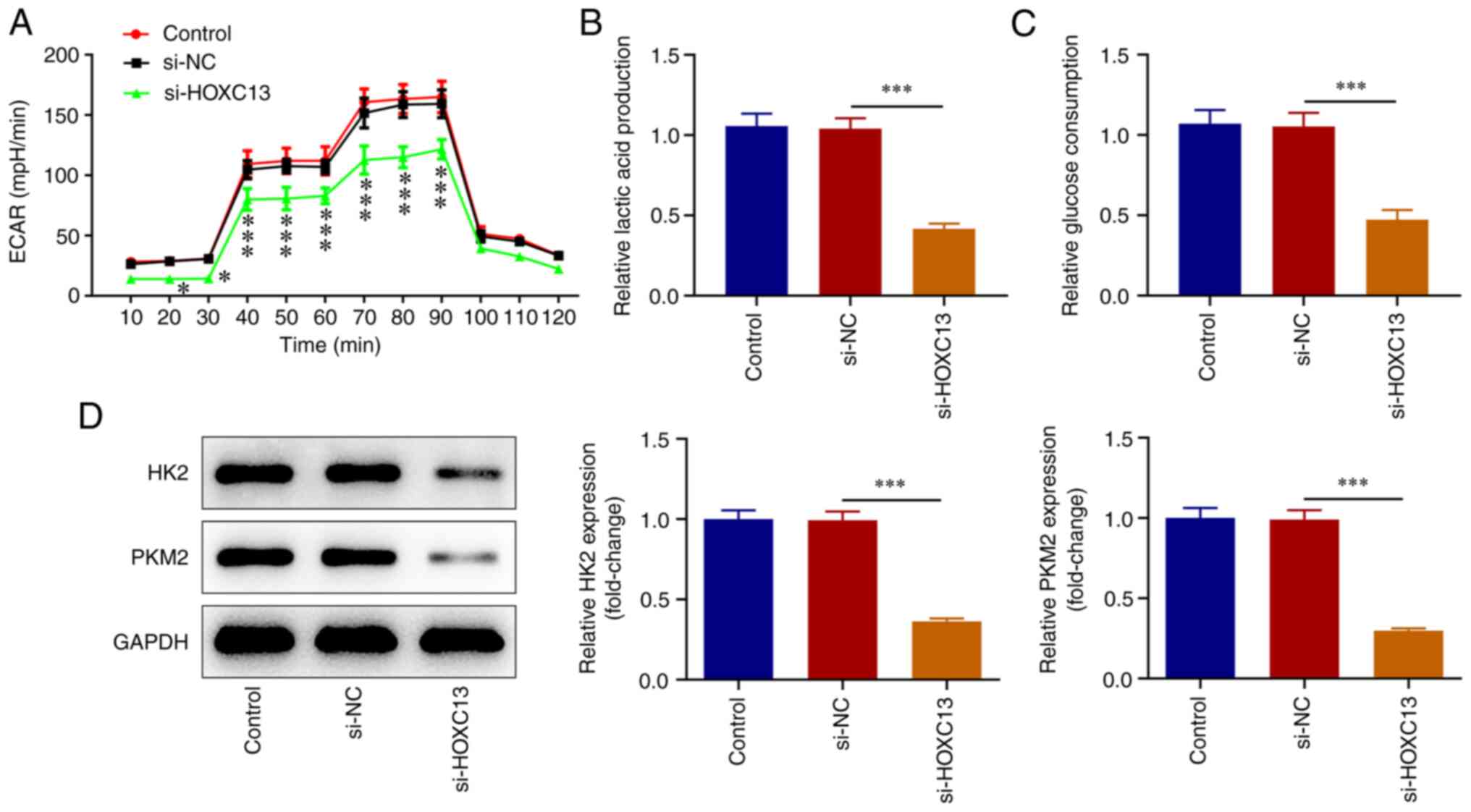
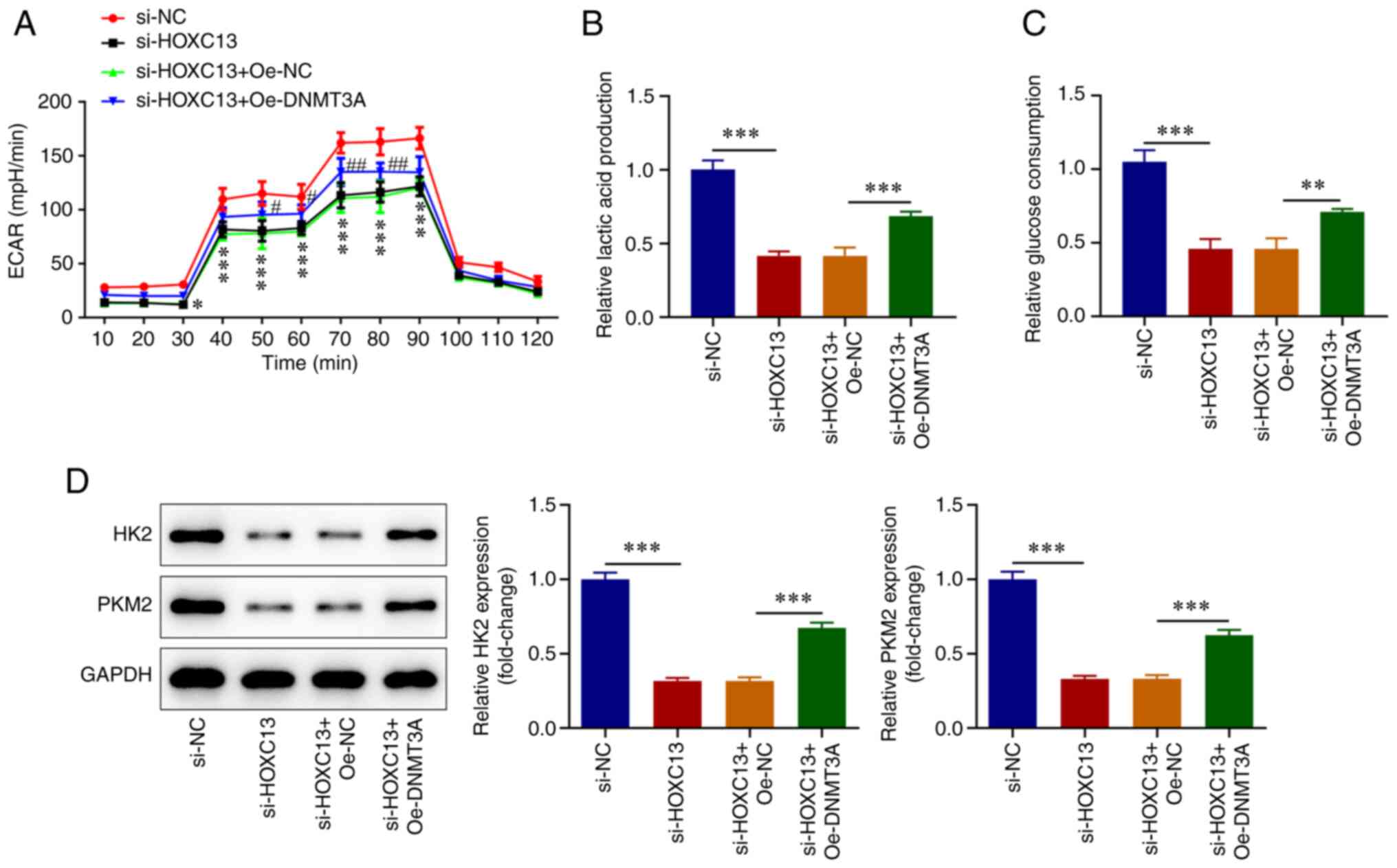
HOXC13 knockdown inhibits glycolysis
of BC cells
XF96 extracellular flux analyzer was used to detect
ECAR. The results of the present study revealed that cell ECAR at
20-90 min were significantly decreased following HOXC13 knockdown
(Fig. 4A). Moreover, lactic acid
production (Fig. 4B), glucose
consumption (Fig. 4C), and the
expression levels of HK2 and PKM2 were significantly decreased
(Fig. 4D) by HOXC13 knockdown.
