Introduction
Prostate cancer (PC) is one of the most common
malignancies worldwide, affecting 1 in 9 male patients >65 years
of age (1-3).
At present, there is no effective treatment for advanced PC, which
has led to it being the second leading cause of cancer-associated
death in men (4). Therefore,
identifying novel endogenous factors responsible for PC cell
viability, migration and invasion will aid the understanding of the
progression of PC and lead to the development of novel approaches
for diagnosis and treatment.
The nuclear lamina, located in the inner layer of
the nuclear membrane, is a protein network composed of numerous
proteins, including lamins (3).
One of the main roles of lamins is to maintain the physiological
balance of cells (5). Lamin B1
(LMNB1) is a key B-type lamin protein that regulates cell
apoptosis, signal transduction and other functions and serves an
important role in the occurrence and development of tumors
(6). Forkhead box D1 (FOXD1)
belongs to the forkhead family transcription factor subfamily,
which can active downstream target genes through transcription and
participates in numerous biological activities, such as stem cell
differentiation, organogenesis, cell cycle regulation and signal
transduction (7). FOXD1 is a
mediator of smooth cell reprogramming via self-renewal and
differentiation (8). FOXD1 is
associated with oncogenicity, tumor progression and metastasis in
numerous types of carcinoma (9-11).
FOXD1 expression has been reported to be upregulated in head and
neck squamous cancer (12). FOXD1
is highly expressed in nasopharyngeal carcinoma and promotes cell
malignancy (13). Fan et al
(14) reported that FOXD1
expression is upregulated in laryngeal squamous cell carcinoma and
promotes cell epithelial-to-mesenchymal transition by targeting
zinc finger protein 532 expression. The aforementioned studies
suggest that FOXD1 may function as a cancer-causing gene multiple
cancer type. Jin et al (15) demonstrated that FOXD1 expression is
upregulated in PR and that FOXD1 silencing reduces the expression
of β-catenin and cyclin D1, which are involved in the Wnt/β-catenin
signaling pathway. The present study aimed to identify the key
functions of FOXD1 in PC and underlying mechanisms.
Materials and methods
Database analysis
The differential expression of FOXD1 in prostate
cancer and adjacent tissues, as well as its expression in PC
tissues at different stages, was analyzed by the Cancer Genome
Atlas (TCGA) database from the University of ALabama at Birmingham
CANcer data analysis Portal (UCLCAN) database (https://ualcan.path.uab.edu/cgi-bin/ualcan-res.pl).
Expression of FOXD1 in PC and para-cancer tissue was assessed. Gene
Expression Profiling Interactive Analysis (GEPIA) database
(gepia.cancer-pku.cn/) is an interactive
web server developed by Peking University that was used to analyze
the differential expression of associated genes and their
association with the survival prognosis of patients with prostate
cancer. The Human Protein Atlas (HPA) database (proteinatlas.org/) provided data on FOXD1 protein
expression in PC tissues (16).
Kaplan-Meier Plotter (kmplot.com/analysis/) was used to analyze the
association between FOXD1 expression and prognosis in PC. The
clinical patient data was from Xiantao (https://www.xiantaozi.com/). The most commonly used
staging for prostate cancer is TNM staging. TNM staging involves
three stages for all prostate cancer, where T refers to the in
situ tumor and describes the extent of the in situ
tumor; N refers to regional lymph nodes, describing whether there
is metastasis in the regional lymph nodes; M refers to distant
metastasis. The binding between LMNB1 and FOXD1 was predicated by
Human Tiny Flash Database (TFDB) (http://bioinfo.life.hust.edu.cn/AnimalTFDB/#!/).
Expression of FOXD1 in normal prostate and PC tissues, the
expression difference of FOXD1 in different grades of PC tissues,
and its association with the survival prognosis of patients with PC
were assessed.
Cell culture
RWPE-1 normal immortalized human prostate epithelial
cells and DU145, PC-3 and LNCaP prostate cancer cells (Shanghai
Institute of Cell Biology) were maintained in DMEM (Nanjing
Biochannel Biotechnology Co., Ltd.) with 10% FBS (Nanjing
Biochannel Biotechnology Co., Ltd.) at 37˚C with 5%
CO2.
Cell transfection and treatment
FOXD1 overexpression vector (pcDNA3.1-FOXD1), small
interfering RNA (siRNA)-LMNB1 and siRNA negative control (NC) were
synthesized by Nanjing Genscript Biotechnology Co., Ltd. The sense
and antisense strands for each siRNA were as follows: siRNA-FOXD1,
5'-TCGCCGAGCTCTGTTCTTAGACTCT-3'; siRNA1-LMNB1,
5'-TCCCGCGTGCGTGTGTGAGTGGGTG-3'; siRNA2-LMNB1,
5'-GGGCAAGTTAGGTTTGCTAGCTGCT-3'; and siRNA negative control (NC),
5'-UUCUCCGAACGUGUCUTT-3'. The siRNA-FOXD1 (500 ng/µl) and
siRNA-LMNB1 (500 ng/µl) was transfected into prostate cancer cells
for 48 h at 37˚C using Lipofectamine 3000 (Invitrogen; Thermo
Fisher Scientific, Inc.) according to the manufacturer's
instructions. Cells were collected 48 h after transfection. The
efficiency of transfection was analyzed using reverse
transcription-quantitative PCR (RT-qPCR).
RT-qPCR analysis
Total RNA of cells was extracted using TRIzol
(Thermo Fisher Scientific, Inc.) according to the manufacturer's
protocol. A total of 2 µl cDNA was synthesized from 2 ng total RNA
with a RT Toolkit (Promega Corporation) according to the
manufacturer's protocol. RT-qPCR was performed using a
SYBR® Premix Ex Taq™ (Takara Bio, Inc.).
