Introduction
Deep vein thrombosis (DVT) is caused by the abnormal
coagulation of blood in the deep veins, thus leading to lumen
stenosis and obstruction of the venous return. DVT is the third
major cardiovascular cause of death and/or disability after
ischemic cardiomyopathy and stroke, and its incidence is increasing
each year. Clinically, 90% of cases of venous thromboembolism (VTE)
are due to DVT (1,2). The onset of DVT is insidious, sudden
and is mostly asymptomatic. However, delayed diagnosis and
treatment may lead to pulmonary embolism and post-thrombotic
syndrome, or even death (3). A
recent study has suggested that conventional pharmacologic
prophylaxis may not be sufficient to prevent the development of VTE
in patients with severe coronavirus infection (4). D-dimer is the clinical biomarker most
commonly used to exclude a diagnosis of DVT, with high sensitivity
but low specificity (5).
Therefore, further studies are needed to explore the mechanisms of
DVT initiation and progression, to provide patients with improved
diagnosis and treatment strategies.
Circular RNAs (circRNAs) belong to a novel class of
regulatory non-coding RNAs. In contrast to linear RNAs, with 5'
terminal caps and 3' tails, the ends of circRNAs are covalently
linked, thus forming a closed circular structure that renders them
more stable than their linear counterparts (6,7).
This molecular structure allows circRNAs to act as microRNA (miRNA)
sponges, by regulating transcription or splicing, interacting with
RNA-binding proteins, and participating in gene expression
regulation. Therefore, circRNAs may potentially have broad
applications in clinical diagnosis and treatment (8-10).
To date, numerous studies have shown that circRNAs
are closely associated with the diagnosis and treatment of
cardiovascular diseases, and certain circRNAs have been identified
as cardiovascular disease biomarkers (11-15).
Recently, circRNAs have been reported to accelerate lower extremity
DVT via sponging miRNA, thus regulating mRNA expression (16,17).
Although circRNAs have been suggested to be involved in the
pathogenesis and regulatory mechanism of DVT, few studies have
examined the global dynamic changes in circRNAs and mRNAs during
DVT progression.
To address this issue, the dynamic changes in
circRNAs and mRNAs during the development of DVT were investigated,
in a rat model by performing RNA sequencing. Differentially
expressed genes encoding circRNAs and mRNAs were detected and it
was predicted how these RNAs may function through bioinformatics
analysis. Short Time Series Expression Miner (STEM) and
circRNA-mRNA regulatory network analyses were also conducted.
Furthermore, the expression of key circRNAs was verified by reverse
transcription-quantitative PCR (RT-qPCR). The identification of
DVT-associated key circRNAs may contribute to DVT diagnosis and
treatment.
Materials and methods
Construction of the DVT rat model
A total of 90 male Sprague-Dawley rats (weight,
250-300 g; age, 10-12 weeks) were obtained from the Experimental
Animal Center of the Medical College of Nantong University
(Nantong, China) [SCXK (Su) 2014-0001]. The rats were kept under a
12-h light/dark cycle, a temperature of 23±3˚C, and a humidity of
55-60%, and were given free access to food and water. The present
experiment was approved (approval no. 20170930-001) by the
experimental animal ethics committee of Nantong University
(Nantong, China). A stasis-induced venous thrombosis model was
implemented as previously described (18,19).
Briefly, the rats were anesthetized by intraperitoneal injection of
0.3% pentobarbital sodium (30 mg/kg) and placed in supine position.
After a midline laparotomy, the intestines were exteriorized and
placed to the left of the animal. The inferior vena cava (IVC) was
carefully separated from the surrounding tissues and partially
ligated just below the renal veins, along with complete ligation of
all visible side branches. The rats were allowed to recover
post-surgery and housed under a 12-h light/dark cycle, a
temperature of 23±3˚C, and a humidity of 55-60%, and were given
free access to food and water during recovery. Finally, the rats
were sacrificed at 0.5, 1, 2, 4, 6, 8, 12 and 24 h, and 2, 3, 7, 14
or 21 days after the operation. Before sacrifice, a total of 2 ml
venous blood was collected from the suprarenal IVC into EDTA-coated
capillary tubes for next-generation sequencing after the model rats
were anesthetized in an isoflurane chamber (3-4%) and maintained
with a face mask using 1-2% isoflurane. All rats were sacrificed by
cervical dislocation under deep anesthesia with 2% isoflurane.
Subsequently, the thrombosed IVC was carefully dissected, and the
thrombus was extracted. The weight (g) and length (cm) of thrombi
harvested from the IVC were measured with an electronic balance
(Shanghai Mettler Toledo Co., Ltd.) and Vernier calipers (Deli Co.,
Ltd.). The IVC plus thrombus was fixed immediately in formalin and
stained with hematoxylin and eosin (H&E). A total of six rats
were included in each time point and the sham group, in which
suture lines were passed, but no blood vessels were ligated. The
main IVC and branches from rats in the sham group were separated by
laparotomy and treated in the same manner as those from the model
rats.
Histological examination
After extraction, the IVCs plus thrombi were
collected in 10% buffered formalin, embedded in paraffin, serially
cross-sectioned at 5 µm, and stained with H&E (20 min in
hematoxylin solution and 60 sec in eosin solution at room
temperature), according to standard procedures. All histological
images were acquired with an inverted microscope (Motic (Xiamen)
Electric Group Co., Ltd.) and analyzed in Motic Images 2.0
software.
Total RNA isolation, library
construction and sequencing
Total RNA from cells obtained from the IVC was
extracted with TRIzol® reagent (cat. no. 15596-026;
Invitrogen; Thermo Fisher Scientific, Inc.) and analyzed with an
Agilent 2100 BioAnalyzer (Agilent Technologies, Inc.) and Qubit
fluorometer (Invitrogen; Thermo Fisher Scientific, Inc.). Samples
with an RNA integrity number >7.0 and a 28S:18S ratio >1.8
were used in subsequent experiments. Libraries were generated and
sequenced by CapitalBio Technology, Inc. A total of 5 µg RNA per
sample was used as input material for RNA sample preparation.
First, ribosomal RNAs were depleted with an Epicentre
Ribo-zero™ rRNA Removal kit (cat. no. RZH1046;
Epicentre; Illumina, Inc.) to obtain rRNA-depleted RNAs, which were
further treated with RNase R (cat. no. RNR07250; Epicentre) and
subjected to TRIzol extraction. Sequencing libraries were then
generated with the rRNA-depleted and RNase R-digested RNAs and a
NEBNext® Ultra™ Directional RNA Library Prep
kit (cat. no. E7530S; New England Biolabs), according to the
manufacturer's recommendations. Briefly, fragmentation was
performed with divalent cations under elevated temperature in
NEBNext First Strand Synthesis Reaction Buffer. First-strand cDNA
was synthesized with random hexameric primers and M-MuLV reverse
transcriptase (RNaseH). Second-strand cDNA was subsequently
synthesized with DNA polymerase I and RNase H. In the reaction
buffer, dNTPs with dTTP were replaced by dUTP. Remaining overhangs
were converted into blunt ends via exonuclease/polymerase activity.
After adenylation of the 3' ends of the DNA fragments, an NEBNext
Adaptor with a hairpin loop structure was ligated for
hybridization. To select cDNA fragments 150-200 bp in length, the
library fragments were purified with an AMPure XP system (Beckman
Coulter, Inc.), then performed treatment with 3 µl USER Enzyme
(cat. no. M5505; New England Biolabs) with size-selected,
adaptor-ligated cDNA at 37°C for 15 min, then 5 min at
95˚C, before PCR. PCR was performed with Phusion High-Fidelity DNA
polymerase, Universal PCR primer, and Index (X) Primer. Finally,
the library was purified (AMPure XP system), and quality was
assessed with an Agilent Bioanalyzer 2100 system. The library
preparations were sequenced on the Illumina Hiseq XT platform, and
300-bp paired-end reads were generated.
RNA-sequencing data analysis
Raw reads (in FASTQ format) were processed with
FASTQ and fastp. Clean reads were then obtained by removal of
adapter-containing, poly-N-containing, and low-quality reads from
the raw data. The Q20, Q30 and GC content of the clean data were
calculated. All downstream analysis was based on the resulting
high-quality clean data. Reference genome and gene annotation data
were downloaded from the NCBI genome website (https://www.ncbi.nlm.nih.gov/pmc/). An index of
the reference genome was built with Bowtie v2.0.6(20), and paired-end clean reads were
aligned to the reference genome with Tophat2. Unmapped reads were
retained, and 20-mers from the 5' and 3' ends of these reads were
extracted and aligned independently to reference sequences with
Tophat2. Anchor sequences were extended with fnd circ, such that
the complete read was aligned, and the breakpoints were flanked by
GU/AG splice sites. The back-spliced reads with at least two
supporting reads were then annotated as circRNAs. Expression levels
of circRNAs were normalized by spliced reads per billion mapped
reads as follows: Normalized expression=(back-spliced reads)
x109/(total reads). Differential expression between
groups was determined with limma (21). The default threshold P-value for
differential expression was set to 0.05 and |log2 (fold
change)|>1.
