Introduction
Asthma is a common complex disease characterized by
chronic respiratory tract inflammation, reversible airflow
obstruction and airway hyper-responsiveness (1). Asthma can be induced by multiple
environmental factors, such as cold and dry air and airborne
substances, including chemical fumes, dust and pollen (1). Additionally, genetics plays an
important role in the onset and the heritability for asthma is as
high as ~80% (1). To discover the
potential genetic contribution to asthma, numerous genome-wide
association studies (GWASs) have been performed, and one
single-nucleotide polymorphism (SNP), rs7130588, located at 11q13.5
was observed to be associated with asthma, including both
adult-onset and childhood-onset, in Caucasian populations (2-6).
It had been suggested that there are ~80 million
genetic variations in human genome (7). However, owing to the space limit of
the microarray, only a small number (usually ~500,000) can be
included and genotyped by GWAS. Therefore, it cannot be discounted
that the real causal SNP(s) for asthma might not be rs7130588, but
the one(s) in linkage disequilibrium (LD) with it. However, this
question has not yet been scrutinized. In addition, since rs7130588
is located at the intergenic region between EMSY transcriptional
repressor, BRCA2 interacting (EMSY; also known as C11orf30)
and leucine-rich repeat-containing 32 (LRRC32; also known as
glycoprotein-A repetitions predominant), with the latter gene
encoding for a protein involved in immune response, it had been
hypothesized that rs7130588 may be associated with asthma by
regulating LRRC32 expression (2). However, owing to the relative long
distance (~110.0 kb) between this SNP and the LRRC32
promoter, this interaction has never been demonstrated. As a
result, this locus was termed as ‘EMSY (or
C11orf30)-LRRC32’ in previous studies (8-14).
The present study attempted to reveal the functional
variation(s) in 11q13.5 associated with asthma and its mechanism.
Through public genotype data analysis, potential SNPs associated
with asthma were identified. Dual-luciferase assay were used to
characterize the causal SNP. Furthermore, the present study used a
functional genomics approach to investigate the underlying
mechanism for asthma.
Materials and methods
1000 Genomes Project (1KGP) data
analysis
The upstream and downstream 500 kb genetic sequence
flanking rs7130588 for three representative populations, including
CEU (Utah Residents with European Ancestry), CHB (Han Chinese in
Beijing) and YRI (Yoruba in Ibadan; Nigeria), were downloaded from
1KGP website (http://www.internationalgenome.org). The LD pattern
was determined using the Genome Variation Server version 150
(http://gvs.gs.washington.edu/GVS150),
with r2≥0.8.
Dual luciferase assay
The rs7130588 and rs6592645 flanking region (~1.5
kb; ~750 bp in each side) was amplified by nested PCR with primers
listed in Table SI, digested by
KpnI and BglII (New England Biolabs) and inserted
into pGL3-promoter vector (Promega Corporation). To avoid
artificial mutations, PCR was performed using Phusion High-Fidelity
DNA Polymerase (Thermo Fisher Scientific, Inc.) with the following
thermocycling conditions: 98˚C for 30 sec; followed by 35 cycles of
98˚C for 10 sec, 56˚C for 30 sec and 72˚C for 30 sec. Following
sequencing by BigDye terminator (Thermo Fisher Scientific, Inc.),
plasmids containing another allele were generated by mutagenesis
using the Q5 Site-directed Mutagenesis Kit (New England Biolabs)
and primers listed in Table SI.
Prior to transfection, all plasmids were sequenced to verify the
sequence and the haplotype orientation.
The human lung bronchial epithelial cell line
Beas-2B was purchased from Conservation Genetics CAS (Chinese
Academy of Science) Kunming Cell Bank (http://www.kmcellbank.com; cat. no. KCB200922YJ) and
maintained in Dulbecco's modified Eagle's medium (high glucose;
HyClone; Cytiva) with 10% FBS (Biological Industries USA, Inc.) in
5% CO2 at 37˚C. Beas-2B Cells (~105
cells/well) were plated into 24-well plate and 475 ng plasmid
constructs were transfected by Lipofectamine® 2000
(Invitrogen; Thermo Fisher Scientific, Inc.) at 37˚C for 12 h.
After transfection, cells were cultured for additional 36 h at 37˚C
and harvested. The luciferase activity was determined by
Dual-Luciferase Reporter Assay System (Promega Corporation). A
total of six independent transfections were performed for each
plasmid. In each transfection, the Renilla luciferase
control plasmid pRL-TK (25 ng; Promega Corporation) plasmid was
cot-transfected and used to normalize the transfection
efficiency.
Chromosome conformation capture
(3C)
Spatial contacts between the enhancers and the
promoters of nearby genes were examined by 3C and quantitative PCR
(qPCR). Briefly, ~108 Beas-2B cells were cross-linked by
formaldehyde (1% final concentration) and lysed by lysis buffer (10
mM Tris-HCl, 10 mM NaCl, 0.2% NP40, pH 8.0), and the chromatin was
digested by restrictive enzyme (HindIII or EcoRI; see
below). After ligation, DNA was purified by standard
phenol-chloroform method (https://www.thermofisher.cn/cn/zh/home/references/protocols/nucleic-acid-purification-and-analysis/dna-extraction-protocols/phenol-chloroform-extraction.html).
Along with the Beas-2B cells, related bacterial artificial
chromosomes (BACs) containing the enhancer and nearby region were
ordered from BACPAC Resources Center (https://bacpacresources.org/), cultured in E.
coli, isolated, digested, ligated and quantified by qPCR with
iQ SYBR green (Bio-Rad Laboratories, Inc.; cat. no. 1725122) and
the primers in Tables SII and
SIII with the following
thermocycling conditions: 96˚C for 10 min; followed by 40 cycles of
96˚C for 10 sec and 60˚C for 30 sec. Owing to the distance between
rs6592645 and the promoter of nearby genes, in addition to the
complex restrictive enzyme map, 3C was performed separately for the
upstream and the downstream regions of rs6592645. For upstream
region 3C, HindIII (New England Biolabs) was chosen to
digest chromatin. The two BACs RP11-269F1 and RP11-795A14 (BACPAC
Resources Center) were mixed with the same amount (10 µg for each)
and used as control. By contrast, EcoRI (New England
Biolabs) and BAC RP11-672A2 were utilized in downstream region 3C.
In the current study, unidirectional primers were designed to
anchor three protein-coding genes [GVQW motif-containing 3
(GVQW3), THAP domain-containing 12 (THAP12) and
EMSY] for the upstream region, and one protein-coding gene
(LRRC32) for the downstream one, this enhancer and several
random genome regions (Tables SII
and SIII). GVQW3 and
THAP12 utilize different strands of the same fragment as the
promoter. Therefore, these two genes occupied the same position in
the assay.
The enrichment for chromatin was evaluated using the
2-ΔΔCq method (15).
All PCR products were verified by sequencing. Three repeats were
performed for each unidirectional anchor primer.