Thermocycling conditions were as follows: Initial denaturation for
10 min at 95˚C, followed by 35 cycles of 95˚C for 5 sec and 72˚C
for 30 sec. The 2-ΔΔCq method (17) was used to calculate relative gene
expression. GAPDH was used to normalize RNA expression. The primer
sequences were as follows: FOXD1 forward,
5'-TGAGCACTGAGATGTCCGATG-3' and reverse,
5'-CACCACGTCGATGTCTGTTTC-3'; LMNB1 forward,
5'-AAGCAGCTGGAGTGGTTGTT-3' and reverse, 5'-TTGGATGCTCTTGGGGTTC-3';
and GAPDH forward, 5'-CCCATGTTCGTCATGGGTGT-3' and reverse,
5'-TGGTCATGAGTCCTTCCACGATA-3'.
Cell proliferation analysis
Cell proliferation was determined using Cell
Counting Kit-8 (CCK-8; Beyotime Institute of Biotechnology).
Briefly, 2x104 cells/well were seeded in 96-well plates
for 24 h and transiently transfected with siRNA-FOXD1, as
aforementioned. A total of 10 µl CCK-8/well was added at 0, 1, 2, 3
and 4 days post-transfection. The plates were cultured at 37˚C for
2 h. The absorbance at 450 nm was measured using a microplate
reader.
Luciferase reporter assay
Luciferase reporter assay was used to detect the
association between FOXD1 and LMNB1. FOXD1 promoter region fragment
containing the potential binding site of LMNB1 was amplified by PCR
and cloned into the pGL3-basic vector (Promega Corporation), as
were the wild-type/mutant sequences (WT/Mut). The PCR was performed
using the following sequences: Forward, GTGTGGTTGGGACTCACGTCGCTTTC
and reverse, TAGCAGAAGGGGGCCTGTCACATGG (Nanjing Genscript
Biotechnology Co., Ltd.). Next, the pGL3-basic vector or pGL3-LMNB1
(Shanghai GenePharma Co., Ltd.) and luciferase reporter vectors
were co-transfected into 293 cells using Lipofectamine 3000
(Invitrogen; Thermo Fisher Scientific, Inc.). A luciferase reporter
assay kit (cat. no. E1910; Promega Corporation) was used for the
luciferase activity measurement 48 h after transfection. Firefly
fluorescence intensity was calculated using Renilla
luciferase activity for normalization.
Western blot analysis
RIPA Lysis Buffer (Beyotime Institute of
Biotechnology) was used to extract total proteins from prostate
cancer tissue and cell samples, and a BCA kit (Beyotime Institute
of Biotechnology) was used to detect the concentration. An equal
quantity of proteins (30 µg) was added in each lane of a 10%
SDS-PAGE gel. Membrane Blocking Solution (MilliporeSigma) was used
to block the PVDF membranes with transferred proteins for 1 h at
room temperature. Membranes were incubated with primary antibodies
against FOXD1 (1:1,000; cat. no. sc-293238), E-cadherin (1:1,000;
cat. no. sc-8426), N-cadherin (1:1,000; cat. no. sc-8424), Vimentin
(1:1,000; cat. no. sc-6260) and GAPDH (1:2,000; cat. no. sc-47724)
(all Santa Cruz Biotechnology, Inc.) overnight at 4˚C.
Subsequently, membranes were incubated with horseradish
peroxidase-conjugated secondary antibody (1:5,000; cat. no.
sc-2357; Santa Cruz Biotechnology, Inc.) at room temperature for 2
h before ECL detection using BeyoECL Plus kit (Beyotime Institute
of Biotechnology). ImageJ software (version 1.53; National
Institutes of Health) was used to analyze density of immune blots.
GAPDH acted as the internal reference protein.
Transwell assay
A total of 3x104 cells were added to
serum-free DMEM and placed in the upper layer of the Transwell
chamber (Corning, Inc.) that had been precoated with Matrigel at
room temperature for 1 h. A total of 600 µl DMEM, including 20% FBS
(Nanjing Biochannel Biotechnology Co., Ltd.), was placed in the
bottom layer at 37˚C. Following 24 h of incubation, 4%
paraformaldehyde was used for 20 min to fix the cells that had
invaded the bottom layer at room temperature. Subsequently, cells
were stained with crystal violet for 20 min at room temperature.
Cells were counted manually under a light microscope (Olympus
Corporation) in five randomly selected fields of view/sample.
Wound healing assay
LNCaP cells in logarithmic growth phase (when the
cell density reached ≥90%) were inoculated into the 6-well plate.
For LNCaP cells in the control, NC, siRNA-FOXD1, siRNA-LMNB1,
siRNA-LMNB1 + pcDNA 3.1 and siRNA-LMNB1 + pcDNA 3.1-FOXD1 groups,
lines were drawn horizontally using a 200-µl pipette tip at the
bottom of the culture plate for reference. When cells were attached
to the surface of the plate, the DMEM (Nanjing Biochannel
Biotechnology Co., Ltd.) was replaced with fresh medium containing
1% FBS (Nanjing Biochannel Biotechnology Co., Ltd.). After 48 h at
37˚C, scratch width was imaged by a light microscope (Olympus
Corporation) observed to calculate wound healing rate as follows:
(Wound width at 0 h-wound width at 48 h)/wound width at 0 h
x100.
Statistical analysis
All experimental results from three independent
experiments were analyzed using GraphPad software (version 5.0;
GraphPad Software, Inc.; Dotmatics). All data are presented as the
mean ± SD. Student's t-test was used for comparisons between two
groups and one-way analysis of variance was used for comparisons
among multiple groups. The receiver operating characteristic (ROC)
curve was compared by log-rank test. P<0.05 was considered to
indicate a statistically significant difference.
Results
FOXD1 is upregulated in PC and
associated with adverse prognosis
Tumor samples from TCGA were analyzed to
characterize FOXD1 expression (Fig.
1A). FOXD1 was upregulated in numerous types of cancer.