STEM
STEM defines a set of distinct and representative
model temporal expression profiles independently of the data, which
correspond to possible profiles of changes in gene expression over
time. The number of genes expected to be assigned to a profile is
estimated by randomly permuting the original time point values,
renormalizing the gene expression values, assigning genes to their
most closely matching model profiles, and repeating the process for
numerous permutations. Statistically significant model profiles
that are similar to one another can be grouped into clusters. A
color fill indicates a significant profile, and each color
indicates a different cluster. The biological significance of the
set of genes assigned to the same profile or the same cluster of
profiles can then be assessed with enrichment analysis. For
identification of genes temporally coregulated and clustering with
the phenotypic parameters, the STEM software tool was used (STEM,
version 1.3.10; Jason Ernst; http://www.cs.cmu.edu/~jernst/stem/). STEM was run
with the ‘normalize data’, option, with all other settings set to
the default.
Gene ontology (GO) annotations and
Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway
analysis
The differential expression of mRNAs was analyzed to
further investigate their biological functions and pathways
according to GO functional annotation and KEGG pathway analysis
(https://www.genome.jp/kegg/) with the
cluster Profiler R package 4.0(22)
GO terms and KEGG pathways with corrected P<0.05 were considered
significantly enriched.
RT-qPCR
Total RNAs in blood samples were extracted with
TRIzol reagent. RNA concentration and purity were measured
spectrophotometrically by absorbance measurements at 260 and 280 nm
using the NanoDrop ND-1000 (Thermo Fisher Scientific, Inc.).
OD260/OD280 ratios between 1.8 and 2.0 were deemed acceptable. RNA
was reverse transcribed into cDNA with a High-Capacity cDNA Reverse
Transcription kit (cat. no. 4368814; Thermo Fisher Scientific,
Inc.) according to the manufacturer's protocol. RT-qPCR was
performed with TaqMan Multiplex Real-Time Solution (cat. no.
446188;1 Thermo Fisher Scientific, Inc.) according to the
manufacturer's protocol, with GAPDH expression serving as an
endogenous control. Briefly, the RT-qPCR cycling protocol consisted
of enzyme activation at 95˚C for 10 min, 40 cycles of denaturation
at 95˚C for 15 sec, and annealing and extension at 60˚C for 1 min,
with melting curve analysis (temperature ramp 2%) at 60-95˚C. On
the basis of the sequences of circRNAs, primers were designed for
the candidate circRNAs across a splice site (back-splice), owing to
the loop forming characteristic of the circRNAs. Primer 5.0
(www.premierbiosoft.com/primerdesign/index.thml)
was used to design specific primers for candidate mRNAs and
circRNAs (primer sequences are listed in Table I). RT-qPCR was performed with an
ABI 7500 Real-Time PCR system (Applied Biosystems; Thermo Fisher
Scientific, Inc.). Each relative RNA expression level was
calculated from three separate experiments. The relative expression
levels of circRNAs and mRNAs were determined with the
2-ΔΔCq method (23).
 | Table IReverse transcription-quantitative
PCR primers for candidate mRNAs and circRNAs. |
Table I
Reverse transcription-quantitative
PCR primers for candidate mRNAs and circRNAs.
| mRNA/circRNA
name | Sequence
(5'-3') |
|---|
| Lgals3bp | F:
TGGTCTGCTCCAACGATTCC |
| | R:
GTCCTGCTGGCTGTCAAAGA |
| Kif18a | F:
CAGCCAAGTTGTCGCGGT |
| | R:
TGTGAGTCTTCCCAGATCCAGT |
| Kif4a | F:
TAAACTCCGAAGGCGCACAT |
| | R:
GCGTTGTTTTGGCTGGTCTT |
| Tuba4a | F:
AGACGTACATCCCAAACTCATC |
| | R:
AAGGAGTCATCCCCTCCACC |
| CBT15_ | F:
CGGGTCTGGCCATTTTTACATC |
| circR_3235 | R:
CAGCAACAGCTCTAACATCGT |
| CBT15_ | F:
TTGGGGATGTCTGCTTGTTGA |
| circR_31289 | R:
GCAGGATGGTGACAGATTGC |
| CBT15_ | F:
CCGCCCACCAATCTCCAAAG |
| circR_4786 | R:
TGCCCAGGTTCATTTCGTCC |
| CBT15_ | F:
GCGACTCTGATGACTCTCTCC |
| circR_33780 | R:
CTGCGGCTCATTCACAATCTC |
| CBT15_ | F:
ATTGACGTCTAGCATGTTTTCCT |
| circR_14797 | R:
TCACCACGATGTTGGAGGG |
| CBT15_ | F:
AGGAGTTGGAGAAAACCTTGGA |
| circR_2763 | R:
GACCACATCCGTAAACTGGG |
| CBT15_ | F:
TCTTTGCAATATGATTGGACTTGAT |
| circR_14565 | R:
AGCAAGTTCTTGGCTGGACT |
| CBT15_ | F:
AAGTCTGTGGGCATCGGAGAG |
| circR_16149 | R:
CAGTGGAGAGGGAAGTGCGTA |
| CBT15_ | F:
ACTATGGCAGCCCACCTAAC |
| circR_40422 | R:
GTGATCAGGTGCAAGGTGTCT |
| CBT15_ | F:
AAAAGGAAGGAGCTACACCGAG |
| circR_22738 | R:
CATCCCACACTGGTGCAAA |
| GAPDH-Rat | F:
AGTGCCAGCCTCGTCTCATA |
| | R:
AACTTGCCGTGGGTAGAGTC |
Databases
To further confirm the presence of the verified
circRNAs, CIRCpedia-V2 (https://www.biosino.org/circpedia/), an updated
comprehensive database containing circRNA annotations across six
different species, was used. To identify human circRNAs that are
homologous to the key circRNA, circBase (http://www.circbase.org) was used. miRNAs binding both
the circRNA and the mRNA were screened with MiRdb (https://mirdb.org) and TargetScan (https://www.targetscan.org/vert_80/).
Statistical analysis
The data were processed in SPSS 22.0 statistical
software (IBM Corp.). Measured data are expressed as the mean ±
standard deviation. Unpaired data with a normal distribution and
homogeneity of variance between two groups were compared with
unpaired t-tests, and multiple groups were compared with one-way
analysis of variance (ANOVA) followed by Tukey's test. P<0.05
was considered to indicate statistically significant
difference.
Results
Dynamic observation of DVT progression
in the rat model
A rat model was successfully established of DVT by
stenosis. Rats were divided into 13 DVT groups (0.5, 1, 2, 4, 6, 8,
12 and 24 h; and 2, 3, 7, 14 and 21 days) and a sham group, with
six rats per time point, except in the 1 h group, which included
eight rats (Table SI). The model
was successfully established in 86 rats, and four rats succumbed
(one each because of excessive bleeding during operation,
anesthetic overdose, sudden death, or intestinal obstruction).