The potential unknown genes were searched in two
lncRNA databases; NONCODE (http://www.noncode.org) and GENCODE (https://www.gencodegenes.org).
Tissue collection and LRRC32
expression quantification
A total of 46 lung tissues were collected from
Department of Respiratory Critical Care Medicine, the First
Affiliated Hospital of Kunming Medical University (Kunming, China)
between January 2016 and June 2016. The detail of the donors can be
found in Table I. All donors were
suspected of lung cancer and thus tissues were sectioned for
pathological examination. After examination, the remaining tissues
were used for research. Among the donors, 22 individuals were
diagnosed as asthma according to global initiative for asthma
(16). None of the patients were
eventually diagnosed with lung cancer and no other comorbidities
were reported. All available tissues (22 asthma and 24 controls)
were included, and no individuals were excluded. All participants
were Han Chinese and provided written informed consent. The present
study was approved by ethics committee of Shaanxi Normal University
(approval number 20170308).
 | Table ICharacteristics of the tissue
donors. |
Table I
Characteristics of the tissue
donors.
| Characteristic | Asthma group | Control group |
|---|
| Sample size | 22 | 24 |
| Sex (M/F) | 12/10 | 14/10 |
| Age
distribution | 51-70 | 47-69 |
RNA was isolated by TRIzol® (Thermo
Fisher Scientific, Inc.). cDNA was generated by reverse
transcription with RevertAid First Strand cDNA Synthesis Kit
(Thermo Fisher Scientific, Inc.). LRRC32 expression was
quantified by qPCR with primer pair 5'-GCATAGCAACGTGCTGATGGAC-3'
and 5'-GATGCTGTTGCAGCTCAGGTCT-3'; GAPDH expression was also
measured as a control with primer pair 5'-GAAGGTGAAGGTCGGAGTC-3'
and 5'-GAAGATGGTGATGGGATTTC-3'. qPCR was performed by relative
standard curve approach (17);
that is, a cDNA mixture was serial diluted and thus a standard
curve was generated. Gene expression was determined by the position
of Cq in standard curve. The reagent and thermocycling condition
were the same as the aforementioned qPCR. A total of three repeats
were performed for each individual.
RNA-seq and expression quantitative
trait locus (eQTL) analysis
RNA-seq data (SRA format) for lymphoblastoid cell
lines (LCL) from two studies (18,19)
were obtained from the SRA database (https://www.ncbi.nlm.nih.gov/sra; accession numbers:
PRJNA357867 and PRJNA122271) and converted into fastq format by SRA
toolkit (https://github.com/ncbi/sra-tools). After alignment
with LRRC32 mRNA sequence (GenBank ID NM_005512.3) by
bowtie2 (http://bowtie-bio.sourceforge.net/bowtie2/index.shtml),
the expression was calculated by eXpress (https://pachterlab.github.io/eXpress) using the
default parameters and reported as fragments per kilobase of
transcript per million fragments mapped (FPKM). The genotype for
LCL was obtained from the HapMap project (https://www.genome.gov/10001688/international-hapmap-project),
and linear regression was performed between genotype and
LRRC32 expression by SPSS 20.0 Statistics (IBM Corp.).
Chromatin immunoprecipitation
(ChIP)
The online program Match (http://www.gene-regulation.com/cgi-bin/pub/programs/match/bin/match.cgi)
was used to identify potential transcription factors. ChIP was
carried out by EZ ChIP Kit (MilliporeSigma) according to the
manufacturer's protocol. Briefly, ~1x107 Beas-2B cells
were cross-linked with formaldehyde (1% final concentration) at
room temperature for 10 min. After washing with phosphate buffered
saline (Beijing Solarbio Science & Technology Co., Ltd.), cells
were scraped, lysed by lysis buffer (MilliporeSigma; cat. no.
20-163), sonicated (2% magnitude, 60 sec at 4˚C) into small
fragments (~400-800 bp) and precleared with 60 µl protein A beads
(MilliporeSigma; cat. no. 16-201C) by centrifuge at 3,000 x g for 1
min at 4˚C. The protein/chromatin complex (1 ml) was captured by
adding 2 µg mouse anti-transcription factor 3 (TCF3) antibody
(Santa Cruz Biotechnology, Inc.; cat. no. sc-133074) or 2 µg normal
mouse IgG (Santa Cruz Biotechnology, Inc.; cat. no. sc-2025) as a
control, and precipitated with 60 µl protein A beads. Following
washing with 1 ml Low Salt, High Salt, LiCl and TE wash buffers
(MilliporeSigma; cat. nos. 20-154, 20-155, 20-156 and 20-157;
respectively), the immunoprecipitated protein/chromatin complex was
dissolved and de-crosslinked by adding 8 µl 5M NaCl and incubating
at 65˚C overnight. Protein was removed by proteinase K (Roche
Diagnostics) digestion and DNA was purified by supplied column in
aforementioned EZ ChIP Kit. In brief, DNA was binding to the spin
filter, washed by reagent A and resolved in reagent B. qPCR was
used to evaluate the enrichment of the obtaining DNA with primer
pair 5'-ATAGCATTAGATTGTTGTTCTGC-3' and 5'-ACGGAGATGATGGGTGAGA-3'.
This primer pair was designed to bind rs6592645 surrounding region.
The qPCR was performed by relative standard curve method (17). The reagent and thermocycling
condition were identical with the aforementioned qPCR. Three
repeats were performed for each experiment.
Electrophoretic mobility shift assay
(EMSA)
Nuclear extracts were prepared and quantified using
a Nuclear and Cytoplasmic Protein Extraction kit (Beyotime
Institute of Biotechnology; cat. no. P0027). The probes for both
alleles of rs6592645 are shown in Table SIV; they were synthesized by
Sangon Biotech Co., Ltd., labeled with biotin using an EMSA probe
biotin labeling kit (Beyotime Institute of Biotechnology, cat. nos.
GS008) and incubated with nuclear extracts (5 µg) of Beas-2B cells
at 37˚C for 30 min. The probe/protein complex was separated by
electrophoresis in nondenaturing polyacrylamide gel (6%) and
transferred to nylon membranes (Beyotime Institute of
Biotechnology). For each allele, two controls were also included.
One control was biotin-labeled probes alone while the other was
probe/protein complex incubating with competitor oligonucleotides
(non-labeled probes). After incubating with streptavidin-HRP
conjugate (Beyotime Institute of Biotechnology; cat. no. GS009),
the membrane was visualized in ECL chemiluminescence
(MilliporeSigma).
Statistical analysis
One-way ANOVA with Tukey's test was used to compare
the data from multiple groups, including luciferase activity, 3C
and partial LRRC32 expression. Independent Student's t-tests
were utilized to compare the difference of two groups, including
gene expression and ChIP results. All statistical analyses were
performed using SPSS 20.0 (IBM Corp.). P#x003C;0.05 was considered
to indicate a statistically significant difference.