Subsequently, FOXD1 expression in PC was assessed based on the
GEPIA2 (Fig. 1B) and TCGA
(Fig. 1C) databases. FOXD1
expression was significantly upregulated in PC samples.
Furthermore, analysis of the association between FOXD1 and
histological grading of patients with PC was performed. FOXD1
expression was positively associated with the Gleason score of the
patients with PC (Fig. 1D).
Additionally, the protein expression of PC based on the HPA
database confirmed the abnormally high expression of FOXD1 in PC
tissue (Fig. 1E).
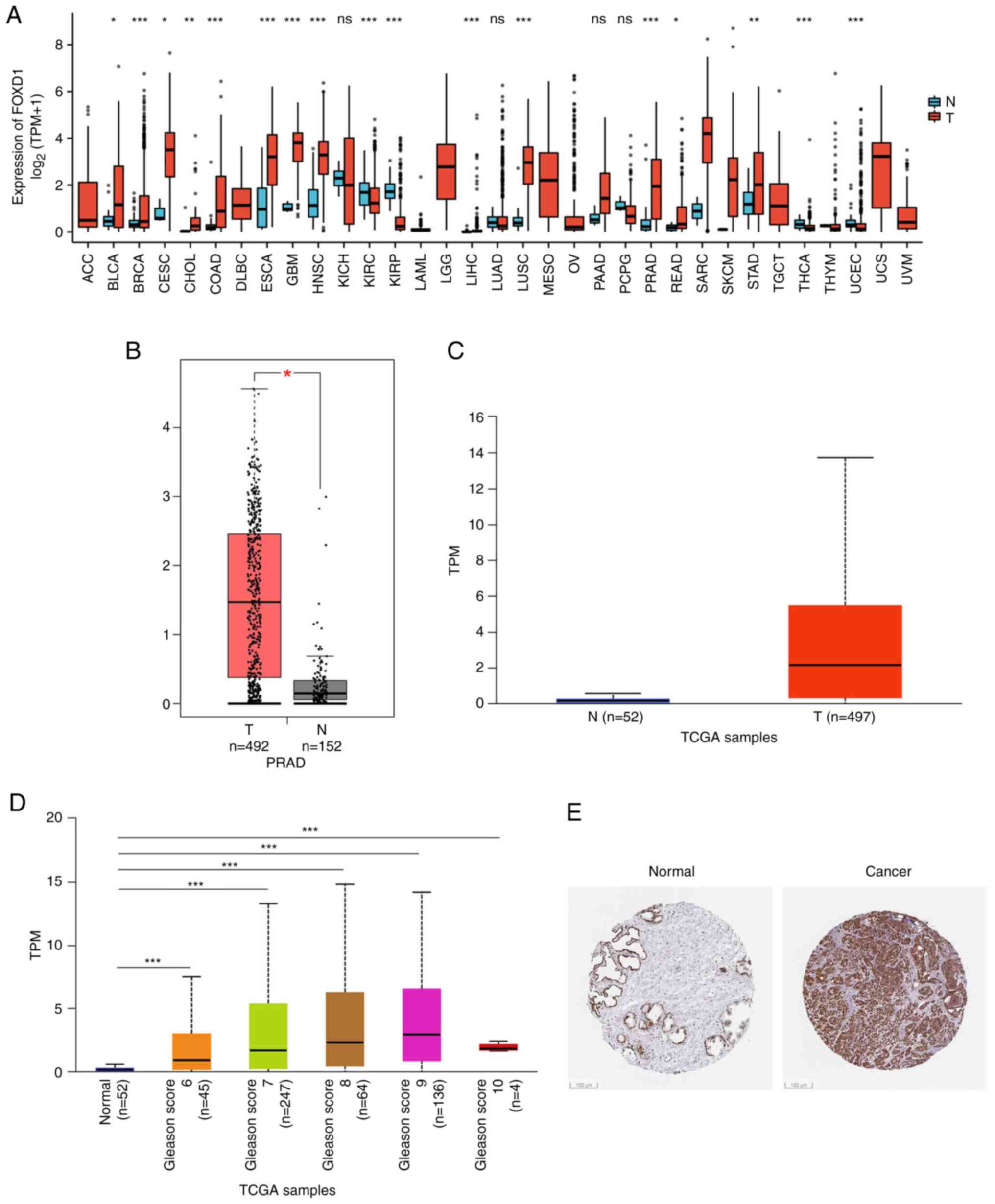 | Figure 1Differential expression of FOXD1 in PC
and normal tissues. (A) Pan-cancer analyses of FOXD1 expression
(P-value, T vs. N). (B) Plot showing that PC tissues (n=152) had
significantly elevated FOXD1 expression levels compared with normal
tissues (n=492) in the GEPIA2 database. (C) Plot showing that PC
tissues (n=497) had significantly elevated FOXD1 expression levels
compared with normal tissues (n=52) in TCGA database. (D)
Expression of FOXD1 in PC based on patient's Gleason score. (E)
Expression of FOXD1 in PC tissues (Human Protein Atlas database;
Scale bar, 100 µm). *P<0.05, **P<0.01
and ***P<0.001. T, tumor; N, normal; TPM, transcripts
per million; TCGA, The Cancer Genome Atlas; PC, prostate cancer;
FOXD1, forkhead box D1; GEPIA, Gene Expression Profiling
Interactive Analysis; PRAD, prostate adenocarcinoma. |
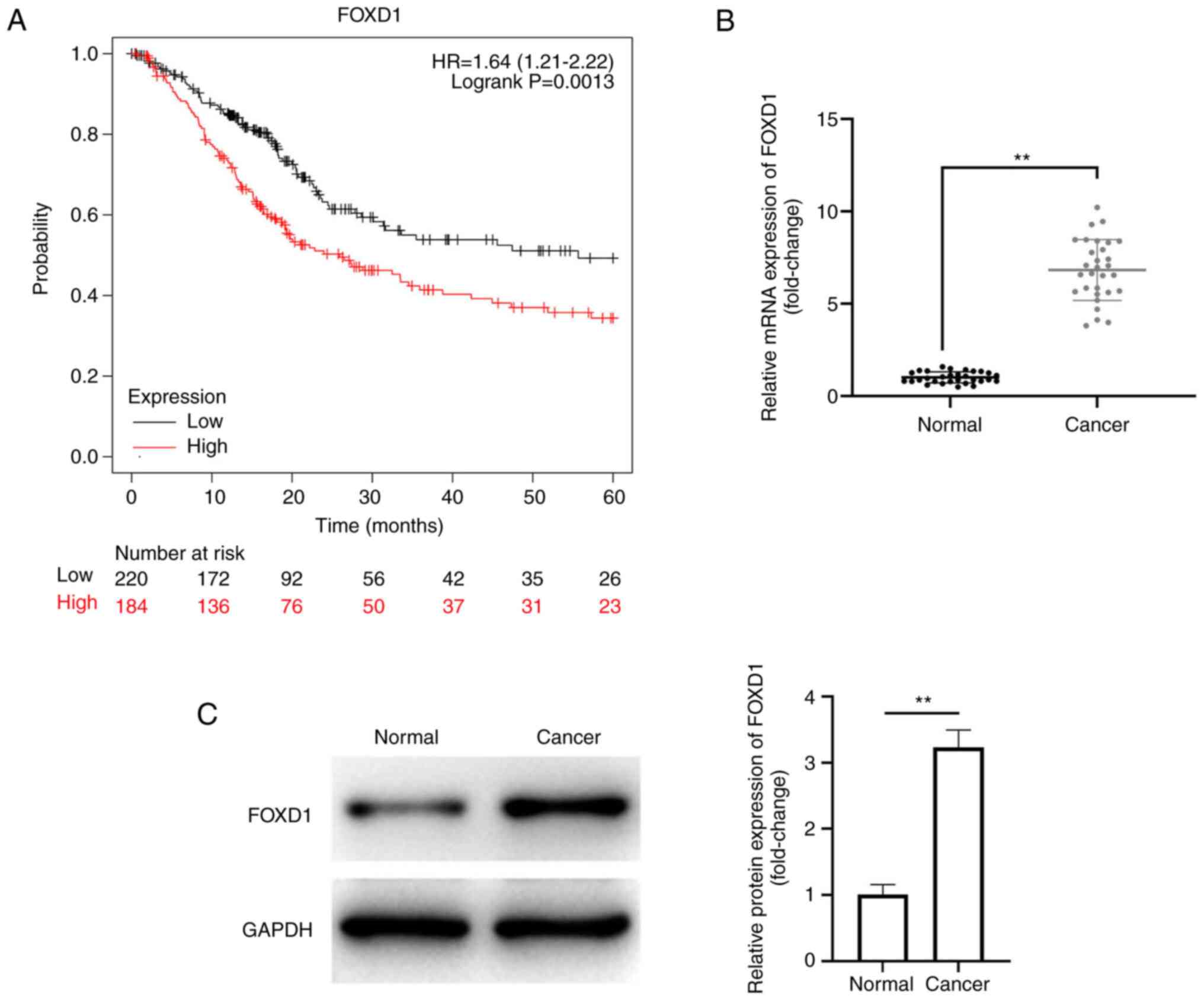
The present study evaluated the relationship between
FOXD1 and prognosis in PC. FOXD1 expression was an independent
prognostic factor for TNM classification (Table I). Patients with low FOXD1
expression exhibited a markedly higher probability of survival at
all time points analyzed (Fig.
2A). Also, relative mRNA expression level of FOXD1 in PC tissue
was significantly higher than that in normal tissue (Fig. 2B). Meanwhile, relative protein
expression of FOXD1 in PC tissue was significantly higher than that
in PC tissues (Fig. 2C). These
results supported the oncogenic role of FOXD1 in PC