Accordingly, HOXC13 silencing impeded the glycolysis of BC
cells.
HOXC13 positively regulates DNMT3A
transcription in BC cells
The JASPAR database predicted the binding between
HOXC13 and promoter sites of DNMT3A (Fig. 5A). RT-qPCR and western blot
analysis revealed that DNMT3A expression was significantly
increased in MDA-MB-231 cells compared with that in MCF-10A cells
(Fig. 5B). Subsequently, the
Oe-HOXC13 plasmid was constructed, and cells were divided into the
Control, Oe-NC and Oe-HOXC13 groups. RT-qPCR and western blot
analysis were used to detect transfection efficiency of HOXC13
overexpression plasmids and it was discovered that HOXC13
expression was significantly elevated following transfection of
Oe-HOXC13 (Fig. 5C). The
dual-luciferase and ChIP assays revealed that HOXC13 bound to the
DNMT3A promoter in MDA-MB-231 cells compared with MCF-10A cells
(Fig. 5D and E). RT-qPCR and western blot analysis
demonstrated that DNMT3A expression was significantly increased in
the Oe-HOXC13 group compared with that in the Oe-NC group. By
contrast, DNMT3A expression was significantly decreased in the
si-HOXC13 group compared with the si-NC group (Fig. 5F). Collectively, HOXC13 was a
transcription activator of DNMT3A in BC cells.
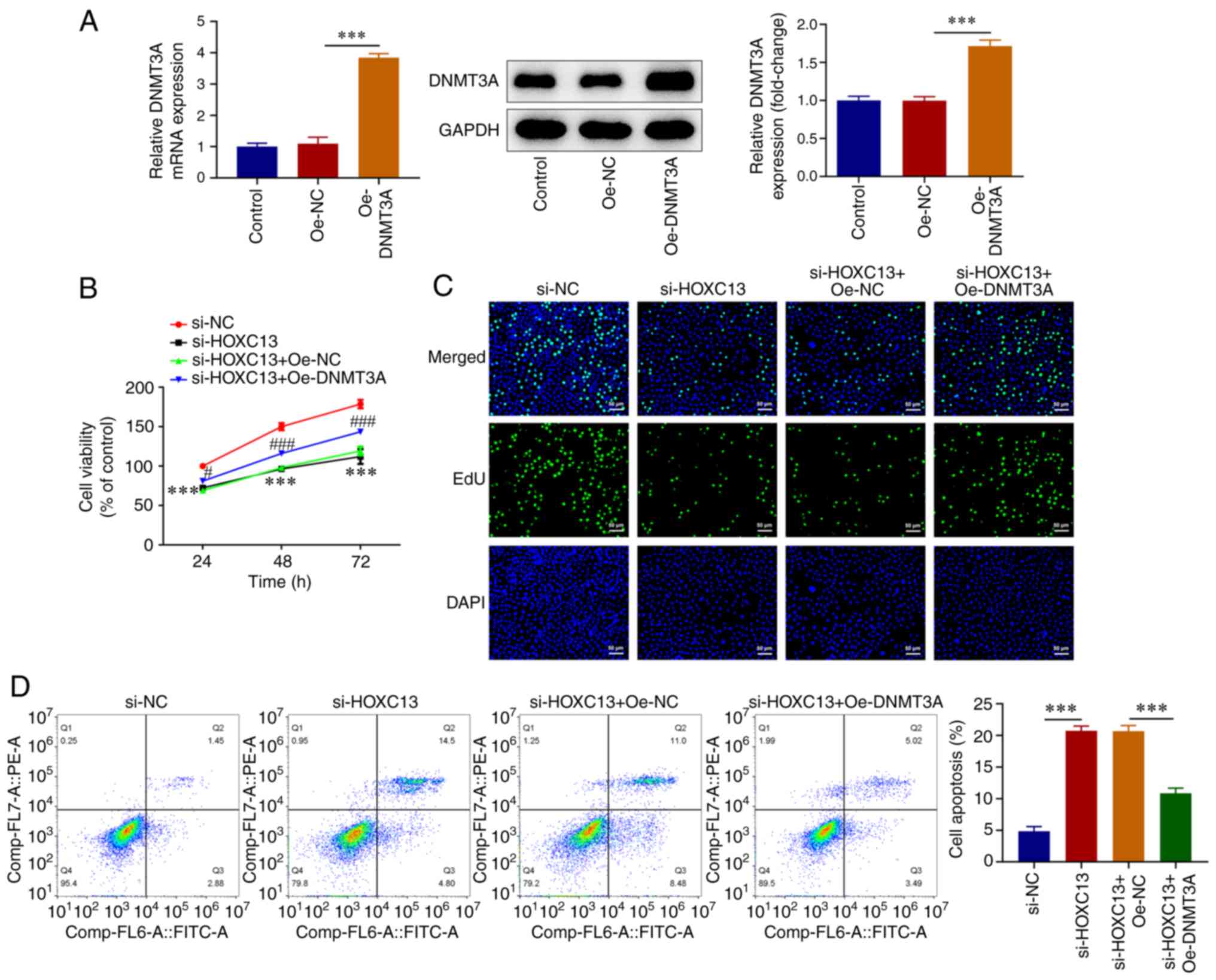
HOXC13 regulates DNMT3A in malignant
progression of BC
To investigate the mechanisms underlying HOXC13
regulation of the malignant progression of BC, an Oe-DNMT3A plasmid