Results
Genetic variations near rs7130588
As the causal SNP(s) for asthma might be those in LD
with rs7130588 (20,21), the present study investigated the
LD pattern in representative 1KGP populations. Within the 500 kb
region surrounding rs7130588, there are 2,253 SNPs for CEU, 2,279
SNPs for CHB and 3,776 SNPs for YRI. Among these SNPs, one was
located ~0.3 kb away, rs6592645, and showed complete LD with
rs7130588 (r2=1 for all three populations; Fig. S1), which indicated that rs6592645
and rs7130588 appear simultaneously. Therefore, rs6592645 should
also present differentially distribution in case and control groups
and be associated with asthma.
Another three SNPs were located ~30.6 kb away,
including rs7926914, rs7927894 and rs7927997, which are in complete
LD with each other (r2=1 for all three populations) and
are associated with Crohn's disease (22) and atopic dermatitis (23), presented a strong LD with rs7130588
in CEU (r2=0.937) but relatively weak LD in CHB
(r2=0.373) and YRI (r2=0.309) populations.
All other SNPs had relatively low LD with rs7130588 in these three
populations (all r2#x003C;0.530). Considering this LD
pattern and that these three SNPs in the microarray (or the tag
one, rs7927894) fail to reach a genome-wide significance level in
GWAS for asthma (24), we
hypothesized that rs7130588 and rs6592645 may be potential causal
SNPs for this disease.
Function of rs7130588 and
rs6592645
Since both rs7130588 and rs6592645 are located in
intergenic region of EMSY and LRRC32, it was
hypothesized that they might regulate gene expression by altering
enhancer activity (21). To
investigate this, the present study cloned the segment containing
rs7130588 and rs6592645 (~1.5 kb), created plasmids with the
corresponding alleles, performed transient transfections and
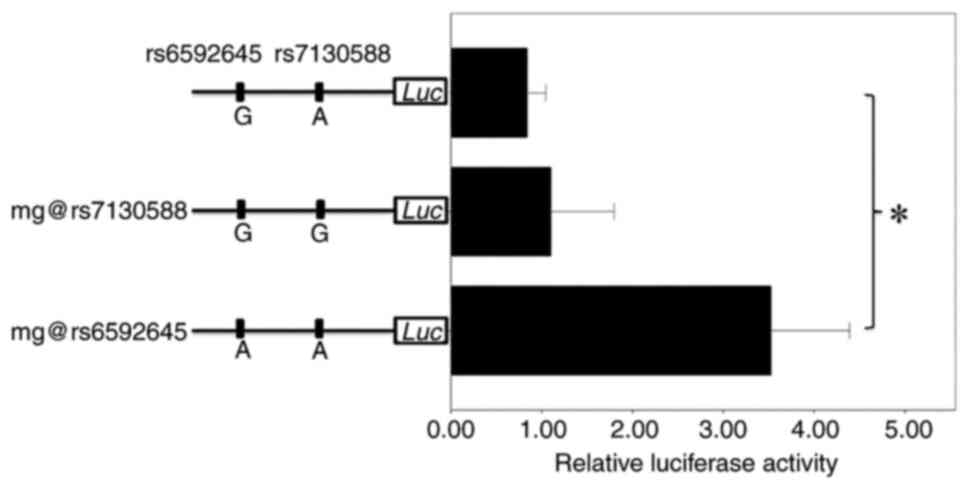
subsequent dual-luciferase assays. ANOVA indicated that there was
significant difference among the luciferase activity of these three
plasmids (P=0.000049; Fig. 1). To
explore the functional SNP, the present study further pairwisely
compared the luciferase activity by ANOVA with Tukey's test. As
shown in Fig. 1, the mutagenesis
at rs7130588 failed to alter the relative luciferase activity when
compared with the original plasmid construct (P=0.839), thus
indicating that rs7130588 is not functional in lung cell and
unlikely to be the causal SNP for asthma. By contrast, the A allele
of rs6592645 presented a significant increase in relative
luciferase activity compared with the G allele (P=0.000093), thus
suggesting that rs6592645 may be a causal SNP for asthma. Since the
risk allele is G of rs7130588 in GWAS (2) and that G of rs7130588 was in complete
LD with A of rs6592645, the causal variation for asthma should be A
of rs6592645.
Gene interaction with the enhancer
containing rs6592645
Since rs6592645 is within the intergenic region of
EMSY and LRRC32, it was hypothesized that this
mutation might be within an enhancer region and may have the
ability to modify enhancer activity (21). In addition, the histone
modification for rs6592645 surrounding region was searched in
ENCODE project (https://www.encodeproject.org), which includes
different ChIP-seq results for multiple cell lines. Since Beas-2B
cell line was not included in ENCODE, lung cancer cell line A549
was searched due to the similar origin of these two cell lines. As
shown in Fig. S2, there are
evident histone 3 lysine 4 (H3K4) monomethylation and H3K27
acetylation signals, which are two common histone modifications on
active enhancers (25),
surrounding rs6592645 region in lung cell. However, the regulatory
target remained undetermined. To investigated this, 3C was
performed to determine the potential spatial contacts. Owing to the
long distance between rs6952645 and nearby gene (EMSY and
LRRC32) promoter, it was hard to perform 3C in one
experiment. Therefore, the nearby region was divided into upstream
and downstream regions and these were investigated separately.
As shown in Fig.
2A, in the upstream region, the GVQW3/THAP12 (the
fifth point on the x-axis, ~179.1 kb away from rs6592645) and the
EMSY (the eighth point on the x-axis, ~115.0 kb away from
rs6592645) promoters failed to show any increases in ligation
efficiency, thus indicating that none of them are the target of
this enhancer. By contrast, the fourth, sixth and seventh point on
the x-axis displayed some increase in ligation frequency (Fig. 2A). However, no known protein-coding
genes are within these three restriction fragments. Previous
studies have suggested that long non-coding RNA (lncRNA; RNA with
length >200 base pairs but without protein-coding function) are
also involved in the onset of asthma (26,27).
Therefore, we hypothesized that there might be a novel lncRNA
within this region. To reveal the potential gene, this segment was
searched in two lncRNA databases; NONCODE and GENCODE. However, no
lncRNAs were observed to be within this segment. Therefore, it was
considered that this interaction in space between this segment and
the enhancer containing rs6592645 might be a random one and without
biological sense, which might result from the affinity between
these two genome regions and the same protein complex.
For the downstream region, the promoter of
LRRC32 showed a strong increase in ligation efficiency (the
seventh point on the x-axis, ~110.0 kb away from rs6592645;
Fig. 2B). Using ANOVA to compare
the interaction efficiency of this point with nearby ones, a
significant difference can be observed (P#x003C;0.001), which
indicated that the putative regulation target of this enhancer may
be LRRC32.
