Introduction
Intrahepatic cholangiocarcinoma (ICC) is a rare
(0.79 cases per 100,000 individuals) but often fatal malignancy
originating from the epithelium of the secondary bile duct and its
branches (1,2). ICC is the second most common primary
liver malignancy after hepatocellular carcinoma (HCC), and the
incidence of ICC has increased globally over the last decades,
whereas the incidence of perihilar cholangiocarcinoma and distal
cholangiocarcinoma has decreased (3-8).
Some of these changes are attributable to the alterations in
disease classification, or to the more advanced diagnostic
modalities that may identify early lesions and biliary malignancies
that were previously undiagnosed (2). Furthermore, the increasing incidence
of ICC may be associated with certain newly recognized strong risk
factors, such as viral hepatitis, non-specific cirrhosis, nonviral
chronic liver diseases and metabolic diseases (9). Due to the rarity, early metastasis
and unclear symptoms of early ICC, only 10-15% of patients can
undergo radical resection (10,11).
The median overall survival (OS) of patients with ICC is 12-18
months, with the 5-year OS being rates <5% (12,13).
Thus, it is necessary to elucidate the precise molecular mechanisms
of ICC pathogenesis for predicting prognosis.
Integrins are a group of integral transmembrane
heterodimers with numerous functions, including cell adhesion in
the extracellular matrix (ECM) and acting as receptors for
recognizing Arg-Gly-Asp (RGD) peptide motifs, laminin, snake
venoms, viruses and other pathogens (14-17).
Given the roles of integrins in multiple fundamental biological
processes, the aberrant expression of integrin family members is
linked to the prognosis of various types of cancer, including
gastric cancer (18), breast
cancer (19), pancreatic carcinoma
(20) and colorectal cancer
(21).
Integrin β5 (ITGB5), which associates with integrin
αV (22), has been indicated to
facilitate cancer cell migration, invasion and transforming growth
factor β (TGF-β)-induced epithelial-mesenchymal transition
(23). Lin et al (24) reported that ITGB5 promoted
tumorigenesis in HCC by interacting with β-catenin. Furthermore,
Wortzel et al (25)
revealed that ITGB5 was enriched in liver metastatic pancreatic
cancer exosomes. ITGB5 is a potential independent prognostic
biomarker and therapeutic target for hepatitis B virus
(HBV)-related HCC and may be useful for its diagnosis (26). These studies have indicated the
potential role of ITGB5 in intercellular communication during tumor
progression and metastasis. However, the role of ITGB5 in ICC
remains largely unknown. The aim of the present study was to
investigate the ITGB5 expression levels in ICC tissues and to
examine whether the expression level of ITGB5 was associated with
the prognosis of patients with ICC.
Materials and methods
Data processing of acquisition and
identification of differentially expressed genes (DEGs)
Microarray data for ICC were obtained from the Gene
Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/). Two expression
profiling datasets, GSE26566(27)
and GSE107943(28), were obtained.
The GSE26566 dataset contained 104 ICC samples and 6 normal
samples. The GSE107943 dataset contained 30 ICC samples and 27
normal samples, as well as clinicopathological information
regarding the tumor samples. All expression profiles were
downloaded and processed using the R package of GEOquery (29). The transcriptome profiles of 32 ICC
samples and 9 normal samples and clinical information of tumor
samples were obtained from The Cancer Genome Atlas (TCGA) database
(https://cancergenome.nih.gov/) and
analyzed using the R package of TCGAbiolinks (30). The GSE26566 and TCGA datasets were
analyzed separately as volcano maps using GraphPad Prism 9
(GraphPad Software; Dotmatics). Log2 fold-change
(FC)>1 and P<0.05 were defined as the screening thresholds.
Common DEGs of the GSE26566 and TCGA datasets were obtained through
the TBtools (31). For the
definition of high or low expression levels of ITGB5 in the
GSE107943 and TCGA datasets, the expression of ITGB5 was divided
into two groups according to the survival status of patients with
ICC, and separate receiver operating characteristic curves were
obtained to determine the ITGB5 cut-off value with area under the
curve >0.8 and P<0.05.