progression.
 | Table IClinical characteristic of
patients. |
Table I
Clinical characteristic of
patients.
| Characteristic | Low expression of
FOXD1 (n=249) | High expression of
FOXD1 (n=250) | P-value |
|---|
| T stage, n/total n
(%) | | |
<0.001a |
|
T2 | 117/492 (23.8) | 72/492 (14.6) | |
|
T3 | 129/492 (26.2) | 163/492 (33.1) | |
|
T4 | 1/492 (0.2) | 10/492 (2.0) | |
| N stage, n/total n
(%) | | | 0.035a |
|
N0 | 180/426 (42.3) | 167/426 (39.2) | |
|
N1 | 30/426 (7.0) | 49/426 (11.5) | |
| M stage, n/total n
(%) | | | 0.118 |
|
M0 | 233/458 (50.9) | 222/458 (48.5) | |
|
M1 | 0/458 (0.0) | 3/458 (0.7) | |
| Median age (IQR),
years | 61 (56-66) | 61.5 (56-66) | 0.609 |
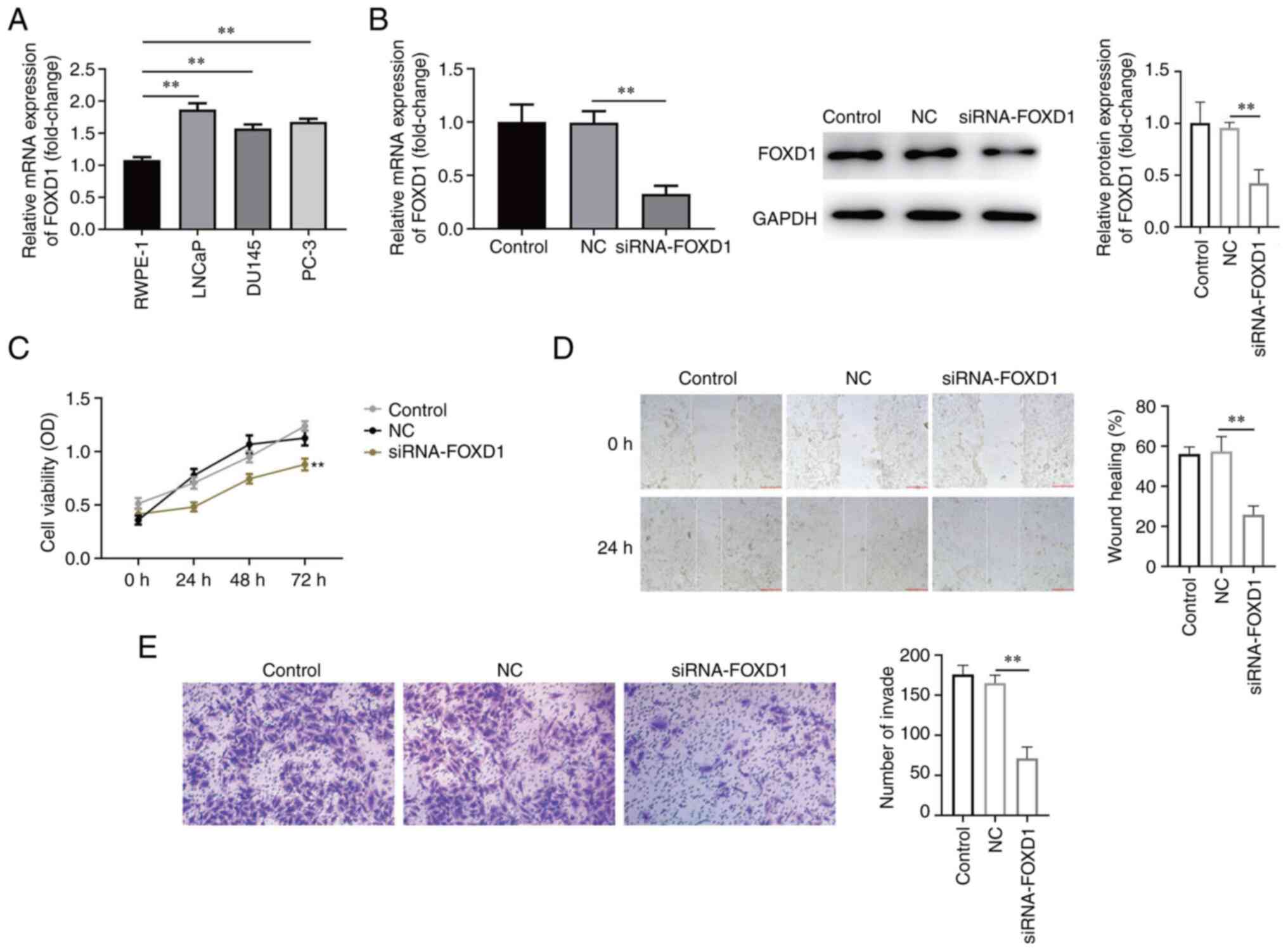
FOXD1 knockdown inhibits PC cell
viability, migration and invasion
FOXD1 expression in PC cell lines was detected by
western blotting. FOXD1 expression was upregulated in all PC cell
lines; LNCaP cells, which exhibited the highest expression levels,
were selected for subsequent experiments (Fig. 3A). siRNA-FOXD1 markedly inhibited
FOXD1 mRNA and protein expression (Fig. 3B). The result of CCK-8 analysis
indicated a significant decrease in the viability of LNCaP cells in
which FOXD1 was knocked down (Fig.
3C). The results of wound healing and Transwell assays revealed
that the knockdown of FOXD1 significantly inhibited LNCaP cell
migration and invasion (Fig. 3D
and E). These results demonstrated
that knockdown of FOXD1 inhibited LNCaP cell viability, invasion
and migration.
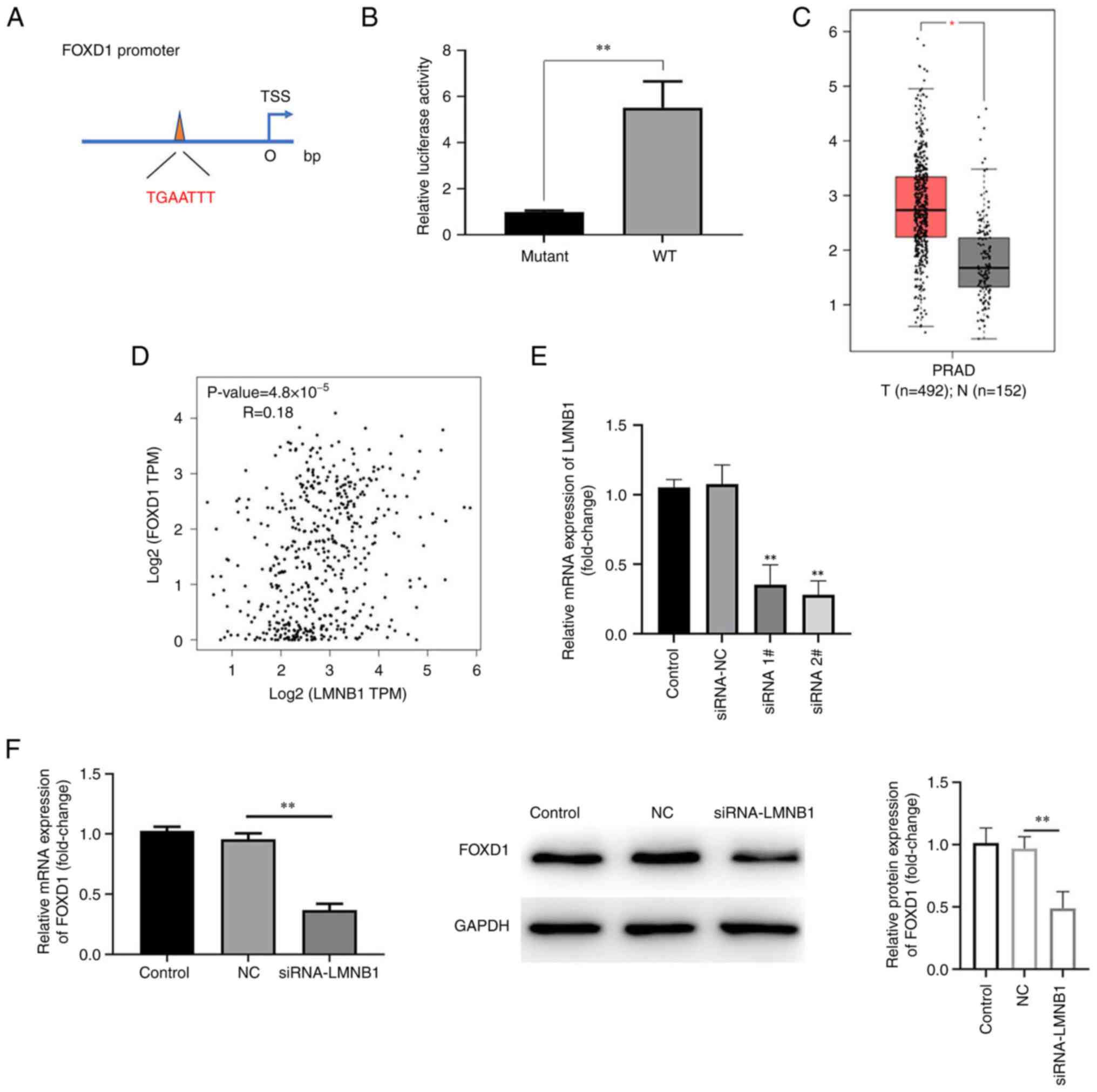
LMNB1 is positively associated with
FOXD1
HumanTFDB analysis revealed that LMNB1 targets FOXD1
mRNA (positions 1,128-1,143; Fig.
4A). The luciferase activity was significantly elevated in
cells co-transfected with LMNB1 WT compared with Mut, indicating
binding between FOXD1 and LMNB1 (Fig.
4B). Subsequently, the present study analyzed TCGA database and
revealed that LMNB1 was highly expressed in patients with prostate
adenocarcinoma (Fig. 4C).
Additionally, the GEPIA database was used to characterize the