LRRC32 expression between asthma and
controls
Results from the luciferase assay indicated that the
putative causal allele, A of rs6592645, may induce high gene
expression. If LRRC32 is indeed involved in asthma onset,
then this gene should be overexpressed in patients with asthma. To
validate this, lung tissues from 22 patients with asthma and 24
healthy controls were collected, and LRRC32 expression
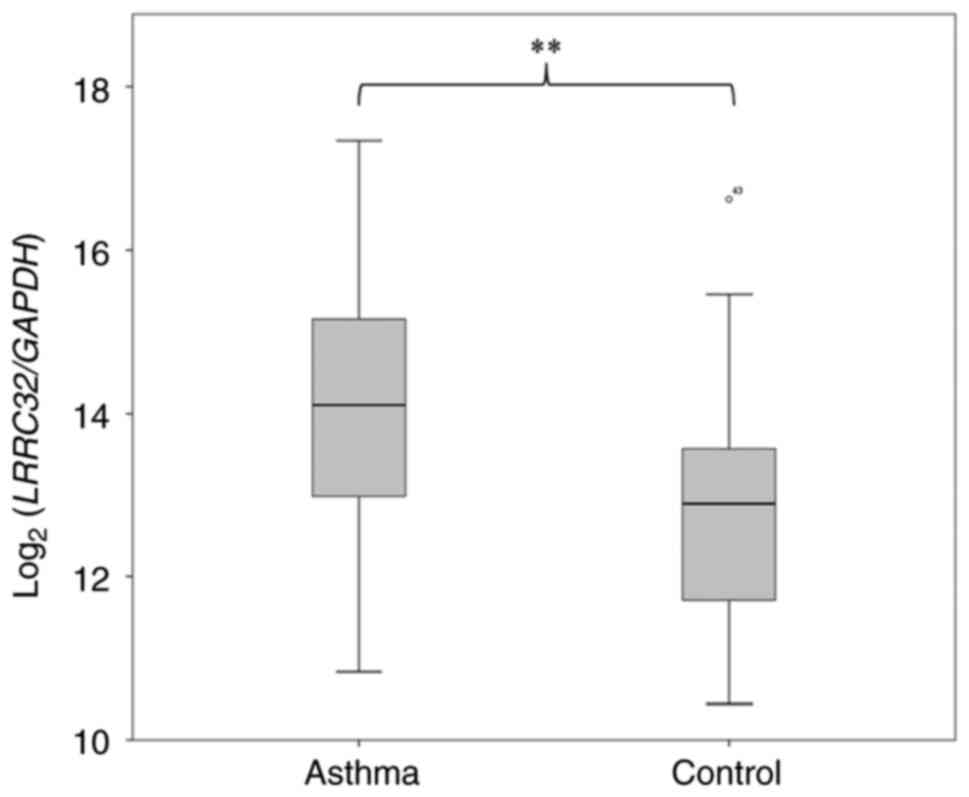
levels were quantified. As shown in Fig. 3, LRRC32 expression in
patients with asthma was significantly higher compared with that in
controls (P=0.0095), thus confirming this hypothesis.
Previous GWAS analysis has suggested that rs7130588
is significantly associated with atopic asthma, thus indicating
that this locus may be a risk factor mainly for allergic asthma
(2). Therefore, it is useful to
compare LRRC32 expression among allergic asthma and
controls. Among the cohort of the present study, only six
individuals were diagnosed with allergic asthma. ANOVA indicated
that there was significant difference among LRRC32
expression of these three groups (P=0.030). Further ANOVA with
Tukey's test indicated that no significant difference was observed
in LRRC32 expression between allergic asthma and
non-allergic asthma (P=0.55) or control (P=0.25; Fig. S3). By contrast, LRRC32
expression in non-allergic asthma was significantly higher compared
with the control patients (P=0.0060; Fig. S3). However, there must be caution
in interpreting this result, as the sample size was too small for
allergic asthma.
LRRC32 expression upon
stimulation
A number of environmental factors, including house
dust mites (HDMs) and rhinoviruses (RVs), can induce asthma. To
investigate whether LRRC32 may be involved in the response
of these two environmental factors, two RNA-seq datasets were
downloaded from SRA database (accession number PRJNA379624 and
PRJNA700069) and LRRC32 expression was analyzed as
aforementioned. In one dataset, the authors collected peripheral
blood mononuclear cells (PBMCs) from a number of individuals and
treated them with HDM extract for 48 h (28). As shown in Fig. S4, LRRC32 expression in HDM
extract treatment group was significantly higher compared with that
in the control group (P=6.40x10-9). In another cohort,
the authors isolated PBMCs and stimulated with RV for 24 h
(29). As shown in Fig. S5, LRRC32 expression in RV
stimulated group was significantly higher compared with that in the
non-stimulated group (P=0.000034). These data indicated that
LRRC32 may be involved in the immune response to HDM or
RV.
eQTL analysis
If rs6592645 can indeed influence LRRC32
expression, this locus could be an eQTL for this gene (21). Since enough lung tissues with known
genotype and expression data were not available for the present
study, the well-established model using LCL (30) cells was used to elucidate this.
Therefore, two published RNA-seq data for LCL were downloaded for
analysis (18,19) and the results are shown in Fig. 4. Since the individuals are not
included in 1KGP and rs6592645 genotype is not available in HapMap
project for one study (19),
rs7130588 genotype was retrieved from HapMap for analysis. This
study includes three populations, CEU, YRI and CHS (Southern Han
Chinese) (19). Since the genotype
for the CHS individuals from that study are not available from 1KGP
or HapMap, only the CEU and YRI patient data were included for the
present study analysis. In CEU, no association was observed between
rs7130588 genotype (A/A and A/G) and LRRC32 expression
(P=0.45; data not shown). In YRI, owing to the low frequency (~20%)
of rs7130588 G allele, no individuals were identified as homozygous
for this allele. Therefore, the present study used independent
samples t-test to compared LRRC32 expression levels between
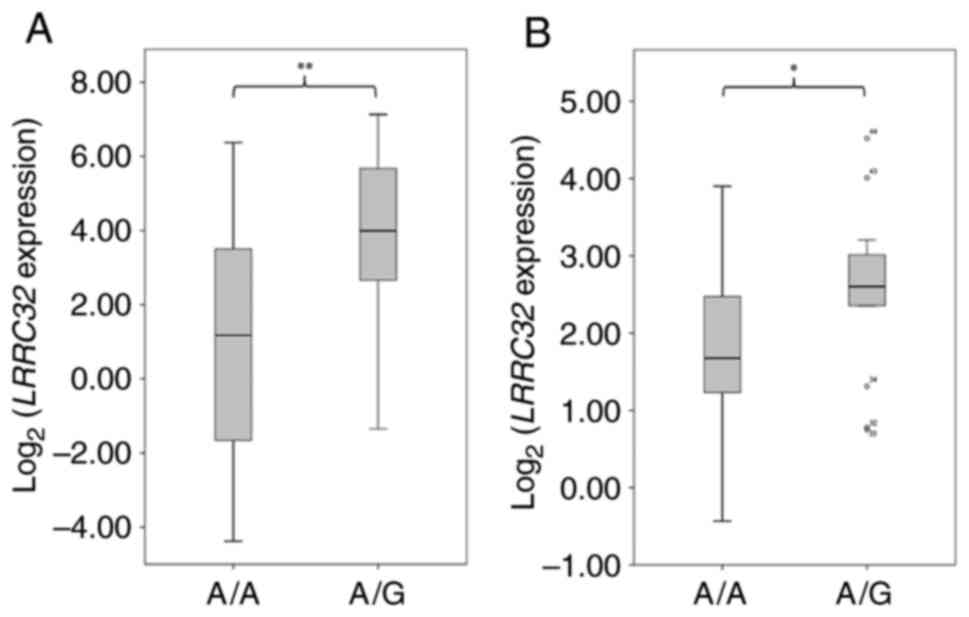
A/A and A/G groups. As shown in Fig.