Patient tissues
The present retrospective study on patient tissues
and data was approved by the Medical Ethics Committee of Taizhou
People's Hospital (approval no. KY 2020-091-01), and was conducted
according to the Declaration of Helsinki. Written informed consent
was received from each patient at the time of surgery. Surgical
resection for ICC was used to treat 34 patients in Taizhou People's
Hospital (Taizhou, China) between January 2014 and December 2020
(Table I). All specimens were
obtained from the Department of Pathology of Taizhou People's
Hospital, and were histologically diagnosed in accordance with the
World Health Organization criteria (32). Clinical features were extracted
from patient medical records. Tumor stages were classified
according to the 8th American Joint Cancer Committee
tumor-node-metastasis (TNM) classification (stages I-IV) (33).
 | Table IClinical characteristics of patients
with intrahepatic cholangiocarcinoma. |
Table I
Clinical characteristics of patients
with intrahepatic cholangiocarcinoma.
| Clinicopathological
characteristic | N (%) |
|---|
| Sex | |
|
Male | 21(62) |
|
Female | 13(38) |
| Age, years | |
|
≤65 | 20(59) |
|
>65 | 14(41) |
| Histological
grade | |
|
High | 5(15) |
|
Medium | 15(44) |
|
Low | 14(41) |
| TNM stage | |
|
I | 7(21) |
|
II | 10(29) |
|
III | 7(21) |
|
IV | 10(29) |
| Serum CA19-9
levels, U/ml | |
|
≤37 | 4(12) |
|
>37 | 30(88) |
| ITGB5
expression | |
|
High | 22(65) |
|
Low | 12(35) |
Gene function enrichment analysis
Gene Ontology (GO) is widely used in bioinformatics,
and covers three aspects of biology, namely biological processes,
molecular functions and cellular components (34). Kyoto Encyclopedia of Genes and
Genomes (KEGG) is a set of high-throughput genes and protein
pathways (35). Metascape is an
online analysis tool suite with the function of annotations and
analyses (36). GO and KEGG
pathway enrichment analyses were performed for the upregulated DEGs
using Metascape analysis. All significant GO and KEGG enrichment
results were visualized with the bubble chart of GraphPad Prism 9
(GraphPad Software; Dotmatics).
Protein-protein interaction (PPI)
network and functional annotations of ITGB5
The Search Tool for the Retrieval of Interacting
Genes/Proteins (STRING) online database (http://string-db.org) could predict and trace out the
PPI network. The top 10 interacting genes associated with ITGB5
were obtained using STRING. Cytoscape version 3.9.0, a free
visualization software, was used to visualize the PPI network
(37). The 210 genes interacting
with ITGB5, as determined using the STRING online database (10
genes) and Gene Expression Profiling Interactive Analysis of GEPIA
(38) (200 genes), were all
inputted into the Metascape (36)
for further functional annotations and analyses. Gene Set
Enrichment Analysis (GSEA) version 4.2.3(39), a free gene analysis software, was
used to reveal the functional pathways of ITGB5 in ICC, using
transcriptional data from the GSE26566 dataset. A 1,000 permutation
test, nominal (NOM) P<0.05 and false discovery rate (FDR)
q<0.25 were used as the screening criteria to identify the most
significantly involved pathways. Patients were divided into two
groups according to the median ITGB5 mRNA expression level. GSEA
was performed to determine whether the identified sets of genes
exhibited significant differences between the two groups based on
the normalized enrichment score (NES) and FDR. Based on correlation
coefficients, pathway analysis associated with ITGB5 using GSEA was
implemented. The aforementioned three methods were validated
against each other to determine the most relevant pathway.
Cell lines and culture
Human cholangiocarcinoma HuCCT1 cells were purchased
from the Shanghai Cell Bank of The Chinese Academy of Science.
Cells were cultured in Dulbecco's Modified Eagle's Medium (DMEM)
(Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% fetal
bovine serum (FBS) (Invitrogen; Thermo Fisher Scientific, Inc.),
100 U/ml penicillin and 100 µg/ml streptomycin. Cells were
maintained at 37˚C in a humidified environment containing 5%
CO2.