association between FOXD1 and LMNB1, which demonstrated a positive
association between them (Fig.
4D). RT-qPCR was used to detect transfection efficiency of the
siRNAs. mRNA expression of LMNB1 was downregulated in both siRNA 1#
and siRNA 2# groups and mRNA expression of LMNB1 in siRNA 2# was
lower than that in siRNA 1# group. Therefore, siRNA 2# was selected
for subsequent experiments (Fig.
4E). Next, western blotting was used to detect the LMNB1
protein levels after knockdown of FOXD1. The results indicated
decreased LMNB1 expression following transfection of siRNA-FOXD1
(Fig. 4F).
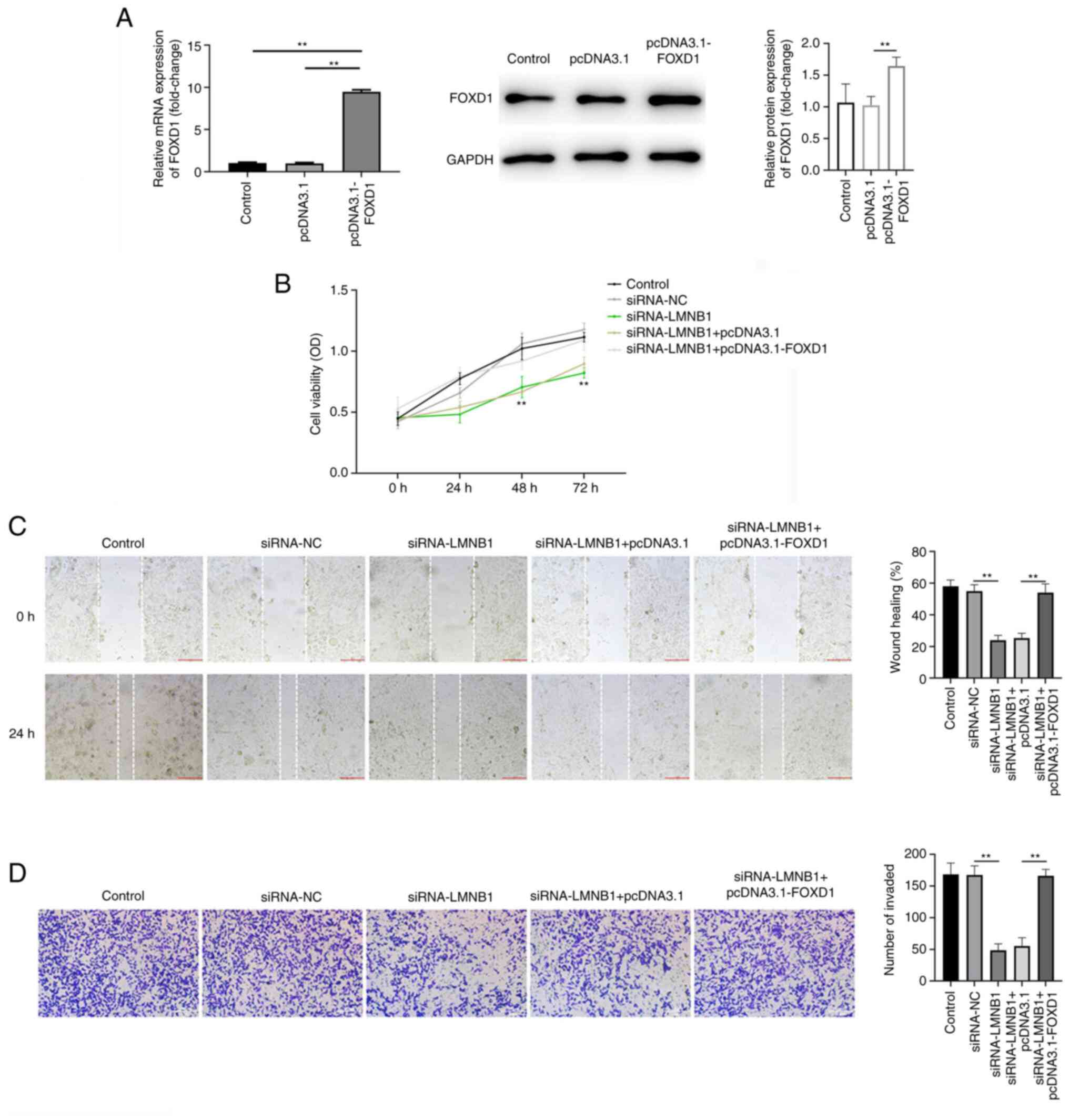
LMNB1 acts directly on FOXD1 to
mediate PC malignant progression
To clarify the mechanism of the influence of LMNB1
on PC cell progression, LNCaP cells were transfected with pcDNA3.1
and pcDNA3.1-FOXD1. RT-qPCR and western blotting demonstrated that
pcDNA3.1-FOXD1 was successfully transfected (Fig. 5A). CCK-8 assay revealed that
siRNA-LMNB1 notably decreased cell viability compared with the
control group, while FOXD1 overexpression counteracted this effect
(Fig. 5B). Similarly, knockdown of
LMNB1 significantly inhibited cell migration and invasion, and
these effects were reversed by FOXD1 overexpression (Fig. 5C and D).
Discussion
The present study investigated the effect of FOXD1
in the occurrence and development of PC and its possible
mechanisms. Utilizing TCGA, the present study demonstrated that
FOXD1 mRNA expression was higher in PC than normal tissues and its
abnormally high expression was associated with poor prognosis.
In vitro experiments revealed that FOXD1 knockdown markedly
inhibited viability and invasion of PC cells. LMNB1 targeted FOXD1
and mediated the PC malignant progression. These findings
highlighted that LMNB1 targeted regulation of FOXD1 to promote the
occurrence and development of PC.
The FOX family is a key complex family of genes that
includes a variety of cell- and tissue-specific ‘wing helix’
transcription factors (18).