4A, LRRC32 expression in A/G group (n=34) was
significantly higher compared with that in the A/A group (n=11;
P=0.003), which indicated that G allele of rs7130588 was associated
with higher LRRC32 expression. Since G of rs7130588 is in
complete LD with A of rs6592645 (Fig.
S1), it can be concluded that A of rs6592645 is also associated
with higher LRRC32 expression, which is consistent with our
luciferase assay (Fig. 1). In
another YRI RNA-seq dataset (18),
LRRC32 expression in A/G group (n=31) was significantly
higher compared with that in A/A group (n=13; P=0.03; Fig. 4B), which verified that this locus
may be an eQTL for LRRC32, at least in YRI population.
Transcription factor binding
rs6592645
Considering the location of rs6592645, it was
hypothesized that this mutation might be within a transcription
factor binding site and thus may alter interaction affinity
(21). Only TCF3 was predicted to
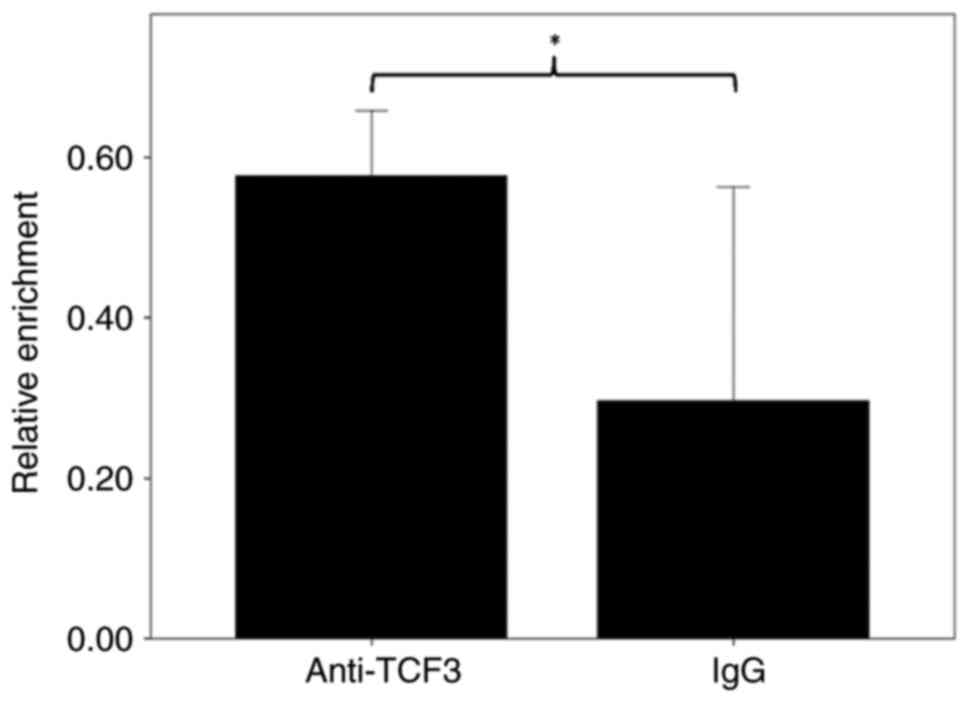
reside in the surrounding region of rs6592645. To verify this, ChIP
was performed with anti-TCF3 antibodies and the relative chromatin
enrichment quantified for rs6592645 surrounding region. As shown in
Fig. 5, the amount of
immunoprecipitated chromatin samples were significantly higher in
the TCF3 antibody experiment compared with IgG (P=0.025), thus
suggesting that TCF3 may interact with rs6592645 surrounding region
in lung cells.
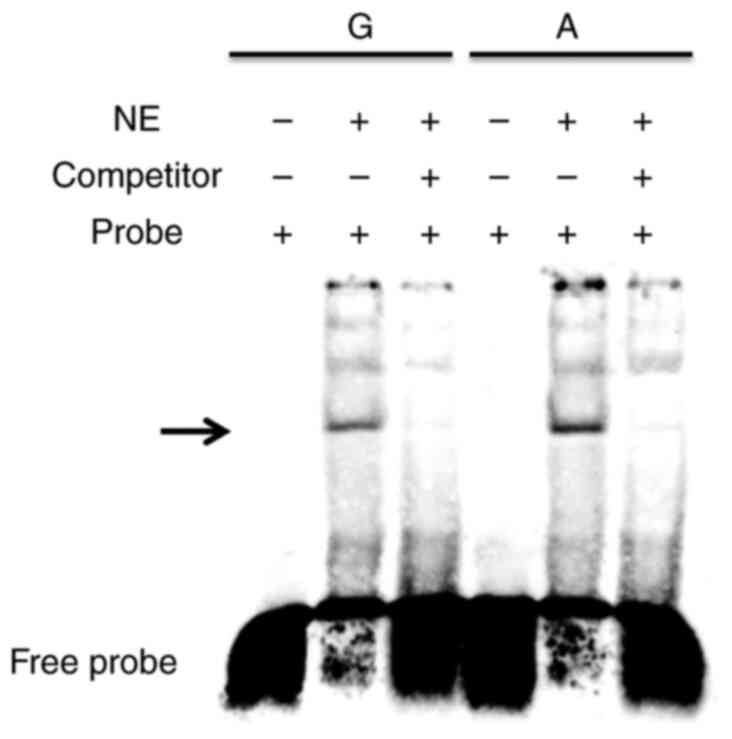
TCF3 binding affinity difference
between rs6592645 alleles
To investigate the binding efficiency between two
alleles of rs6592645, EMSA was performed. As shown in Fig. 6, the two alleles of rs6592645
showed bands with evidently different density, which indicated
apparent different affinity with nuclear proteins from Beas-2B
cells. The high expression allele, A of rs6592645, possesses a much
higher band density, which suggested a stronger binding
affinity.