Immunohistochemical (IHC)
staining
Paraffin-embedded sections of tissues were
deparaffinized, hydrated and incubated with 0.3% hydrogen peroxide
to block endogenous peroxidase. Microwave heating (microwave oven
for 30 min at 250 W) was used for antigen retrieval. The sections
were first incubated in a 2% bovine serum albumin buffer
(Sigma-Aldrich; Merck KGaA) at 37˚C for 30 min and then at 4˚C
overnight with anti-ITGB5 rabbit polyclonal antibody (1:100; cat.
no. ab15459; Abcam). For the antibody binding, The sections were
then washed three times with 0.5% Tween, 0.1 M Tris-base, 0.9%
NaCl, (TBS-T; pH 7.6) for 5 min each wash and incubated in
biotinylated goat anti-rabbit IgG (1:100; cat. no. ab172730; Abcam)
at 37˚C for 30 min. Positive reactions were visualized using
diaminobenzidine solution followed by counterstaining with
hematoxylin at room temperature for 8 min. Tissue sections were
observed using an AX10-Imager A1 light microscope (Zeiss GmbH), and
all images were captured using AxioVision microscopy software
(version 4.7; Zeiss GmbH). All IHC staining was independently
evaluated by two experienced gastrointestinal pathologists. IHC
scores were calculated by multiplying the intensity of staining (0:
Negative; 1: Light yellow; 2: Yellowish brown; and 3: Brown) by the
percentage of positive cells (1: <5%; 2: 6-25%; 3: 26-70%; and
4: >70%), and finally interpreted as high or low expression
levels. If the final score was ≥4, the ITGB5 expression level was
considered high; otherwise, it was considered low.
Western blot analysis
Whole-cell lysates were prepared by lysing HuCCT1
(2x106 cells/ml) pellets in RIPA lysis buffer. Following
centrifugation at 1,000 x g for 30 min at 4˚C to remove all debris,
and the protein levels were estimated using a Super-Bradford
Protein Assay kit (CoWin Biosciences Co., Ltd.), according to the
manufacturer's protocol. Each 40-µg aliquot of total protein was
loaded on a 10% SDS-PAGE gel (25 µg) and separated at 100 V for 1.5
h. After electrophoresis, the proteins were transferred onto PVDF
membranes (EMD Millipore) and then blocked with 5% skimmed milk for
60 min at room temperature. After washing with TBST three times,
membranes were co-incubated with the primary antibodies against
ITGB5 (1:500 dilution; cat. no. ab184312) and β-actin (1:1,000
dilution; cat. no. ab115777; both from Abcam) overnight at 4˚C in
TBST. After incubation with horseradish peroxidase-conjugated goat
anti-rabbit immunoglobulin G (1:5,000 dilution; cat. no. BA1055;
Beyotime Institute of Biotechnology) in TBST at room temperature
for 60 min, bands were detected using BeyoECL Plus (Beyotime
Institute of Biotechnology) and captured using an Image Quant
LAS-4000 (FUJIFILM Wako Pure Chemical Corporation). The expression
of ITGB5 protein was normalized to β-actin expression.
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA from harvested cells was extracted using
an RNA isolation kit with genomic DNA filter columns (BioTeke
Corporation) according to the manufacturer's protocol. After RNA
quantification and quality control using spectrophotometry with the
optical density (OD) OD260/OD280 ratio controlled at 1.8-2.0, RNA
samples were reverse transcribed into cDNA using a reverse
transcriptase kit (Takara Biotechnology Co., Ltd.), followed by PCR
with SYBR® Green RT-PCR Master mix (Takara Bio, Inc.)
according to the manufacturer's protocols. The relative levels of
target gene mRNA transcripts to the control β-actin in individual
samples were determined in an ABI PRISM 7000 Sequence Detection
System (Applied Biosystems; Thermo Fisher Scientific, Inc.) under
the following thermocycling conditions: 50˚C for 2 min, 9˚C for 10
min, 40 cycles at 95˚C for 15 sec and 60˚C for 1 min. Data were
normalized to the control β-actin and analyzed using the
2-ΔΔCq method (40).