FOXD1, a newly identified FOX family transcription factor, serves
as an oncogene in multiple types of cancer (19-21).
For example, Sun et al (19) indicated that FOXD1 promoted
progression of metastatic melanoma by regulating CTGF expression.
Zong et al (20) suggested
that FOXD1 was a biomarker of colorectal cancer, and Li et
al (21) revealed that FOXD1
was associated with the development of primary oral squamous cell
carcinoma.
Current studies showed that the expression of FOXD1
was upregulated in PC (22,23).
Furthermore, the present study analyzed the association between
abnormal FOXD1 expression and PC grade and prognosis using
bioinformatics. Further experiments using PC cell lines revealed
that FOXD1 expression was aberrantly high in PC cells, as expected.
Additionally, knockdown of FOXD1 markedly inhibited PC cell
viability, migration and invasion.
To determine the potential molecular mechanism of
LMNB1, the present study predicted its possible target, FOXD1.
LMNB1 is a protein component of the nucleoskeleton (24). LMNB1 possesses numerous biological
functions. In addition to maintaining shape and integrity of the
nucleus, LMNB1 regulates cell proliferation and senescence, DNA
replication and gene expression, DNA damage repair and chromosome
distribution and aggregation (25,26).
Additionally, LMNB1 is associated with the development of
neurological disease and tumors (27,28).
Previous studies and network analyses have demonstrated the
biomarker utility of LMNB1 in human cancer (7). To the best of our knowledge, the only
recent report states that LMNB1 upregulation is associated with
cancer metastasis and adverse survival outcomes in patients with
primary PC (28). Furthermore, the
present results indicated that LMNB1 positively regulated FOXD1.
The effects of knockdown of LMNB1 on PC cells were reversed by
FOXD1 overexpression.
In conclusion, the present study provided evidence
that FOXD1 is key in malignant progression of PC. The function may
be positively regulated through LMNB1. However, the number of
clinical samples in the present study was small and further