Association between rs11236797
genotype and LRRC32 expression
In the ~10.6-31.1 kb downstream region of rs6592645,
there was another LD block containing 16 SNPs (rs2212434,
rs61893460, rs7126418, rs7110818, rs7114362, rs7936070, rs7936312,
rs7936323, rs7936434, rs4494327, rs11236791, rs10160518, rs2155219,
rs11236797, rs7931483 and rs7930763; pairwise
r2>0.85; Fig. S1)
in CEU. This LD block displays a moderate LD with rs6592645
(r2#x003C;0.62 for all SNPs; Fig. S1) in CEU. In recent GWAS for
Caucasians, this LD block is proposed to be associated with grass
sensitization, self-reported allergy, allergic sensitization,
eosinophilic esophagitis, alopecia areata, giant cell arteritis,
atopic march, gut microbiome, food allergy, asthma and the
aforementioned autoimmune disorders (https://www.ebi.ac.uk/gwas/variants/rs11236797).
Through functional genomics work, rs11236797 was proposed to be a
causal SNP that can also regulate LRRC32 expression in
Treg cells (31), which
is similar with the function of rs6592645. Considering the LD
pattern and the 28.6 kb distance between rs6592645 and rs11236797,
rs11236797 may be within an independent cis-regulatory
element for LRRC32 with rs6592645. However, eQTL analysis
was not conducted in a previous study (31). Notably, the GTEx search also failed
to verify the association between this locus and LRRC32
expression (data not shown).
Therefore, the present study also used the
aforementioned LCL RNA-seq datasets to perform eQTL analysis. Since
the genotype of rs11236797 is not available in HapMap, the genotype
of tag SNP rs2155219 was retrieved for analysis. In one cohort
(19), there was no significant
association between rs2155219 genotype and LRRC32 expression
in CEU (P=0.776) or YRI (P=0.332; data not shown). In another
cohort (18), rs2155219 genotype
is strongly associated with LRRC32 expression (linear
regression analysis, r=0.895, P#x003C;1x10-6; Fig. S6). In addition, the risk allele, T
of rs2155219 (or A in reverse strand; see GWAS catalog), is
associated with a higher LRRC32 expression (Fig. S6), which is similar to the results
of the present study.
Discussion
The present study identified a potential causal SNP
for asthma at 11q13.5 by population genetics and dual luciferase
assay. Using multiple functional genomics approaches, the
regulation target and underlying mechanism of the putative enhancer
were identified. These data established a putative connection
between a genetic marker in this locus and asthma
susceptibility.
TGF-β is a pleiotropic cytokine involved in immune
response, cell proliferation and differentiation, apoptosis,
carcinogenesis and other physiological processes (32). Considering the role of immune
response in asthma onset, TGF-β has been suggested to be a risk
factor and therapeutic target for asthma (33). LRRC32 is a transmembrane cell
surface protein and the docking receptor for latent TGF-β
(non-covalently bound of mature TGF-β and latency-associated
peptide; the latent form of TGF-β cannot bind with TGF-β receptor)
(34). Upon binding, LRRC32 can
tether latent TGF-β and enhance the activation of mature TGF-β
(29). Overexpression of LRRC32
can enhance TGF-β bioactivity, especially in regulatory T
lymphocytes (Treg) (35-37).
These data suggest an essential role of LRRC32 in immune system
(38,39). In this locus, the risk allele A of
rs6592645, can induce a significantly higher LRRC32
expression level, which may result in an exaggerated response and
further asthma susceptibility. In this regard, it was reported that
rs7130588 is significantly associated with serum IgE level
(14), which was consistent with
the hypothesis of the present study.
eQTL analysis from the present study indicated that
this locus is significantly associated with LRRC32
expression only in YRI population. Therefore, we hypothesized that
there might be some negative regulatory element(s) in CEU but not
in YRI population which can attenuate the association. Notably, a
search in GTEx database (https://gtexportal.org) and one previous study
(40) also failed to verify this
association (data not shown), which might be due to the origin of
tissues in the cohorts. Indeed, only European individuals were used
to investigate the association between rs7130588 and LRRC32
or EMSY expression (40).
A number of asthma-related loci have been suggested
to produce their effect by influencing the immune system and are
thus proposed to be associated with some other autoimmune diseases,
such as allergic diseases and atopic dermatitis (26,41,42).
In recent GWAS, the rs7130588 locus has been reported to be
associated with increased white cell count or percentage (43-45),
inflammatory bowel disease (4,46-48),
Crohn's disease (46-49),
ulcerative colitis (46-49),
eczema or atopic dermatitis (4,42,50)
and allergic rhinitis (4,51), which probably result from the
cis-regulation of LRRC32.
Supplementary Material
Linkage disequilibrium pattern in the
rs7130588 surrounding region. Results for population (A) CEU, (B)
CHB and (C) YRI, respectively. Each square denotes one SNP. The
colors from white to black correspond to r2-values
between 0 and 1. The vertical arrows indicate the position of
rs7130588. CEU, Utah Residents with European Ancestry; CHB, Han
Chinese in Beijing; YRI, Yoruba in Ibadan.
Histone modification in lung cancer
cell line A549 for the segment surrounding rs6592645. The yellow
lines indicate the location of rs6592645. Each part represents one
type of histone modification. The x-axis indicates genome
coordinate in chromosome 11 and the start and end are shown below.
The y-axis denotes the relative enrichment of chromatin
immunoprecipitation followed by sequencing. H3K4me1, histone 3
lysine 4 (H3K4) monomethylation; H3K27Ac, histone 3 lysine 27
acetylation.
LRRC32 expression distribution
between allergic asthma, patients with non-allergic asthma and
normal controls. The data are from the quantitative PCR of the
present study and shown as a box plot. **P<0.01.
LRRC32, leucine-rich repeat-containing 32.
Boxplot of LRRC32 expression
(log transformed) between HDM extract treatment and control group
in peripheral blood mononuclear cells. Data from (28) and shown as
a box plot. **P<0.01. HDM, house dust mite;
LRRC32, leucine-rich repeat-containing 32.
Boxplot of LRRC32 expression
(log transformed) between RV stimulation and control group in
peripheral blood mononuclear cells. Data from 29 and shown as a box
plot. **P<0.01. LRRC32, leucine-rich
repeat-containing 32; RV, rhinoviruses.
Association between rs2155219 genotype
and relative LRRC32 expression in YRI population from
literature. Data from (18) and shown as a box plot. As the data are
in log-transformed form, some individuals display negative
expression value. **P<0.01. LRRC32,
leucine-rich repeat-containing 32.
Primer sequences for plasmid
construction and mutagenesis.
Primer sequences for chromosome
conformation capture-quantitative PCR in the upstream region of
rs6592645.
Primer sequences for chromosome
conformation capture-quantitative PCR in downstream region of
rs6592645.
Probe sequences for the different
alleles of rs6592645 used in electrophoretic mobility shift
assays.
Acknowledgements
Not applicable.
Funding
Funding: The present study was supported by The National Natural
Science Foundation of China (grant no. 31370129).