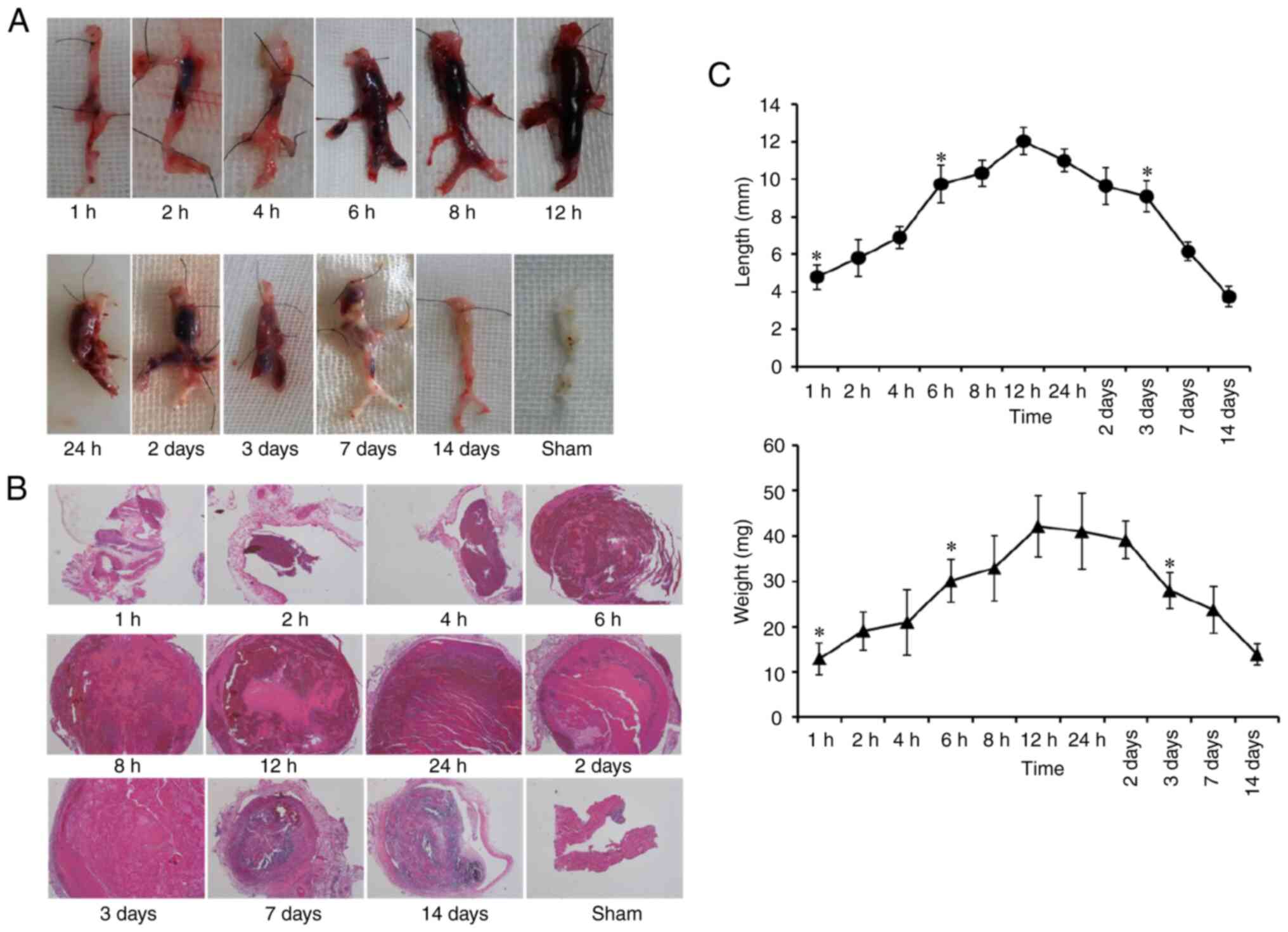
At 2 h post-operation, the vein distal to the
partial ligation appeared dark red, and a coagulation-like
substance formed in the lumen, which moved when light pressure was
applied to the tube wall. At 6 h post-operation, the color of the
ligated vein had deepened, the vein wall became tough and less
elastic, and the hardness of the clot increased. At 12 h, the
partial ligated distal vein was black and red, the internal
thrombus was tough, the activity of the model vein decreased, and
the mesenteric vein was dilated. After 3 days, careful separation
and exposure of the model segment vein revealed that the blood
vessel and embolus were dark and stiff, the color of the tube wall
was pale, and the clot in the lumen adhered to the tube wall
(Fig. 1A). In the sham-operated
group, the activities of both hind limbs were normal at all time
points, the diameter of the IVC remained unchanged, and the blood
was dark red and flowed smoothly. The vascular wall was soft and
elastic, the mesenteric veins were uniform in thickness with no
abnormal expansion, and no thrombosis was observed.
HE staining indicated that the IVC was not
completely thrombosed at 1 h after model establishment. Over time,
the blood flow stagnated, and the thrombus grew an embolus that
completely blocked the vessel by 6 h, thus indicating wetting,
smoothness, softness, and elasticity. With prolongation of the
modeling time from 6-48 h, the thrombus dried out, and developed
cracks and fibrosis, although it still filled the entire vessel.
Thrombus organization was observed on day 3, and HE staining showed
clear organization, desiccation, pyknosis, and disintegration of
the thrombus on days 7 and 14 (Fig.
1B).
Gross observation and HE staining of the thrombi
revealed differences in the percentage thrombus formation at each
time point, with 0% (0/6 at 0.5 h; 75% (6/8) at 1 h; 100% (48/48)
at 2, 4, 6, 8, 12 and 24 h, and 2 and 3 days; 83.3% (10/12) at 7
and 14 days; and 50.0% (3/6) at 21 days (Table SI).
Thrombus length and weight are important parameters
in thrombus research. As shown in Fig.
1C, the initial stage of thrombosis occurred from 1 to 6 h, the
stable stage occurred from 6 to 48 h, and the thrombus diminished
or became organized after 3 days. Thrombus formation began at 1 h
after model establishment, and no significant differences in
thrombus length or weight were observed at 2 and 4 h (P>0.05).
Thrombus length and weight increased significantly at 6 h and were
significantly higher at 6-48 h than 2-4 h (P<0.05). However, no
significant differences were observed among thrombi at 6, 8, 12, 24
and 48 h (P>0.05). A total of three days after the operation,
the thrombus had clearly diminished, and the length and weight of
thrombus had decreased significantly with respect to those at 48 h
(P<0.05). Thrombus resolution continued, and by day 7, no
thrombus was observed in 1 of the 6 DVT rats.
Given that 1, 6 and 12 h, and 3 days appeared to be
the key time points of thrombosis progression, blood samples were
collected from the model rats at those time points and
next-generation sequencing of circRNAs and mRNAs was performed.
Global expression profiles of circRNAs
and mRNAs in distinct developmental stages of DVT
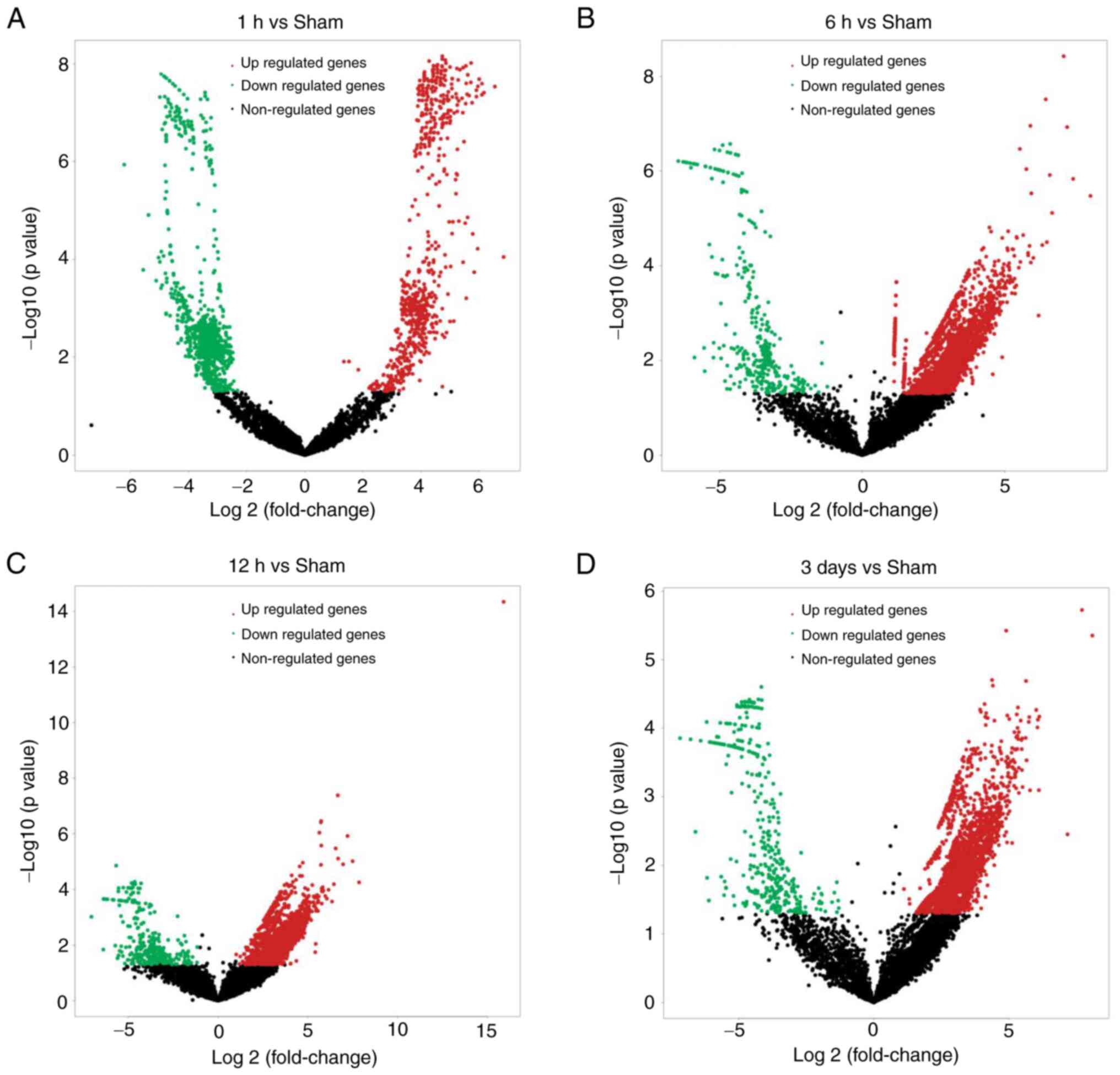
CircRNA expression profiles are presented as a
volcano plot (Fig. 2A-D). A total
of 1,680, 4,018, 3,724 and 3,036 circRNAs were differentially
expressed in model rats at 1, 6 and 12 h, and 3 days, respectively,
compared with sham group rats (fold change >2.0 or <-2.0,
P<0.05), of which 736, 3,691, 3,406 and 2,639 were upregulated,
and 944, 327, 318 and 397 were downregulated. The most common type
of circRNA was exon type. The numbers of in-gene, intron, and
intron and exon type circRNAs were lowest (4, 21 and 5,
respectively). Of the upregulated circRNAs, 85 were detected at all
time points, according to Venn analysis. Among the downregulated
circRNAs, 138 were detected at all time points, according to Venn
analysis (Fig. S1A-D).