was constructed (Fig. 6A). Cells
were divided into the si-NC, si-HOXC13, si-HOXC13 + Oe-NC and
si-HOXC13 + Oe-DNMT3A groups. The CCK-8 and EdU assays demonstrated
that cell viability was markedly increased in the si-HOXC13 +
Oe-DNMT3A group compared with that in the si-HOXC13 + Oe-NC group
(Fig. 6B and C). Flow cytometry revealed that apoptosis
was significantly decreased in the si-HOXC13 + Oe-DNMT3A group
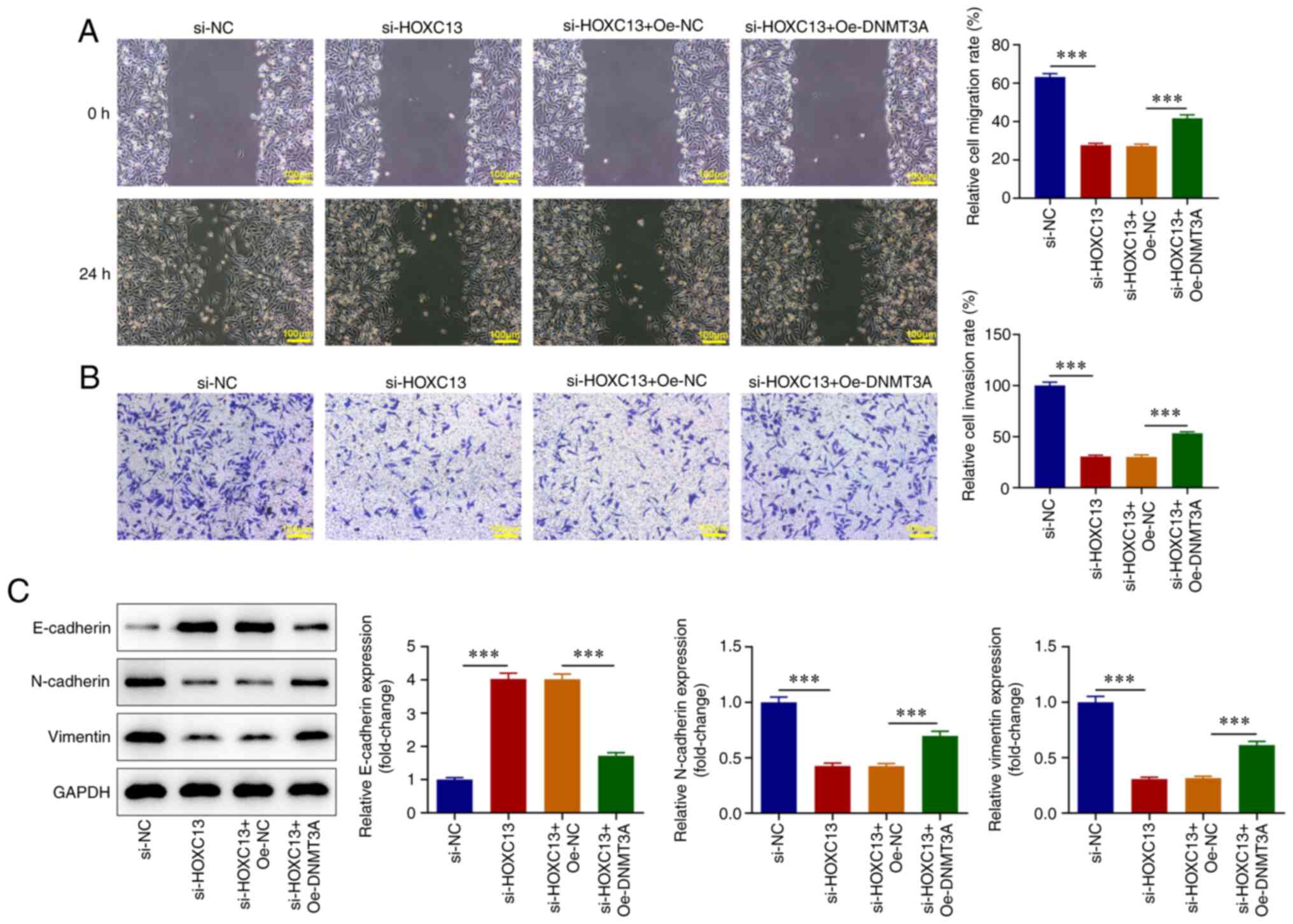
compared with that in the si-HOXC13 + Oe-NC group (Fig. 6D). Wound healing and Transwell
assays revealed that DNMT3A overexpression significantly reversed
the inhibitory effects of HOXC13 knockdown on BC cell invasion and
migration (Fig. 7A and B). Western blot analysis demonstrated
that E-cadherin expression levels were significantly decreased, and
N-cadherin and Vimentin expression levels were significantly
increased in the si-HOXC13 + Oe-DNMT3A group compared with those in
the si-HOXC13 + Oe-NC group (Fig.
7C). Results obtained using the XF96 extracellular flux
analyzer demonstrated that in the si-HOXC13 + Oe-DNMT3A group, ECAR
at 30-80 min, lactate levels (Fig.
8A and B), glucose consumption
(Fig. 8C), and HK2 and PKM2
expression levels were significantly increased (Fig. 8D) compared with in the si-HOXC13 +
Oe-NC group. Taken together, HOXC13 functioned in the aggressive
behaviors of BC cells by regulating DNMT3A.
Discussion
Previous studies have demonstrated that HOXC13 is