Availability of data and materials
The luciferase activity, 3C, LRRC32 expression,
ChIP and genotype from 1KGP datasets generated and/or analyzed
during the current study are available in the Jianguoyun
repository, https://www.jianguoyun.com/p/DSjM-N4Q_cv3BRj5r4QFIAA.
All other datasets used and/or analyzed during the current study
are available from the corresponding author on reasonable
request.
Authors' contributions
CS and WPF conceived and designed the present study
and wrote the manuscript. YKL, YC, XQS performed luciferase, 3C and
ChIP experiments. YKL analyzed the data. HYW, XXZ and KL performed
LRRC32 expression and EMSA experiments. CS performed eQTL
analysis and database search. YKL and HYW confirm the authenticity
of all the raw data. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
The present study was reviewed and approved by the
ethics committee of Shaanxi Normal University (approval no.
20170308; Xi'an, China). Informed consent was obtained from all
subjects involved in the study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Martinez FD and Vercelli D: Asthma.
Lancet. 382:1360–1372. 2013.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Ferreira MA, Matheson MC, Duffy DL, Marks
G, Hui J, Souëf PL, Danoy P, Baltic S, Nyholt DR, Jenkins M, et al:
Identification of IL6R and chromosome 11q13.5 as risk loci for
asthma. Lancet. 378:1006–1014. 2011.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Ferreira MAR, Mathur R, Vonk JM, Szwajda
A, Brumpton B, Granell R, Brew BK, Ullemar V, Lu Y, Jiang Y, et al:
Genetic architectures of childhood- and adult-onset asthma are
partly distinct. Am J Hum Genet. 104:665–684. 2019.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Donertas HM, Fabian DK, Valenzuela MF,
Partridge L and Thornton JM: Common genetic associations between
age-related diseases. Nat Aging. 1:400–412. 2021.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Demenais F, Margaritte-Jeannin P, Barnes
KC, Cookson WOC, Altmüller J, Ang W, Barr RG, Beaty TH, Becker AB,
Beilby J, et al: Multiancestry association study identifies new
asthma risk loci that colocalize with immune-cell enhancer marks.
Nat Genet. 50:42–53. 2018.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Valette K, Li Z, Bon-Baret V, Chignon A,
Bérubé JC, Eslami A, Lamothe J, Gaudreault N, Joubert P, Obeidat M,
et al: Prioritization of candidate causal genes for asthma in
susceptibility loci derived from UK Biobank. Commun Biol.
4(700)2021.PubMed/NCBI View Article : Google Scholar
|
|
7
|
The 1000 Genomes Project Consortium. Auton
A, Brooks LD, Durbin RM, Garrison EP, Kang HM, Korbel JO, Marchini
JL, McCarthy S, McVean GA and Abecasis GR: A global reference for
human genetic variation. Nature. 526:68–74. 2015.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Binia A and Kabesch M: Respiratory
medicine-genetic base for allergy and asthma. Swiss Med Wkly.
142(w13612)2012.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Meyers DA, Bleecker ER, Holloway JW and
Holgate ST: Asthma genetics and personalised medicine. Lancet
Respir Med. 2:405–415. 2014.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Kim KW and Ober C: Lessons learned from
GWAS of asthma. Allergy Asthma Immunol Res. 11:170–187.
2019.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Willis-Owen SAG, Cookson WOC and Moffatt
MF: The genetics and genomics of asthma. Annu Rev Genomics Hum
Genet. 19:223–246. 2018.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Vicente CT, Revez JA and Ferreira MAR:
Lessons from ten years of genome-wide association studies of
asthma. Clin Transl Immunology. 6(e165)2017.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Ortiz RA and Barnes KC: Genetics of
allergic diseases. Immunol Allergy Clin North Am. 35:19–44.
2015.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Li X, Ampleford EJ, Howard TD, Moore WC,
Li H, Busse WW, Castro M, Erzurum SC, Fitzpatrick AM, Gasto B, et
al: The C11orf30-LRRC32 region is associated with total serum IgE
levels in asthmatic patients. J Allergy Clin Immunol. 129:575–578,
578 e571-579. 2012.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Global Initiative for Asthma: Global
Strategy for Asthma Management and Prevention (2018 update). 2018
(https://ginasthma.org/wp-content/uploads/2019/01/2018-GINA.pdf).
|
|
17
|
Green MR and Sambrook J: Constructing a
standard curve for real-time polymerase chain reaction (PCR)
experiments. Cold Spring Harb Protoc. 2018:836–842. 2018.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Pickrell JK, Marioni JC, Pai AA, Degner
JF, Engelhardt BE, Nkadori E, Veyrieras JB, Stephens M, Gilad Y and
Pritchard JK: Understanding mechanisms underlying human gene
expression variation with RNA sequencing. Nature. 464:768–772.
2010.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Jadhav B, Monajemi R, Gagalova KK, Ho D,
Draisma HHM, van de Wiel MA, Franke L, Heijmans BT, van Meurs J,
Jansen R, et al: RNA-Seq in 296 phased trios provides a
high-resolution map of genomic imprinting. BMC Biol.
17(50)2019.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Edwards SL, Beesley J, French JD and
Dunning AM: Beyond GWASs: Illuminating the dark road from
association to function. Am J Hum Genet. 93:779–797.
2013.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Spitz F and Furlong EE: Transcription
factors: From enhancer binding to developmental control. Nat Rev
Genet. 13:613–626. 2012.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Barrett JC, Hansoul S, Nicolae DL, Cho JH,
Duerr RH, Rioux JD, Brant SR, Silverberg MS, Taylor KD, Barmada MM,
et al: Genome-wide association defines more than 30 distinct
susceptibility loci for Crohn's disease. Nat Genet. 40:955–962.
2008.PubMed/NCBI View
Article : Google Scholar
|
|
23
|
Esparza-Gordillo J, Weidinger S,
Folster-Holst R, Bauerfeind A, Ruschendorf F, Patone G, Rohde K,
Marenholz I, Schulz F, Kerscher T, et al: A common variant on
chromosome 11q13 is associated with atopic dermatitis. Nat Genet.
41:596–601. 2009.PubMed/NCBI View
Article : Google Scholar
|
|
24
|
Marenholz I, Bauerfeind A,
Esparza-Gordillo J, Kerscher T, Granell R, Nickel R, Lau S,
Henderson J and Lee YA: The eczema risk variant on chromosome 11q13
(rs7927894) in the population-based ALSPAC cohort: A novel
susceptibility factor for asthma and hay fever. Hum Mol Genet.
20:2443–2449. 2011.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Calo E and Wysocka J: Modification of
enhancer chromatin: What, how, and why? Mol Cell. 49:825–837.
2013.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Li YK, Zhang XX, Yang Y, Gao J, Shi Q, Liu
SD, Fu WP and Sun C: Convergent evidence supports TH2LCRR as a
novel asthma susceptibility gene. Am J Respir Cell Mol Biol.
66:283–292. 2022.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Zhu Y, Mao D, Gao W, Han G and Hu H:
Analysis of lncRNA expression in patients with eosinophilic and
neutrophilic asthma focusing on LNC_000127. Front Genet.