Expression profiles of mRNA are presented as a
scatterplot (Fig. 3A-D). A total
of 400, 1176, 373, and 573 mRNAs were differentially expressed in
model rats at 1, 6 and 12 h, and 3 days, respectively, compared
with the sham group (fold change >2.0 or <-2.0, P<0.05),
of which 169, 885, 298 and 500 were upregulated, and 231, 291, 75
and 73 were downregulated. The top 20 up- and downregulated genes
at 1, 6 and 12 h, and 3 days are shown in Table SII. Of the upregulated mRNAs, 69
were expressed at all time points, according to Venn analysis. None
of the downregulated mRNAs were expressed at all time points,
according to Venn analysis (Fig.
S2A-C).
Enrichment analysis of differentially
expressed mRNAs at different stages of DVT
To further investigate the roles of mRNAs in DVT
development, functional enrichment analyses of genes was performed,
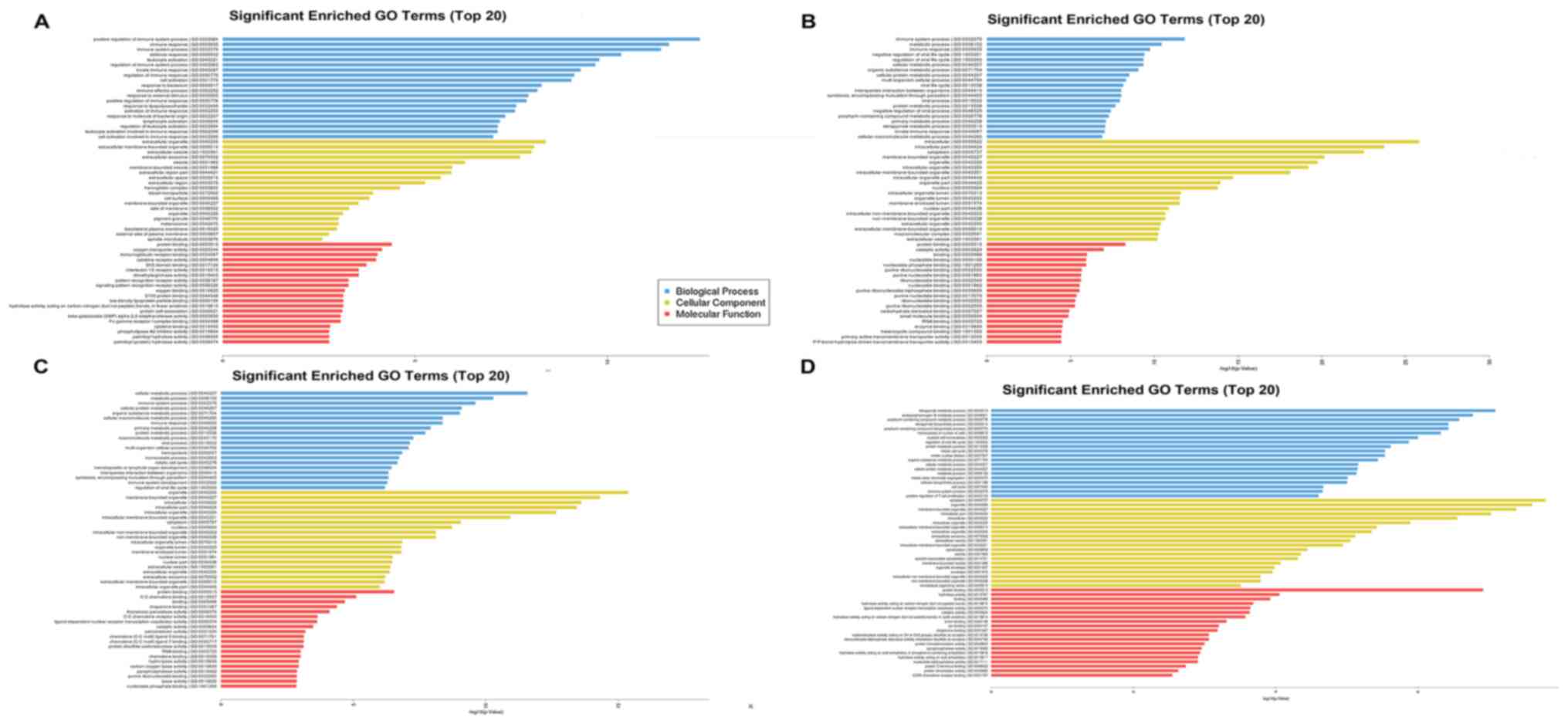
including GO and KEGG analyses. As shown in Fig. 4A-D, GO analysis of differentially
expressed mRNAs in the 1 h group compared with the sham group
indicated that the top 20 significantly enriched biological
processes (BPs), cellular components (CCs), and molecular functions
(MFs) included positive regulation of immune system process, immune
response, and cell activation. Additionally, these BPs were
enriched 6 and 12 h, and 3 days after operation. For annotations of
CCs, the terms of organelle, cytoplasm, extracellular
membrane-bounded organelle and blood microparticle were enriched.
For MF terms, annotations for protein binding, oxygen transporter
activity and catalytic activity were highly enriched.
KEGG analysis of differentially expressed mRNAs in
the 1, 6, and 12 h, and 3-day groups compared with the sham group
indicated that the most enriched pathways were influenza A,
metabolic pathways, phagosome and metabolic pathways (Fig. 5A-D).
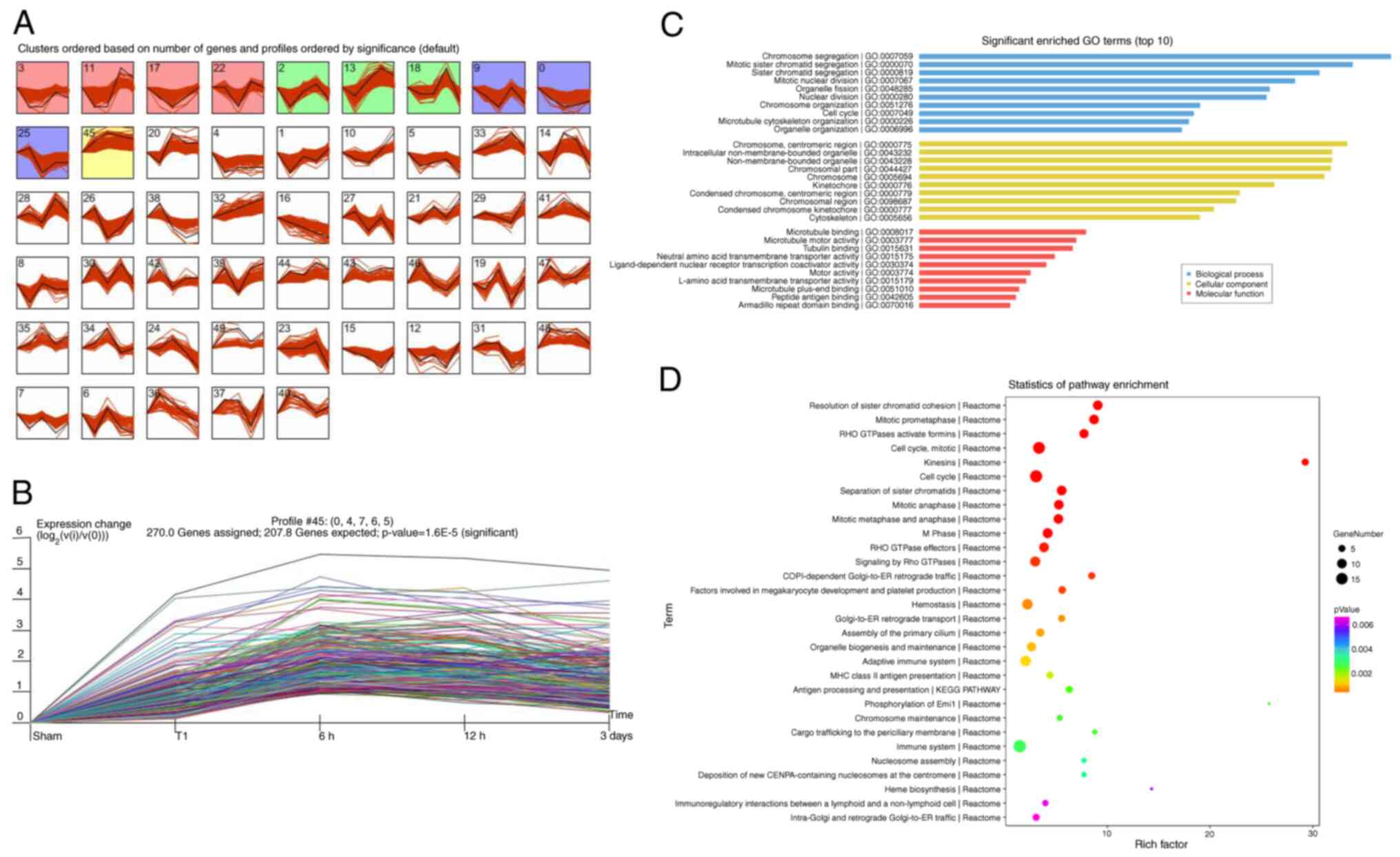
STEM analysis of differentially
expressed circRNAs and mRNAs and STEM-Enrichment analysis
To address the circRNA and mRNA expression patterns
at different stages of DVT, STEM analysis was performed.