research is required.
Acknowledgements
Not applicable.
Funding
Funding: This study was supported in part by grants from the
Guizhou Province Department of Education Project [grant no. QJH KY
(2020) 063] and the Anshun University/Innovation Center for
Efficient Agricultural of Guizhou Mountain Characteristics/Branch
of learning in Agricultural Resources and Environment.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
YH contributed to the conception of the study,
performed the data analyses and wrote the manuscript. LZ performed
the experiments. TL and EL contributed significantly to data
analysis and manuscript preparation. All the authors have read and
approved the final manuscript. YH, LZ, TL and EL confirm the
authenticity of all the raw data.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Pernar CH, Ebot EM, Wilson KM and Mucci
LA: The epidemiology of prostate cancer. Cold Spring Harb Perspect
Med. 8(a030361)2018.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Catalona WJ: Prostate cancer screening.
Med Clin North Am. 102:199–214. 2018.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Boyle HJ, Alibhai S, Decoster L,
Efstathiou E, Fizazi K, Mottet N, Oudard S, Payne H, Prentice M,
Puts M, et al: Updated recommendations of the international society
of geriatric oncology on prostate cancer management in older
patients. Eur J Cancer. 116:116–136. 2019.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Swami U, McFarland TR, Nussenzveig R and
Agarwal N: Advanced prostate cancer: Treatment advances and future
directions. Trends Cancer. 6:702–715. 2020.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Charar C and Gruenbaum Y: Lamins and
metabolism. Clin Sci (Lond). 131:105–111. 2017.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Qin H, Lu Y, Du L, Shi J, Yin H, Jiang B,
Chen W, Diao W, Ding M, Cao W, et al: Pan-cancer analysis
identifies LMNB1 as a target to redress Th1/Th2 imbalance and
enhance PARP inhibitor response in human cancers. Cancer Cell Int.