associated with the occurrence and development of hair and nails
(21,22). Moreover, other studies have
reported that HOX13 is significantly highly expressed in
ameloblastoma, odontogenic tumor, melanoma and liposarcoma compared
with healthy tissues (23-26).
In addition, HOXC13 knockdown significantly inhibits the
proliferation of colon cancer cells and induces cell cycle arrest
(27). As a transcription factor,
HOXC13 regulates the expression of numerous key genes, thus
affecting the occurrence and development of tumors. Zinc finger
protein 521, a proto-oncogene in B cells that causes leukemia, is
co-regulated by HOXC13(28). A
previous study demonstrated that HOXC13 plays a role in promoting
lung cancer via upregulation of cyclin D1 and cyclin E1 expression
(29). Using the UALCAN database,
the present study demonstrated that HOXC13 was highly expressed in
patients with BC, and this was associated with a poor prognosis and
tumor staging of BC. Therefore, the present study aimed to
determine the function and mechanism of HOXC13 in BC. BC and
healthy mammary epithelial cells were used and the results were
consistent with those predicted using the UALCAN database. Notably,
the present study demonstrated that HOXC13 expression was
significantly increased in BC cell lines. These results indicated
the role of HOXC13 in BC; further investigations into the specific
functions and mechanisms are required.