Samples were assayed in triplicate in three independent
experiments. The primers for the amplification of the indicated
genes were as follows: ITGB5, forward 5'-ACCTGGAACAACGGTGGAGA-3'
and reverse 5'-AAAAGATGCCGTGTCCCCAA-3'; and β-actin, forward
5'-CAAGAGATGGCCACGGCTGCT-3' and reverse
5'-TCCTTCTGCATCCTGTCGGCA-3'.
Small interfering RNA (siRNA)
transfection
siRNA against ITGB5 and a negative control siRNA
were designed and synthesized by Shanghai GenePharma Co., Ltd.
HuCCT1 cells were transfected with siRNA (800 µg/ml) using
Lipofectamine® 3000 (Invitrogen; Thermo Fisher
Scientific, Inc.). Cells were plated in six-well plates at a
density of 2x106 cells/well. Lipofectamine®
3000 and siRNA were mixed together in Opti-MEM (Invitrogen; Thermo
Fisher Scientific, Inc.). The cell culture medium was replaced with
Opti-MEM at 37˚C for 6 h. The cells were cultured in normal medium
for 48 h before the subsequent experiments. The ITGB5 siRNA
sequences were: Sense 5'-GGAGGUUACUGAAUGACAAAC-3' and antisense
5'-UUGUCAUUCAGUAACCUCCUA-3'. The sequences of the control
non-targeting siRNA were: Sense 5'-UUCUCCGAACGUGUCACGUTT-3' and
antisense 5'-ACGUGACACGUUCGGAGAATT-3'. Knocked down expression was
confirmed using RT-qPCR or western blotting.
Cell Counting Kit-8 (CCK-8) and
Transwell assays
CCK-8 assay (Nanjing KeyGen Biotech Co., Ltd.) was
used to evaluate cell proliferation and viability. Cells
(~1x105) were seeded in 100 µl DMEM per well in a
96-well plate. Subsequently, 100 µl CCK-8 solution was added to
each well and incubated at 37˚C for additional 2 h. The absorbance
at 450 nm was measured on a spectrophotometric plate reader. Each
group was repeated in three different wells.
The invasiveness of cholangiocarcinoma cells was
detected using 24-well Transwell plates (8-µm pore size; Corning,
Inc.). The bottom of each well insert was precoated with 50 µg
Matrigel (BD Biosciences) to simulate matrix barriers at 37˚C for 4
h. Cells (1x104) in 200 µl serum-free medium were added
to each upper chamber, and the lower compartments were filled with
600 µl medium containing 10% FBS. Following incubation for 24 h at
37˚C, the invasive cells in the lower chamber were fixed with 4%
paraformaldehyde and stained with 0.1% crystal violet at room
temperature for 15 min. The stained cells were counted under a
light microscope (Olympus Corporation) in five random fields.
Statistical analysis
Statistical analyses were performed using SPSS
version 26.0 (IBM Corp.) and GraphPad Prism version 9 (GraphPad
Software; Dotmatics). For the tumor tissue and the normal tissue
adjacent to the tumor samples in the GSE107943 dataset, the ITGB5
mRNA expression levels were analyzed using a paired Student's
t-test. Two-sided Fisher's exact test was used to reveal the
association between the expression levels of ITGB5 and
clinicopathological features. Clinicopathological variables with
P<0.05 in univariate Cox regression analysis were further
analyzed using multivariate Cox regression. Survival data were
analyzed using Kaplan-Meier survival curves with a log-rank test.
Experimental data (≥3 independent replicates) are expressed as the
mean ± standard deviation. One-way ANOVA followed by Tukey's
post-hoc test was used to reveal the invasiveness of tumor cells,
while two-way ANOVA followed by Tukey's post-hoc test was used to
evaluate the proliferation and viability of tumor cells. P<0.05
was considered to indicate a statistically significant
difference.