10(141)2019.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Altman MC, Whalen E, Togias A, O'Connor
GT, Bacharier LB, Bloomberg GR, Kattan M, Wood RA, Presnell S,
LeBeau P, et al: Allergen-induced activation of natural killer
cells represents an early-life immune response in the development
of allergic asthma. J Allergy Clin Immunol. 142:1856–1866.
2018.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Murray LM, Thillaiyampalam G, Xi Y,
Cristino AS and Upham JW: Whole transcriptome analysis of high and
low IFN-α producers reveals differential response patterns
following rhinovirus stimulation. Clin Transl Immunology.
10(e1356)2021.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Sie L, Loong S and Tan EK: Utility of
lymphoblastoid cell lines. J Neurosci Res. 87:1953–1959.
2009.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Nasrallah R, Imianowski CJ,
Bossini-Castillo L, Grant FM, Dogan M, Placek L, Kozhaya L, Kuo P,
Sadiyah F, Whiteside SK, et al: A distal enhancer at risk locus
11q13.5 promotes suppression of colitis by Treg cells. Nature.
583:447–452. 2020.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Morikawa M, Derynck R and Miyazono K:
TGF-β and the TGF-β family: Context-dependent roles in cell and
tissue physiology. Cold Spring Harb Perspect Biol.
8(a021873)2016.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Al-Alawi M, Hassan T and Chotirmall SH:
Transforming growth factor β and severe asthma: A perfect storm.
Respir Med. 108:1409–1423. 2014.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Wang R, Zhu J, Dong X, Shi M, Lu C and
Springer TA: GARP regulates the bioavailability and activation of
TGFβ. Mol Biol Cell. 23:1129–1139. 2012.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Tran DQ, Andersson J, Wang R, Ramsey H,
Unutmaz D and Shevach EM: GARP (LRRC32) is essential for the
surface expression of latent TGF-beta on platelets and activated
FOXP3+ regulatory T cells. Proc Natl Acad Sci USA. 106:13445–13450.
2009.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Metelli A, Wu BX, Fugle CW, Rachidi S, Sun
S, Zhang Y, Wu J, Tomlinson S, Howe PH, Yang Y, et al: Surface
expression of TGFbeta docking receptor GARP promotes oncogenesis
and immune tolerance in breast cancer. Cancer Res. 76:7106–7117.
2016.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Probst-Kepper M, Geffers R, Kroger A,
Viegas N, Erck C, Hecht HJ, Lünsdorf H, Roubin R,
Moharregh-Khiabani D, Wagner K, et al: GARP: A key receptor
controlling FOXP3 in human regulatory T cells. J Cell Mol Med.
13:3343–3357. 2009.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Metelli A, Salem M, Wallace CH, Wu BX, Li
A, Li X and Li Z: Immunoregulatory functions and the therapeutic
implications of GARP-TGF-β in inflammation and cancer. J Hematol
Oncol. 11(24)2018.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Weissler KA and Frischmeyer-Guerrerio PA:
Genetic evidence for the role of transforming growth factor-β in
atopic phenotypes. Curr Opin Immunol. 60:54–62. 2019.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Li X, Hastie AT, Hawkins GA, Moore WC,
Ampleford EJ, Milosevic J, Li H, Busse W, Erzurum SC, Kaminski N,
et al: eQTL of bronchial epithelial cells and bronchial alveolar
lavage deciphers GWAS-identified asthma genes. Allergy.
70:1309–1318. 2015.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Zhu Z, Lee PH, Chaffin MD, Chung W, Loh
PR, Lu Q, Christiani DC and Liang L: A genome-wide cross-trait
analysis from UK Biobank highlights the shared genetic architecture
of asthma and allergic diseases. Nat Genet. 50:857–864.
2018.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Weidinger S, Willis-Owen SA, Kamatani Y,
Baurecht H, Morar N, Liang L, Edser P, Street T, Rodriguez E,
O'Regan GM, et al: A genome-wide association study of atopic
dermatitis identifies loci with overlapping effects on asthma and
psoriasis. Hum Mol Genet. 22:4841–4856. 2013.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Chen MH, Raffield LM, Mousas A, Sakaue S,
Huffman JE, Moscati A, Trivedi B, Jiang T, Akbari P, Vuckovic D, et
al: Trans-ethnic and ancestry-specific blood-cell genetics in
746,667 individuals from 5 global populations. Cell. 182:1198–1213
e1114. 2020.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Vuckovic D, Bao EL, Akbari P, Lareau CA,
Mousas A, Jiang T, Chen MH, Raffield LM, Tardaguila M, Huffman JE,
et al: The polygenic and monogenic basis of blood traits and
diseases. Cell. 182:1214–1231 e1211. 2020.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Astle WJ, Elding H, Jiang T, Allen D,
Ruklisa D, Mann AL, Mead D, Bouman H, Riveros-Mckay F, Kostadima
MA, et al: The allelic landscape of human blood cell trait
variation and links to common complex disease. Cell. 167:1415–1429
e1419. 2016.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Liu JZ, van Sommeren S, Huang H, Ng SC,
Alberts R, Takahashi A, Ripke S, Lee JC, Jostins L, Shah T, et al:
Association analyses identify 38 susceptibility loci for
inflammatory bowel disease and highlight shared genetic risk across
populations. Nat Genet. 47:979–986. 2015.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Jostins L, Ripke S, Weersma RK, Duerr RH,
McGovern DP, Hui KY, Lee JC, Schumm LP, Sharma Y, Anderson CA, et
al: Host-microbe interactions have shaped the genetic architecture
of inflammatory bowel disease. Nature. 491:119–124. 2012.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Peyrot WJ and Price AL: Identifying loci
with different allele frequencies among cases of eight psychiatric
disorders using CC-GWAS. Nat Genet. 53:445–454. 2021.PubMed/NCBI View Article : Google Scholar
|
|
49
|
de Lange KM, Moutsianas L, Lee JC, Lamb
CA, Luo Y, Kennedy NA, Jostins L, Rice DL, Gutierrez-Achury J, Ji
SG, et al: Genome-wide association study implicates immune
activation of multiple integrin genes in inflammatory bowel
disease. Nat Genet. 49:256–261. 2017.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Sliz E, Huilaja L, Pasanen A, Laisk T,
Reimann E and Mägi R: FinnGen; Estonian Biobank Research Team.
Hannula-Jouppi K, Peltonen S, et al: Uniting biobank resources
reveals novel genetic pathways modulating susceptibility for atopic
dermatitis. J Allergy Clin Immunol. 149:1105–1112 e1109.
2022.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Li Y, Chen J, Rui X, Li N, Jiang F and
Shen J: The association between sixteen genome-wide association
studies-related allergic diseases loci and childhood allergic
rhinitis in a Chinese Han population. Cytokine. 111:162–170.
2018.PubMed/NCBI View Article : Google Scholar
|