Results from UALCAN database in the current study
demonstrated that HOXC13 was highly expressed in BC, and was
associated with patient prognosis and tumor stage. Notably, cell
proliferation, invasion and metastasis may be indicative of the
malignant potential of tumors. Numerous studies have demonstrated
that inhibition of the aforementioned functions may impact the
malignant progression of tumor cells (30-32).
In addition, glycolysis is a key metabolic characteristic of cells
in the process of tumor development (33). Not only does glycolysis provide
rapid energy for tumor cells, intermediate metabolites generated in
the process are also crucial precursors for alternate metabolic
pathways and glycolysis may provide raw materials for the synthesis
of various biological macromolecules (34). Therefore, inhibition of aerobic
glycolysis may be used in the clinical treatment of tumors. The
present study demonstrated that the proliferation, invasion,
migration and EMT of BC cells were decreased, and glycolysis was
inhibited following HOXC13 knockdown. These results indicated that
HOXC13 serves a role in promoting BC; further investigations into
the specific mechanisms are required.
Transcription factors serve a key role in tumor
development. The covalent binding domains of transcription factors
and DNA either inhibit or enhance gene transcription (35). As a member of the HOX gene family,
HOXC13 contains homologous domains that facilitate transcription
factor functions (9). The promoter
binding sites of transcription factors HOXC13 and DNMT3A were
predicted using the JASPAR database. Moreover, the regulatory
association between HOXC13 and DNMT3A in BC cells was verified
using dual-luciferase reporter gene and ChIP assays. The present
study demonstrated that DNMT3A expression was significantly
increased in BC cell lines. A previous study demonstrated that
lysine methyltransferase 2C deficiency promotes small cell lung
cancer metastasis through DNMT3A-mediated epigenetic reprogramming
(36). Moreover, DNMT3A
epigenetically regulates key microRNAs (miRs) involved in EMT in
prostate cancer (37). Ginsenoside
20 (S)-Rg3 that is identified as an active saponin monomer derived
from red ginseng inhibits DNMT3A expression in SKOV3 ovarian cancer
cells, reversing the DNMT3A-mediated methylation of the miR-532-3p
host gene promoter. This subsequently increases miR-532-3p levels,
and inhibits HK2 and PKM2 expression levels to inhibit the Warburg
effect (26). The aforementioned
results indicate that DNMT3A exerts a regulatory effect on tumor
proliferation, metastasis and glycolysis. The present study
demonstrated that DNMT3A overexpression in BC cells reversed the
inhibitory effects of HOXC13 knockdown on the viability,
proliferation, migration, invasion, EMT and glycolysis of tumor
cells.
The present study had the limitation that it did not
investigate the expression of HOXC13 in patients and animals with
BC. In future, HOXC13 expression in patients with BC will be
explored.
In conclusion, HOXC13 promoted cell viability,
proliferation, migration, invasion, EMT and glycolysis in BC by
regulating DNMT3A.
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
HL and LQ conceived and designed the study, analyzed
data, and wrote and revised the manuscript. PG, HC, JZ, XZ, GL and
LW performed the experiments. HL and LQ confirm the authenticity of
all the raw data. All authors have read and approved the final
manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Katsura C, Ogunmwonyi I, Kankam HK and
Saha S: Breast cancer: Presentation, investigation and management.
Br J Hosp Med (Lond). 83:1–7. 2022.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global cancer statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249.
2021.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Bose S and Le A: Glucose metabolism in
cancer. Adv Exp Med Biol. 1063:3–12. 2018.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Shiraishi T, Verdone JE, Huang J, Kahlert
UD, Hernandez JR, Torga G, Zarif JC, Epstein T, Gatenby R,
McCartney A, et al: Glycolysis is the primary bioenergetic pathway
for cell motility and cytoskeletal remodeling in human prostate and
breast cancer cells. Oncotarget. 6:130–143. 2015.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Wu Z, Wu J, Zhao Q, Fu S and Jin J:
Emerging roles of aerobic glycolysis in breast cancer. Clin Transl
Oncol. 22:631–646. 2020.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Krumlauf R: Hox genes, clusters and
collinearity. Int J Dev Biol. 62:659–663. 2018.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Ishii Y, Taguchi A and Kukimoto I: The
homeobox transcription factor HOXC13 upregulates human
papillomavirus E1 gene expression and contributes to viral genome
maintenance. FEBS Lett. 594:751–762. 2020.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Li C, Cui J, Zou L, Zhu L and Wei W:
Bioinformatics analysis of the expression of HOXC13 and its role in
the prognosis of breast cancer. Oncol Lett. 19:899–907.