Results
Identification and analysis of DEGs,
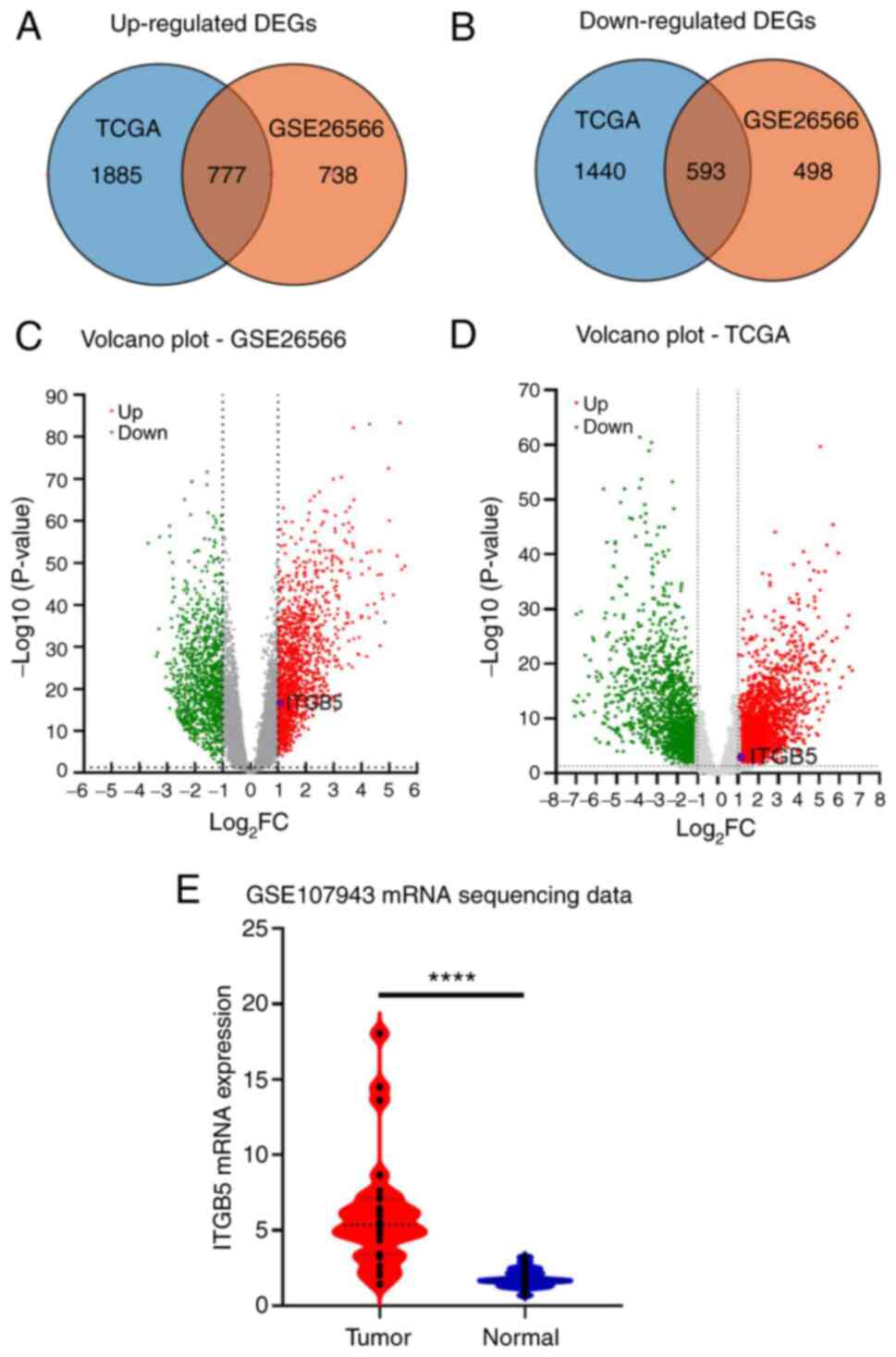
and upregulation of ITGB5 in ICC tissues
The GSE26566 and TCGA datasets were analyzed
separately as volcano maps using GraphPad Prism 9 (GraphPad
Software; Dotmatics). A total of 4,695 DEGs (log2
FC>1, corrected P<0.05) were obtained, including 2,662
significantly upregulated and 2,033 significantly downregulated
DEGs. After standardization of the microarray results, 2,606 DEGs
from the GSE26566 dataset were obtained, including 1,515
upregulated genes and 1,091 downregulated genes. A total of 1,370
common DEGs (777 upregulated and 593 downregulated) in the two
datasets were obtained through the TBtools (31) (Fig.