Consequently, 50 profiles of circRNAs and mRNA were identified.
Among them, 10 circRNA profiles and 11 mRNA profiles were
significantly expressed (P<0.05). The black line in each profile
represents the expression pattern of the genes at different times
(Figs. 6A and 7A). Of note, among all profiles
determined, profile 45 involved 366 circRNAs (Fig. 6B), and profile 45 of involved 270
mRNAs (Fig. 7B), in agreement with
the observed trend in thrombus weight/length.
To identify the major functions and signaling
pathways of profile 45 of mRNA, GO annotation and KEGG pathway
analyses were performed. The most significantly enriched BP, CC and
MF terms of mRNAs in profile 45 were chromosome segregation,
mitotic sister chromatid segregation, cell cycle and
ligand-dependent nuclear receptor transcription coactivator
activity (Fig. 7C). Pathway
analysis of profile 45 indicated enrichment in platelet-associated
pathways (factors involved in megakaryocyte development and
platelet production, platelet degranulation, response to elevated
platelet cytosolic Ca2+, platelet activation, signaling
and aggregation, and platelet activation); immune-associated
pathways (adaptive immune system, immune system, autoimmune thyroid
disease, regulation of innate immune responses to cytosolic DNA,
cytokine signaling in immune system and innate immune system); and
inflammation-relation pathways (inflammation mediated by chemokine
and cytokine signaling pathway, herpes simplex infection, HTLV-I
infection and Epstein-Barr viral infection involved in inflammatory
pathways; Fig. 7D).
Screening of candidate differential
expression circRNAs and mRNAs
Because platelets have been found to be a major
factor in the formation of thrombosis (24), platelet-associated pathways in the
enrichment analysis annotated by mRNA profile 45 were selected and
searched for their input genes (Table
II). The input genes in the platelet-associated pathway
intersected with the annotated genes in profile 45, and four genes
were obtained: α4A-tubulin (Tuba4a), kinesin-8 (Kif18a), kinesin-4
(Kif4a) and galactose-binding protein (Lgals3 bp). According to the
mRNA-circRNA interaction network of the four genes, 328 circRNAs
interacting with the four mRNAs were obtained (Fig. 8A). The circRNAs in the early
formation of DVT were first observed. The upregulated circRNAs at 1
and 6 h after model establishment, according to Venn analysis, and
eight circRNAs were obtained (Fig.
8B). Two other circRNAs (CBT15_circR_40422 and CBT15_circR_
22738) were found to interact with Kif18a, Kif4a, and Lgals3 bp.
Therefore, these two circRNAs are also listed as candidate
circRNAs. On the basis of the aforementioned analysis, a total of
ten circRNAs and four mRNAs were selected as candidates for further
RT-qPCR verification (Table
III).
 | Table IIPlatelet-related pathways and input
gene. |
Table II
Platelet-related pathways and input
gene.
| Term | Input gene |
|---|
| Factors involved in
megakaryocyte development and platelet production | Kif22, Kif18a,
Racgap1, Kif4a, Kif2c |
| Platelet
degranulation | Lgals3bp,
Tuba4a |
| Response to
elevated platelet cytosolic Ca2+ | Lgals3bp,
Tuba4a |
| Platelet
activation, signaling and aggregation | Lgals3bp,
Tuba4a |
| Platelet
activation | Itga2 |
 | Table IIIDetails of the 10 candidate
circRNAs. |
Table III
Details of the 10 candidate
circRNAs.
| Running number | LogFC | P-value | Regulation | Chromosome | Length, bp |
|---|
|
CBT15_circR_3235* | 5.55 | 0.000624151 | Up | Chr1:
255608044-255612190 | 4,146 |
|
CBT15_circR_31289 | 4.43 |
1.31112x10-8 | Up | Chr5:
152377689-152378376 | 687 |
|
CBT15_circR_4786 | 4.35 |
2.11468x10-7 | Up | Chr10:
106731880-106738104 | 6,224 |
|
CBT15_circR_33780 | 3.83 | 0.000489196 | Up | Chr6:
21980222-21980905 | 683 |
|
CBT15_circR_14797 | 2.39 | 0.035099441 | Up | Chr15:
52308036-52308580 | 544 |
|
CBT15_circR_2763 | 2.21 | 0.042581092 | Up | Chr1:
235759863-235761819 | 1,956 |
|
CBT15_circR_14565 | 2.47 | 0.031028548 | Up | Chr15:
41976569-41976792 | 223 |
|
CBT15_circR_16149 | 4.51 | 1.90551E-05 | Up | Chr16:
73687145-73690537 | 3,392 |
|
CBT15_circR_40422 | 0.31 | 0.764794424 | No | Chr8:
90482738-90483077 | 339 |
|
CBT15_circR_22738 | -0.311230182 | 0.796548004 | No | Chr2:
229005941-29006321 | 380 |
Validation of the differential
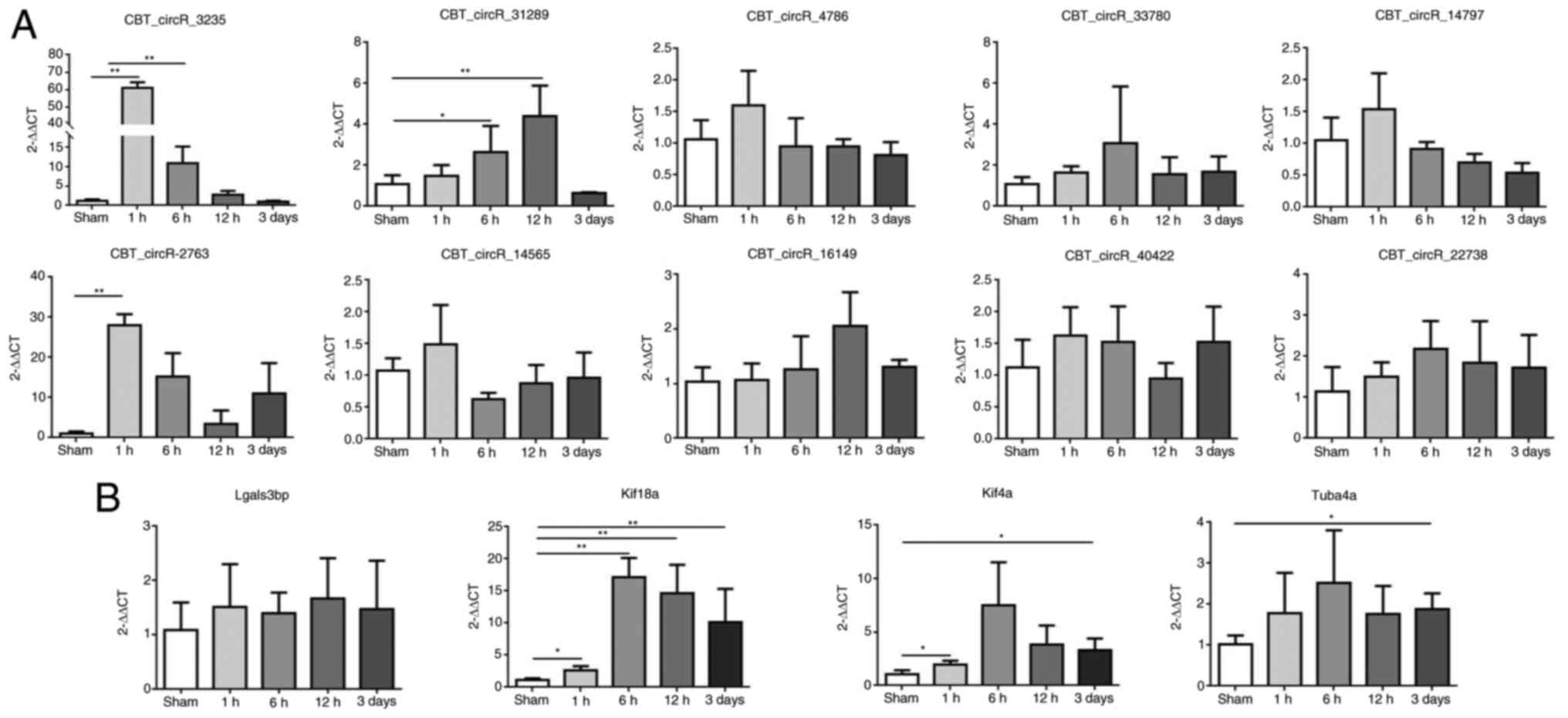
expression of candidate circRNAs and mRNAs by RT-qPCR
On the basis of the sequences of circRNAs, we
designed primers for the candidate circRNAs across a splice site
(back-splice), owing to the loop forming characteristic of the
circRNAs. Primer 5.0 was used to design specific primers for
candidate mRNAs and circRNAs (primer sequences are listed in
Table I.