2020.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Luo J, Wang Z, Huang J, Yao Y, Sun Q, Wang
J, Shen Y, Xu L and Ren B: HOXC13 promotes proliferation of
esophageal squamous cell carcinoma via repressing transcription of
CASP3. Cancer Sci. 109:317–329. 2018.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Liu B, Li J, Li JM, Liu GY and Wang YS:
HOXC-AS2 mediates the proliferation, apoptosis, and migration of
non-small cell lung cancer by combining with HOXC13 gene. Cell
Cycle. 20:236–246. 2021.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Dai M, Song J, Wang L, Zhou K and Shu L:
HOXC13 promotes cervical cancer proliferation, invasion and Warburg
effect through β-catenin/c-Myc signaling pathway. J Bioenerg
Biomembr. 53:597–608. 2021.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Okano M, Bell DW, Haber DA and Li E: DNA
methyltransferases Dnmt3a and Dnmt3b are essential for de novo
methylation and mammalian development. Cell. 99:247–257.
1999.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Yang X, Han H, De Carvalho DD, Lay FD,
Jones PA and Liang G: Gene body methylation can alter gene
expression and is a therapeutic target in cancer. Cancer Cell.
26:577–590. 2014.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Lu J, Zhen S, Tuo X, Chang S, Yang X, Zhou
Y, Chen W, Zhao L and Li X: Downregulation of DNMT3A attenuates the
warburg effect, proliferation, and invasion via promoting the
inhibition of miR-603 on HK2 in ovarian cancer. Technol Cancer Res
Treat. 21(15330338221110668)2022.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Zhou Y, Yang Z, Zhang H, Li H, Zhang M,
Wang H, Zhang M, Qiu P, Zhang R and Liu J: DNMT3A facilitates
colorectal cancer progression via regulating DAB2IP mediated
MEK/ERK activation. Biochim Biophys Acta Mol Basis Dis.
1868(166353)2022.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Li Y, Jiang B, He Z, Zhu H, He R, Fan S,
Wu X, Xie L and He X: circIQCH sponges miR-145 to promote breast
cancer progression by upregulating DNMT3A expression. Aging (Albany
NY). 12:15532–15545. 2020.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Chandrashekar DS, Bashel B, Balasubramanya
SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi BVSK and
Varambally S: UALCAN: A portal for facilitating tumor subgroup gene
expression and survival analyses. Neoplasia. 19:649–658.
2017.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Fornes O, Castro-Mondragon JA, Khan A, van
der Lee R, Zhang X, Richmond PA, Modi BP, Correard S, Gheorghe M,
Baranasic D, et al: JASPAR 2020: Update of the open-access database
of transcription factor binding profiles. Nucleic Acids Res.
48(D1):D87–D92. 2020.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Wang S, Li F, Liu J, Zhang Y, Zheng Y, Ge
W, Qu L and Wang X: Integrative analysis of methylome and
transcriptome reveals the regulatory mechanisms of hair follicle
morphogenesis in Cashmere Goat. Cells. 9(969)2020.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Fernandez-Guerrero M, Yakushiji-Kaminatsui
N, Lopez-Delisle L, Zdral S, Darbellay F, Perez-Gomez R, Bolt CC,
Sanchez-Martin MA, Duboule D and Ros MA: Mammalian-specific
ectodermal enhancers control the expression of Hoxc genes in
developing nails and hair follicles. Proc Natl Acad Sci USA.