1A and B). In addition to
those published genes of minichromosome maintenance complex
component 6 (MCM6) (41) and
tripartite motif containing 59 (TRIM59) (42), a small number of significant DEGs
were highlighted, including ITGB5. In TCGA and GSE26566 datasets,
the volcano map indicated that ITGB5 was significantly
overexpressed in ICC (Fig. 1C and
D), which was consistent with the
ITGB5 mRNA expression levels in ICC in the GSE107943 dataset
(Fig. 1E). These results indicated
that ITGB5 was significantly overexpressed in ICC tumor tissue
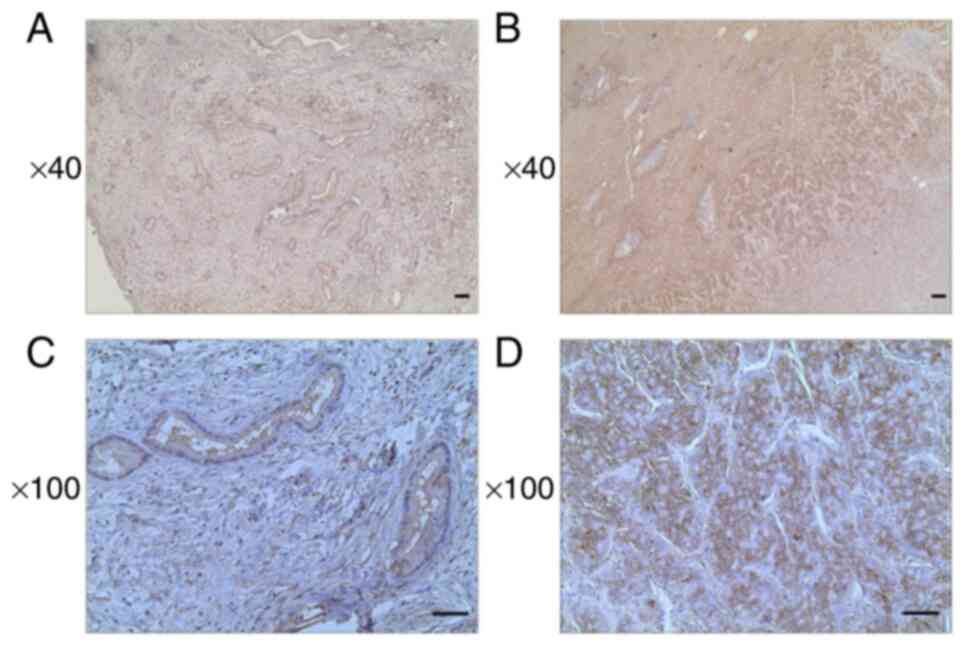
compared with adjacent normal tissue. IHC analysis of tumor tissue
samples of 34 patients with ICC revealed that the ITGB5 expression
levels were significantly increased in 12 patients (Fig. 2).