The expression of the ten circRNAs and four mRNAs
was verified by RT-qPCR. The expression of nine of the ten circRNAs
and all four mRNAs was consistent with the sequencing results, and
CBT15_circR_2763 was discarded due to the large discrepancy between
samples and the instability of the results (Fig. 9).
Discussion
Previous studies have suggested that surgical trauma
and venous stasis are the main causative factors of DVT (25,26).
Venous blood in the legs is prone to stasis in patients undergoing
long-term bed rest or immobilization, thus resulting in hypoxia of
the venous sinus, whereas blood stasis and hypoxia promote each
other. Activation of venous endothelial cells and exogenous
coagulation pathways ultimately leads to thrombosis. In the present
study, a stenosis-induced rat model DVT was used to reflect the
common clinical causes of DVT, such as postoperative
immobilization, clinical trauma, or limb dysfunction. Brill et
al (18) reported thrombus
formation at 2 h in the stenosis model, and a thrombus-formation
rate of 100% at 2 h in the DVT model rats of the present study was
observed. Therefore, in order to observe further the DVT formation,
two time points of 1 and 0.5 h were assessed. No clear clots at 1 h
after surgery were observed but microthrombi in the partial ligated
vessels after separation and washing were detected, as confirmed by
H&E staining. The total thrombus-formation rate was 75% at 1 h.
However, there was no evidence of any thrombosis at the 0.5-h time
point. The difference in thrombus-initiation times between studies
may have been associated with differences in manipulation and
surgical instruments among laboratories, and differences in the
immune status of the rats.
In the present study, the rate of thrombus formation
in the model rats was 100% from 2 h to 3 days. On day 3, the
thrombi became hard and began to organize, and clear organization
was observed by day 7. By day 21, some thrombi had disappeared, and
the IVC had reopened. Thus, the present study revealed the entire
process of thrombus development from initiation, through stability,
to regression, and additionally analyzed thrombus weight and
length. No thrombi were found in the sham group. These results
indicated that the IVC thrombosis model was established
successfully and displayed the entire process of thrombus
evolution.
Moreover, it was revealed that microthrombi began to
form in the ligated vessels at 1 h after operation. The thrombi
formed rapidly in 1-6 h, were almost completely formed at 6 h, and
reached a peak at 12 h after surgery. Subsequently, drying and
recanalization of the thrombi were observed at 3 d after operation.
Hence, the DVT model rats were divided into four groups to explore
in an improved way the roles of circRNAs and mRNAs in the different
stages, and their related functions in the development of DVT.
To observe the dynamic changes in circRNAs and mRNAs
in the development of the DVT rat model, RNA sequencing was
performed to determine the circRNAs and mRNAs significantly
differentially expressed in peripheral blood in the 1, 6 and 12 h,
and 3-day groups compared with the sham group. GO and KEGG pathway
analyses were then performed to predict their biological functions.
The circRNA-mRNA regulatory network was established. Subsequently,
RT-qPCR was conducted, and the results were consistent with the RNA
sequencing results.
Studies have shown that circRNAs are closely
associated with cardiovascular diseases (27-30).
In the present study, the entire progression of circRNAs in a DVT
animal model, was effectively observed. According to the sequencing
results, 1,680, 4,018, 3,724 and 3,036 circRNAs were differentially
expressed in model rats at 1, 6 and 12 h, and 3 days, respectively,
with respect to rats in the sham group. The number of upregulated
circRNAs increased from 736 at 1 h to 3,691 at 6 h, then decreased
slightly to 3,406 concurrently with thrombus stabilization at 12 h,
and decreased further to 2,639 at 3 days, when the thrombi began to
organize. During the entire process, the number of downregulated
circRNAs was markedly lower than that of upregulated circRNAs at
the three time points (6 h: 327; 12 h: 318; 3 days: 397). In view
of this phenomenon, it was hypothesized that the upregulated
circRNAs may have strongly responded to the stimulus of the surgery
used to establish the model. This result also demonstrated that
circRNAs are involved in the occurrence and development of DVT.
Previous studies revealed that PAI-1 (SERPINE1 gene)
and other fibrinolytic genes, such as PLG, PLAT, PLAU, SERPINF2,
SERPINB2 and CPB2, are associated with VTE risk in humans (31,32).
Anticoagulation and coagulation genes are natural candidate genes
for VTE (33). Moreover, Mendelian
randomization analysis indicated causal associations of vWF
(34), SERPINE1, EPHB4, TYRO3,
TNFRSF11A, BOC and CHI3L1 with VTE risk. In a recent study, authors
have validated that thrombomodulin is involved in the evolution of
DVT and can be used as a dynamic biomarker to measure disease
activity (35). However, the
present study globally investigated the dynamic expression of mRNA
in the development of DVT. A total of 400, 1,176, 373 and 573
differentially expressed mRNAs were identified in the 1, 6, and 12
h, and 3-day groups, respectively, vs. the sham group. The results
indicated that 169, 885, 298 and 500 mRNAs were upregulated, and
231, 291, 75 and 73 mRNAs were downregulated. The sum of 69
upregulated mRNAs were identified at four time points, but no
downregulated mRNAs were identified, according to Venn analysis.
Certain targets were associated with DVT pathogenesis, such as vWF,
ICAM-1, VCAM-1, ROS, HIF1a, VEGFB, C-reactive protein, TF, CXCL12,
MDM2 and MMP13. This discovery may provide a foundation for future
mechanistic and clinical studies to advance understanding of the
molecular etiology of DVT.
STEM analysis can cluster, compare and visualize
gene expression data for short time series, and reveal their
expression trends at different times, to help identify
significantly expressed genes. From a biological perspective, genes
with similar expression patterns may have common characteristics
(36). STEM analysis to determine
the temporal trends of the differentially expressed circRNAs and
mRNAs was utilized. The STEM trends for mRNAs (profile 45) were
consistent with those for circRNAs (also profile 45), consistently
with the development of thrombosis in DVT rats, thereby confirming
a positive interaction between circRNAs and mRNAs (37,38).
Enrichment analysis annotated by profile 45
identified several distinct pathways closely associated with
thrombosis, including the platelet-associated pathway,
inflammation-associated pathway, and immune-associated pathway.
Platelets are a known component of venous thrombi (39). A previous study examining the role
of platelets in venous thrombosis by using an in vivo model
of venous valvular stasis has suggested that platelets adhere,
activate, and subsequently promote thrombus growth beyond the valve
sinus and toward the volumetric flow (40). In the present study, blood stasis
was successfully induced, and multiple pathways associated with
platelets were screened with enrichment assays, whose results were
consistent with previous findings. To this day, increasing evidence
supports the role of inflammation in thromboembolism (41). In patients with idiopathic DVT,
inflammatory markers have been observed to persist (42). Increased membrane ICAM-1,
P-selectin, and vWF, and impaired anticoagulant function have been
reported to favor the recruitment of leukocytes and platelets, thus
improving thrombus formation (43). vWF is expressed early in DVT and is
key for platelet recruitment and the initiation of thrombosis in
flow disturbance-driven thrombosis models (18). In the present study,
inflammatory-associated pathways and differentially expressed
inflammatory cytokines were also identified. The discovery of
multiple pathways further demonstrated the results of multiple
factors of thrombosis (44,45).
However, the important roles and mechanisms of key circRNAs and
mRNAs in these pathways in DVT progression, and whether the network
relationship between these pathways promotes DVT development, have
to be further studied in the future.
Because platelet activation is directly associated
with thrombosis, four key genes were discovered during the first
study of the platelet-associated pathway-Tuba4a, Kif18a, Kif4a and
Lgals3bp-directly or indirectly associated with platelet production
or activation, thrombosis, or cardiovascular diseases. Notably,
Tuba4a plays an important role in platelet biosynthesis and
cardiovascular diseases (46-51).
To increase the likelihood of screening functional
circRNAs, candidate circRNAs was identified according to
mRNA-circRNA interaction relationships, on the basis of mRNA
enrichment analysis, rather than simply relying on the expression
fold change of the circRNAs. With RT-qPCR, nine out of ten circRNAs
were verified; and it was further established that three of the
circRNAs were annotated in CIRCpedia-V2, a rat circRNA database,
thus confirming the presence of the three circRNAs. However, the
lack of validation with western blotting and functional
confirmation is a limitation of the present study.