117:30509–30519. 2020.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Li J, Zhang B, Wang B and Zhang X: LncRNA
HOXC-AS5 affects the proliferation, invasion and cell cycle of
ameloblastoma cells by acting on the target gene HOXC13. Cell Mol
Biol (Noisy-le-grand). 68:124–134. 2022.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Hong YS, Wang J, Liu J, Zhang B, Hou L and
Zhong M: Expression of HOX C13 in odontogenic tumors. Shanghai Kou
Qiang Yi Xue. 16:587–591. 2007.PubMed/NCBI(In Chinese).
|
|
25
|
Cantile M, Scognamiglio G, Anniciello A,
Farina M, Gentilcore G, Santonastaso C, Fulciniti F, Cillo C,
Franco R, Ascierto PA and Botti G: Increased HOX C13 expression in
metastatic melanoma progression. J Transl Med.
10(91)2012.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Cantile M, Galletta F, Franco R, Aquino G,
Scognamiglio G, Marra L, Cerrone M, Malzone G, Manna A, Apice G, et
al: Hyperexpression of HOXC13, located in the 12q13 chromosomal
region, in well-differentiated and dedifferentiated human
liposarcomas. Oncol Rep. 30:2579–2586. 2013.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Zhou Y, Zheng X, Lu J, Chen W, Li X and
Zhao L: Ginsenoside 20(S)-Rg3 inhibits the warburg effect via
modulating DNMT3A/MiR-532-3p/HK2 pathway in ovarian cancer cells.
Cell Physiol Biochem. 45:2548–2559. 2018.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Yu M, Al-Dallal S, Al-Haj L, Panjwani S,
McCartney AS, Edwards SM, Manjunath P, Walker C, Awgulewitsch A and
Hentges KE: Transcriptional regulation of the proto-oncogene Zfp521
by SPI1 (PU.1) and HOXC13. Genesis. 54:519–533. 2016.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Yao Y, Luo J, Sun Q, Xu T, Sun S, Chen M,
Lin X, Qian Q, Zhang Y, Cao L, et al: HOXC13 promotes proliferation
of lung adenocarcinoma via modulation of CCND1 and CCNE1. Am J
Cancer Res. 71820–1834. (eCollection 2017)2017.PubMed/NCBI
|
|
30
|
Liu Z, Zhang L, Chen W, Yuan F, Yang Z,
Liu S and Le F: miR-195-5p regulates cell proliferation, apoptosis,
and invasion of thyroid cancer by targeting telomerase reverse
transcriptase. Bioengineered. 12:6201–6209. 2021.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Fan C, Wang Q, van der Zon G, Ren J,
Agaser C, Slieker RC, Iyengar PV, Mei H and Ten Dijke P: OVOL1
inhibits breast cancer cell invasion by enhancing the degradation
of TGF-β type I receptor. Signal Transduct Target Ther.
7(126)2022.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Fu Y, Zhang X, Liu X, Wang P, Chu W, Zhao
W, Wang Y, Zhou G, Yu Y and Zhang H: The DNMT1-PAS1-PH20 axis
drives breast cancer growth and metastasis. Signal Transduct Target
Ther. 7(81)2022.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Abbaszadeh Z, Cesmeli S and Biray Avci C:
Crucial players in glycolysis: Cancer progress. Gene.
726(144158)2020.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Abdel-Wahab AF, Mahmoud W and Al-Harizy
RM: Targeting glucose metabolism to suppress cancer progression:
Prospective of anti-glycolytic cancer therapy. Pharmacol Res.
150(104511)2019.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Lambert M, Jambon S, Depauw S and
David-Cordonnier MH: Targeting transcription factors for cancer
treatment. Molecules. 23(1479)2018.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Na F, Pan X, Chen J, Chen X, Wang M, Chi
P, You L, Zhang L, Zhong A, Zhao L, et al: KMT2C deficiency
promotes small cell lung cancer metastasis through DNMT3A-mediated
epigenetic reprogramming. Nat Cancer. 3:753–767. 2022.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Mancini M, Grasso M, Muccillo L, Babbio F,
Precazzini F, Castiglioni I, Zanetti V, Rizzo F, Pistore C, De
Marino MG, et al: DNMT3A epigenetically regulates key microRNAs
involved in epithelial-to-mesenchymal transition in prostate
cancer. Carcinogenesis. 42:1449–1460. 2021.PubMed/NCBI View Article : Google Scholar
|