22(101)2022.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Golson ML and Kaestner KH: Fox
transcription factors: From development to disease. Development.
143:4558–4570. 2016.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Koga M, Matsuda M, Kawamura T, Sogo T,
Shigeno A, Nishida E and Ebisuya M: Foxd1 is a mediator and
indicator of the cell reprogramming process. Nat Commun.
5(3197)2014.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Chen C, Xu ZQ, Zong YP, Ou BC, Shen XH,
Feng H, Zheng MH, Zhao JK and Lu AG: CXCL5 induces tumor
angiogenesis via enhancing the expression of FOXD1 mediated by the
AKT/NF-κB pathway in colorectal cancer. Cell Death Dis.
10(178)2019.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Gao YF, Liu JY, Mao XY, He ZW, Zhu T, Wang
ZB, Li X, Yin JY, Zhang W, Zhou HH and Liu ZQ: LncRNA FOXD1-AS1
acts as a potential oncogenic biomarker in glioma. CNS Neurosci
Ther. 26:66–75. 2020.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Li D, Fan S, Yu F, Zhu X, Song Y, Ye M,
Fan L and Lv Z: FOXD1 promotes cell growth and metastasis by
activation of vimentin in NSCLC. Cell Physiol Biochem.
51:2716–2731. 2018.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Huang J, Liang B and Wang T: FOXD1
expression in head and neck squamous carcinoma: A study based on
TCGA, GEO and meta-analysis. Biosci Rep.
41(BSR20210158)2021.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Ren D, Lu J, Han X, Xiong W, Jiang H, Wei
Y and Wang Y: LINC00641 contributes to nasopharyngeal carcinoma
cell malignancy through FOXD1 upregulation at the
post-transcriptional level. Biochem Cell Biol. 99:750–758.
2021.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Fan L, Wang J, Deng P, Wang Y, Zhang A,
Yang M and Zeng G: Foxhead box D1 promotes the partial
epithelial-to-mesenchymal transition of laryngeal squamous cell
carcinoma cells via transcriptionally activating the expression of
zinc finger protein 532. Bioengineered. 13:3057–3069.
2022.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Jin Y, Liang Z and Lou H: The emerging
roles of fox family transcription factors in chromosome
replication, organization, and genome stability. Cells.
9(258)2020.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Uhlén M, Fagerberg L, Hallström BM,
Lindskog C, Oksvold P, Mardinoglu A, Sivertsson Å, Kampf C,
Sjöstedt E, Asplund A, et al: Proteomics. Tissue-based map of the
human proteome. Science. 347(1260419)2015.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Quintero-Ronderos P and Laissue P: The
multisystemic functions of FOXD1 in development and disease. J Mol
Med (Berl). 96:725–739. 2018.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Sun Q, Novak D, Hüser L, Poelchen J, Wu H,
Granados K, Federico A, Liu K, Steinfass T, Vierthaler M, et al:
FOXD1 promotes dedifferentiation and targeted therapy resistance in
melanoma by regulating the expression of connective tissue growth
factor. Int J Cancer. 149:657–674. 2021.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Zong Y, Miao Y, Li W, Zheng M, Xu Z, Gao
H, Feng W, Xu Z, Zhao J, Shen L and Lu A: Combination of FOXD1 and
Plk2: A novel biomarker for predicting unfavourable prognosis of
colorectal cancer. J Cell Mol Med. 26:3471–3482. 2022.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Li Z, Yan T, Wu X, Zhang W, Li J, Wang L
and Yang J: Increased expression of FOXD1 is associated with
cervical node metastasis and unfavorable prognosis in oral squamous
cell carcinoma. J Oral Pathol Med. 49:1030–1036. 2020.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Cai K, Chen S, Zhu C, Li L, Yu C, He Z and
Sun C: FOXD1 facilitates pancreatic cancer cell proliferation,
invasion, and metastasis by regulating GLUT1-mediated aerobic
glycolysis. Cell Death Dis. 13(765)2022.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Donmez C and Konac E: Silencing effects of
FOXD1 inhibit metastatic potentials of the PCa via
N-cadherin-Wnt/β-catenin crosstalk. Gene.
836(146680)2022.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Li J, Sun Z, Cui Y, Qin L, Wu F, Li Y, Du
N and Li X: Knockdown of LMNB1 inhibits the proliferation of lung
adenocarcinoma cells by inducing DNA damage and cell senescence.
Front Oncol. 12(913740)2022.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Saed L, Jeleń A, Mirowski M and
Sałagacka-Kubiak A: Prognostic significance of HMGA1 expression in
lung cancer based on bioinformatics analysis. Int J Mol Sci.
23(6933)2022.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Evangelisti C, Rusciano I, Mongiorgi S,
Ramazzotti G, Lattanzi G, Manzoli L, Cocco L and Ratti S: The wide
and growing range of lamin B-related diseases: From laminopathies
to cancer. Cell Mol Life Sci. 79(126)2022.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Luo F, Han J, Chen Y, Yang K, Zhang Z and
Li J: Lamin B1 promotes tumor progression and metastasis in primary
prostate cancer patients. Future Oncol. 17:663–673. 2021.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Hua Y, He Z and Zhang X: A pan-cancer
analysis based on weighted gene co-expression network analysis
identifies the biomarker utility of lamin B1 in human tumors.
Cancer Biomark. 34:23–39. 2022.PubMed/NCBI View Article : Google Scholar
|