Lou et al (16) have found that hsa_circ_000455 is a
new circRNA biomarker for DVT in patients, according to microarray
profiling. Their findings have indicated that certain circRNAs can
be used as indicators of DVT. At present, the expression of the
circRNA in patients with DVT was also observed, by identifying its
human homolog according to the circBase database. The expression of
the human homologous circRNA in the peripheral blood of 21 patients
with DVT was detected by RT-qPCR. The results revealed
significantly higher human homologous circRNA expression levels in
the DVT group than the control group. Receiver Operating
Characteristic analysis of 21 patients with identified circRNA
expression revealed an area under the curve of 0.897 with a
sensitivity of 83.3% and a specificity of 95.4% (data not shown).
These results showed preliminarily favorable diagnostic
performance. In addition, because the identified circRNAs in the
present study are the circRNAs at 1and 6 h after model
establishment, which are at early stage in the evolution of
thrombosis, it is considered that the circRNAs could be used as
indicators of DVT at the early stage, although the results need to
be further verified by expanding the sample.
In summary, in the present study a rat model of DVT
through the stenosis method was successfully established. This
model demonstrated the entire evolution of DVT. Through RNA
sequencing, the circRNA and mRNA expression profile trends were
thoroughly observed. This preliminarily study explored the global
expression profiles of circRNAs and mRNAs in the progression of
DVT. Competing endogenous RNA (ceRNA) is not a specific type of RNA
but a regulatory mechanism, wherein miRNA binds target mRNA and
consequently engages in post-transcriptional gene regulation by
inhibiting the mRNA's translation or degradation. CircRNA binds
miRNA through microRNA response elements, thus relieving the
inhibitory effect of miRNA on mRNA and indirectly regulating mRNA
expression. Therefore, circRNAs positively indirectly regulate gene
expression and function through ceRNA mechanisms (52-54).
Future studies on the ceRNA relationships between
circRNAs and mRNAs in vitro and in vivo are needed.
The potential network regulation mechanism of circRNA-miRNA-mRNA
was confirmed in the identified circRNA and mRNA analysis in DVT.
According to the MiRdb and Targetscan databases, miRNAs binding
both the circRNA and the mRNA were screened. In future studies, RNA
fluorescence in situ hybridization assays will be performed
to identify the subcellular localization of the circRNA and miRNA
in human umbilical vein endothelial cells and dual-luciferase
assays to verify the target relationship between the circRNA and
the miRNA, and the miRNA and the mRNA. In addition, further studies
are also necessary to discover the functions of the key circRNAs
and their regulatory mechanisms in DVT development.
Supplementary Material
Analysis of circRNAs differential
expression. (A) CircRNAs differentially expressed in rats withdeep
vein thrombosis at 1, 6 and 12 h and 3 days after ligation compared
with sham group. (B) Types of differentially expressed circRNAs.
(C) Venn analysis showed common upregulated circRNAs at 1, 6 and 12
h and 3 days after ligation compared with sham group. (D) Venn
revealed showed common downregulated circRNAs at 1, 6 and 12 h, and
3 days after ligation compared with sham group. circRNA, circular
RNA.
Analysis of mRNAs differential
expression. (A) mRNAs differentially expressed in rats with deep
vein thrombosis at 1, 6 and 12 h, and 3 days after ligation
compared with sham group. (B) Venn analysis showed common
upregulated mRNAs at 1, 6 and 12 h and 3 days after ligation
compared with sham group. (C) Venn analysis revealed common
downregulated mRNAs at 1, 6 and 12 h and 3 days after ligation
compared with sham group.
Thrombus formation rate of DVT rats at
each time point.
The top 20 up-and downregulated genes
at each time point.
Acknowledgements
Not applicable.
Funding
Funding: The present study was supported by the Health
Department Project of Jiangsu (grant no. Z2019003), the Nature
Science Foundation of Nantong, Jiangsu (grant no. MS12021003), the
Graduate Research and Practice Innovation Program of Jiangsu (grant
no. KYCX20_2799) and the Jiangsu Provincial Medical Key Discipline
(grant no. ZDXK202240).
Availability of data and materials
Transcriptome sequencing datasets generated and/or
analyzed during the current study are available in the NCBI
database with accession code GSE148333 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE148333).
The datasets used and/or analyzed during the current study are
available from the corresponding author on reasonable request.
Authors' contributions
BS, XC and YQZ designed the experiments. BS, XC, MZ,
QS and XXZ performed the experiments. BS, XC, XW and YQZ take
responsibility for analysis accuracy of the whole experimental
study. XW revised the manuscript. BS wrote the first draft of the
manuscript and all authors commented on previous versions of the
manuscript. BS and XC confirm the authenticity of all the raw data.
All authors read and approved the final version of the
manuscript.
Ethics approval and consent to
participate
Animal experiments were conducted following the
Chinese law on the protection of animals used for scientific
purposes. Furthermore, the study was specifically approved
(approval no. 20170930-001) by the Ethical Committee of Animal
Experiments of Nantong University (Nantong, China). The study was
carried out in compliance with the ARRIVE guidelines.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Chopard R, Albertsen IE and Piazza G:
Diagnosis and treatment of lower extremity venous thromboembolism:
A review. JAMA. 324:1765–1776. 2020.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Heit JA, Spencer FA and White RH: The
epidemiology of venous thromboembolism. J Thromb Thrombolysis.
41:3–14. 2016.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Wendelboe AM and Raskob GE: Global burden
of thrombosis: Epidemiologic aspects. Circ Res. 118:1340–1347.
2016.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Maatman TK, Jalali F, Feizpour C, Douglas
A II, McGuire SP, Kinnaman G, Hartwell JL, Maatman BT, Kreutz RP,
Kapoor R, et al: Routine venous thromboembolism prophylaxis may be
inadequate in the hypercoagulable state of severe coronavirus
disease 2019. Crit Care Med. 48:e783–e790. 2020.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Alexander M and Burbury K: A systematic
review of biomarkers for the prediction of thromboembolism in lung
cancer-results, practical issues and proposed strategies for future
risk prediction models. Thromb Res. 148:63–69. 2016.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Memczak S, Jens M, Elefsinioti A, Torti F,
Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer
M, et al: Circular RNAs are a large class of animal RNAs with
regulatory potency. Nature. 495:333–338. 2013.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Zhang Y, Zhang XO, Chen T, Xiang JF, Yin
QF, Xing YH, Zhu S, Yang L and Chen LL: Circular intronic long
noncoding RNAs. Mol Cell. 51:792–806. 2013.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Ashwal-Fluss R, Meyer M, Pamudurti NR,
Ivanov A, Bartok O, Hanan M, Evantal N, Memczak S, Rajewsky N and
Kadener S: circRNA biogenesis competes with pre-mRNA splicing. Mol
Cell. 56:55–66. 2014.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Liu Y, Yang Y, Wang Z, Fu X, Chu XM, Li Y,
Wang Q, He X, Li M, Wang K, et al: Insights into the regulatory
role of circRNA in angiogenesis and clinical implications.
Atherosclerosis. 298:14–26. 2020.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Chen S, Yang X, Yu C, Zhou W, Xia Q, Liu
Y, Chen Q, Chen X, Lv Y and Lin Y: The potential of circRNA as a
novel diagnostic biomarker in cervical cancer. J Oncol.
2021(5529486)2021.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Aufiero S, Reckman YJ, Pinto YM and
Creemers EE: Circular RNAs open a new chapter in cardiovascular
biology. Nat Rev Cardiol. 16:503–514. 2019.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Wang K, Long B, Liu F, Wang JX, Liu CY,
Zhao B, Zhou LY, Sun T, Wang M, Yu T, et al: A circular RNA
protects the heart from pathological hypertrophy and heart failure
by targeting miR-223. Eur Heart J. 37:2602–2611. 2016.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Ma C, Gu R, Wang X, He S, Bai J, Zhang L,
Zhang J, Li Q, Qu L, Xin W, et al: circRNA CDR1as promotes
pulmonary artery smooth muscle cell calcification by upregulating
CAMK2D and CNN3 via sponging miR-7-5p. Mol Ther Nucleric Acids.
22:530–541. 2020.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Zhao Z, Li X, Gao C, Jian D, Hao P, Rao L
and Li M: Peripheral blood circular RNA hsa_circ_0124644 can be
used as a diagnostic biomarker of coronary artery disease. Sci Rep.
7(39918)2017.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Yi YY, Yi Y, Zhu X, Zhang J, Zhou J, Tang
X, Lin J, Wang P and Deng ZQ: Circular RNA of vimentin expression
as a valuable predictor for acute myeloid leukemia development and
prognosis. J Cell Physiol. 234:3711–3719. 2019.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Lou Z, Li X, Li C, Li X, Du K, Zhang F and
Wang B: Microarray profile of circular RNAs identifies
hsa_circ_000455 as a new circular RNA biomarker for deep vein
thrombosis. Vascular. 30:577–589. 2022.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Lou Z, Ma H, Li X, Zhang F, Du K and Wang
B: Hsa_circ_0001020 accelerates the lower extremity deep vein
thrombosis via sponging miR-29c-3p to promote MDM2 expression.
Thromb Res. 211:38–48. 2022.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Brill A, Fuchs TA, Chauhan AK, Yang JJ, De
Meyer SF, Köllnberger M, Wakefield TW, Lämmle B, Massberg S and
Wagner DD: von Willebrand factor-mediated platelet adhesion is
critical for deep vein thrombosis in mouse models. Blood.
117:1400–1407. 2011.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Diaz JA, Hawley AE, Alvarado CM, Berguer
AM, Baker NK, Wrobleski SK, Wakefield TW, Lucchesi BR and Myers DD
Jr: Thrombogenesis with continuous blood flow in the inferior vena
cava. A novel mouse model. Thromb Haemost. 104:366–375.
2010.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Langmead B, Trapnell C, Pop M and Salzberg
SL: Ultrafast and memory-efficient alignment of short DNA sequences
to the human genome. Genome Biol. 10(R25)2009.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW,
Shi W and Smyth GK: limma powers differential expression analyses
for RNA-sequencing and microarray studies. Nucleic Acids Res.
43(e47)2015.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Wu T, Hu E, Xu S, Chen M, Guo P, Dai Z,
Feng T, Zhou L, Tang W, Zhan L, et al: clusterProfiler 4.0: A
universal enrichment tool for interpreting omics data. Innovation
(Camb). 2(100141)2021.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Panova-Noeva M, Wagner B, Nagler M, Koeck
T, Ten Cate V, Prochaska JH, Heitmeier S, Meyer I, Gerdes C, Laux
V, et al: Comprehensive platelet phenotyping supports the role of
platelets in the pathogenesis of acute venous
thromboembolism-results from clinical observation studies.
EBioMedicine. 60(102978)2020.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Bovill EG and van der Vliet A: Venous
valvular stasis-associated hypoxia and thrombosis: What is the
link? Annu Rev Physiol. 73:527–545. 2011.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Mackman N: New insights into the
mechanisms of venous thrombosis. J Clin Invest. 122:2331–2336.
2012.PubMed/NCBI View
Article : Google Scholar
|
|
27
|
Vilades D, Martínez-Camblor P,
Ferrero-Gregori A, Bär C, Lu D, Xiao K, Vea À, Nasarre L, Sanchez
Vega J, Leta R, et al: Plasma circular RNA hsa_circ_0001445 and
coronary artery disease: Performance as a biomarker. FASEB J.
34:4403–4414. 2020.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Ryu J, Kwon DH, Choe N, Shin S, Jeong G,
Lim YH, Kim J, Park WJ, Kook H and Kim YK: Characterization of
circular RNAs in vascular smooth muscle cells with vascular
calcification. Mol Ther Nucleic Acids. 19:31–41. 2020.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Su Q and Lv X: Revealing new landscape of
cardiovascular disease through circular RNA-miRNA-mRNA axis.
Genomics. 112:1680–1685. 2020.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Ge X, Meng Q, Zhuang R, Yuan D, Liu J, Lin
F, Fan H and Zhou X: Circular RNA expression alterations in
extracellular vesicles isolated from murine heart post
ischemia/reperfusion injury. Int J Cardiol. 296:136–140.
2019.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Lindström S, Wang L, Smith EN, Gordon W,
van Hylckama Vlieg A, de Andrade M, Brody JA, Pattee JW, Haessler
J, Brumpton BM, et al: Genomic and transcriptomic association
studies identify 16 novel susceptibility loci for venous
thromboembolism. Blood. 134:1645–1657. 2019.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Klarin D, Busenkell E, Judy R, Lynch J,
Levin M, Haessler J, Aragam K, Chaffin M, Haas M, Lindström S, et
al: Genome-wide association analysis of venous thromboembolism
identifies new risk loci and genetic overlap with arterial vascular
disease. Nat Genet. 51:1574–1579. 2019.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Zöller B, Svensson PJ, Dahlbäck B,
Lind-Hallden C, Hallden C and Elf J: Genetic risk factors for
venous thromboembolism. Expert Rev Hematol. 13:971–981.
2020.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Yuan S, Titova OE, Zhang K, Gou W,
Schillemans T, Natarajan P, Chen J, Li X, Åkesson A, Bruzelius M,
et al: Plasma protein and venous thromboembolism: Prospective
cohort and mendelian randomisation analyses. Br J Haematol.
201:783–792. 2023.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Cheng X, Sun B, Liu S, Li D, Yang X and
Zhang Y: Identification of thrombomodulin as a dynamic monitoring
biomarker for deep venous thrombosis evolution. Exp Ther Med.
21(142)2021.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Ernst J and Bar-Joseph Z: STEM: A tool for
the analysis of short time series gene expression data. BMC
Bioinformatics. 7(191)2006.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Navarro E, Mallén A, Cruzado JM, Torras J
and Hueso M: Unveiling ncRNA regulatory axes in atherosclerosis
progression. Clin Transl Med. 9(5)2020.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Li M, Duan L, Li Y and Liu B: Long
noncoding RNA/circular noncoding RNA-miRNA-mRNA axes in
cardiovascular diseases. Life Sci. 233(116440)2019.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Stone J, Hangge P, Albadawi H, Wallace A,
Shamoun F, Knuttien MG, Naidu S and Oklu R: Deep vein thrombosis:
Pathogenesis, diagnosis, and medical management. Cardiovasc Diagn
Ther. 7 (Suppl 3):S276–S284. 2017.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Lehmann M, Schoeman RM, Krohl PJ, Wallbank
AM, Samaniuk JR, Jandrot-Perrus M and Neeves KB: Platelets drive
thrombus propagation in a hematocrit and glycoprotein VI-dependent
manner in an in vitro venous thrombosis model. Arterioscler Thromb
Vasc Biol. 38:1052–1062. 2018.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Saghazadeh A, Hafizi S and Rezaei N:
Inflammation in venous thromboembolism: Cause or consequence? Int
Immunopharmacol. 28:655–665. 2015.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Poredos P and Jezovnik MK: In patients
with idiopathic venous thrombosis, interleukin-10 is decreased and
related to endothelial dysfunction. Heart Vessels. 26:596–602.
2011.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Budnik I and Brill A: Immune factors in
deep vein thrombosis initiation. Trends Immunol. 39:610–623.
2018.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Parakh RS and Sabath DE: Venous
thromboembolism: Role of the clinical laboratory in diagnosis and
management. J Appl Lab Med. 3:870–882. 2019.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Najem MY, Couturaud F and Lemarié CA:
Cytokine and chemokine regulation of venous thromboembolism. J
Thromb Haemost. 18:1009–1019. 2020.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Strassel C, Magiera MM, Dupuis A,
Batzenschlager M, Hovasse A, Pleines I, Guéguen P, Eckly A, Moog S,
Mallo L, et al: An essential role for α4A-tubulin in platelet
biogenesis. Life Sci Alliance. 2(e201900309)2019.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Feng L, Yang X, Asweto CO, Wu J, Zhang Y,
Hu H, Shi Y, Duan J and Sun Z: Genome-wide transcriptional analysis
of cardiovascular-related genes and pathways induced by
PM2.5 in human myocardial cells. Environ Sci Pollut Res
Int. 24:11683–11693. 2017.PubMed/NCBI View Article : Google Scholar
|
|
48
|
DeRoo EP, Wrobleski SK, Shea EM, Al-Khalil
RK, Hawley AE, Henke PK, Myers DD Jr, Wakefield TW and Diaz JA: The
role of galectin-3 and galectin-3-binding protein in venous
thrombosis. Blood. 125:1813–1821. 2015.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Tang F, Pan MH, Wan X, Lu Y, Zhang Y and
Sun SC: Kif18a regulates Sirt2-mediated tubulin acetylation for
spindle organization during mouse oocyte meiosis. Cell Div.
13(9)2018.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Kim S, Cho YB, Song CU, Eyun S and Seo YJ:
Kinesin family member KIF18A is a critical cellular factor that
regulates the differentiation and activation of dendritic cells.
Genes Genomics. 42:41–46. 2020.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Nunes Bastos R, Gandhi SR, Baron RD,
Gruneberg U, Nigg EA and Barr FA: Aurora B suppresses microtubule
dynamics and limits central spindle size by locally activating
KIF4A. J Cell Bio. 202:605–621. 2013.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Hansen TB, Jensen TI, Clausen BH, Bramsen
JB, Finsen B, Damgaard CK and Kjems J: Natural RNA circles function
as efficient microRNA sponges. Nature. 495:384–388. 2013.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Jeck WR, Sorrentino JA, Wang K, Slevin MK,
Burd CE, Liu J, Marzluff WF and Sharpless NE: Circular RNAs are
abundant, conserved, and associated with ALU repeats. RNA.
19:141–157. 2013.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Wilusz JE and Sharp PA: Molecular biology.
A circuitous route to noncoding RNA. Science. 340:440–441.
2013.PubMed/NCBI View Article : Google Scholar
|