Association of ITGB5 with
clinicopathological characteristics and OS in patients with
ICC
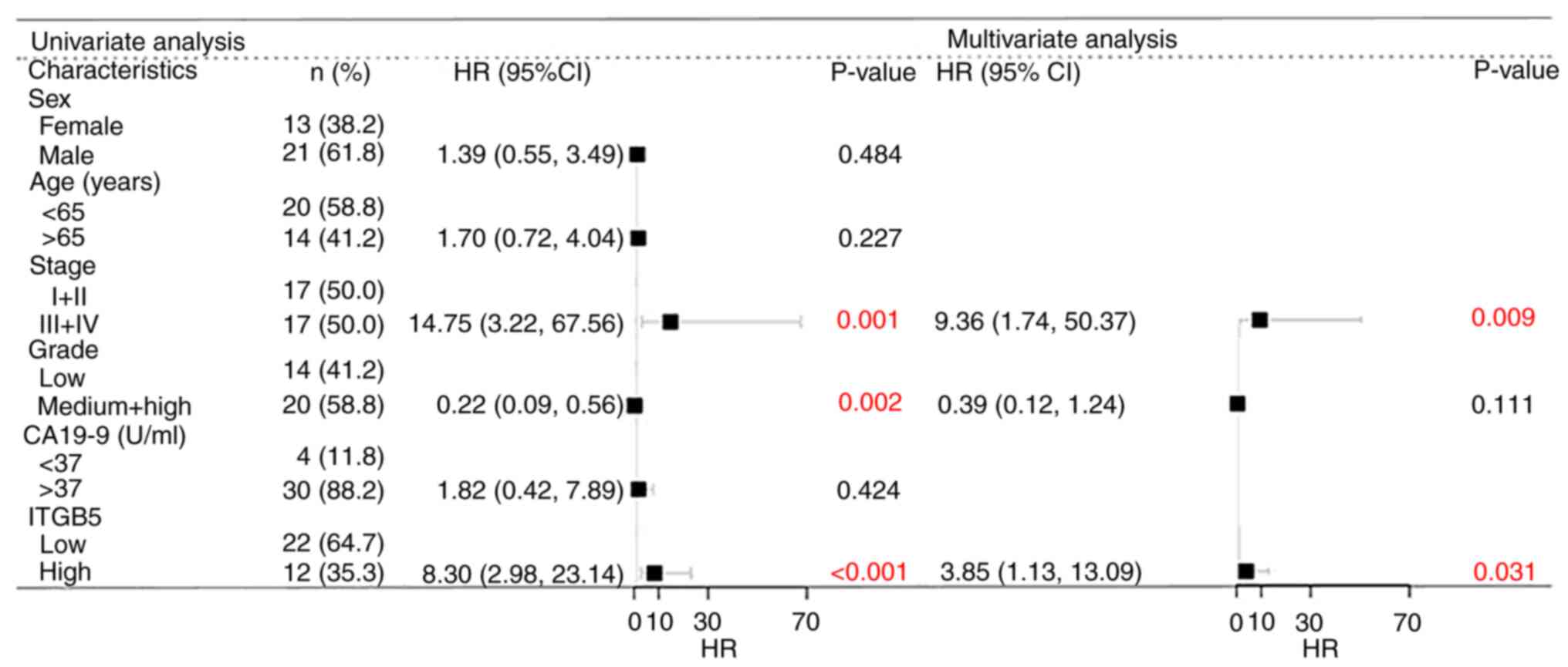
Two-sided Fisher's exact test of clinicopathological
characteristics analysis indicated that the expression of ITGB5 was
significantly associated with histological grade and TNM stage, but
not with clinicopathological indicators such as sex, age or serum
carbohydrate antigen (CA)19-9 level (Table II). In addition, to the best of
our knowledge, jaundice had no effect on ITGB5 expression levels.
Therefore, there was no further discussion regarding the
association between ITGB5 and jaundice. The present study followed
up 34 patients with ICC, and the difference in survival between the
high and low ITGB5 expression groups was investigated using the
Kaplan-Meier method. Multiple factors affecting survival in
patients with ICC were analyzed using the Cox model. Univariate
analysis indicated that sex, age and serum CA19-9 level did not
significantly affect the survival of patients with ICC, while a low
histological grade, late TNM stage and high expression levels of
ITGB5 were risk factors for a reduced survival rate. Multivariate
analysis suggested that a low histological grade was not an
independent risk factor affecting the survival of patients with
ICC, while high expression levels of ITGB5 and a late TNM stage
were independent risk factors for a reduced survival rate (Fig. 3).
 | Table IIAssociation between ITGB5 expression
levels and intrahepatic cholangiocarcinoma characteristics. |
Table II
Association between ITGB5 expression
levels and intrahepatic cholangiocarcinoma characteristics.
| | ITGB5 | |
|---|
| Characteristic | Low | High | P-value |
|---|
| Sex | | | 0.727 |
|
Male | 13 | 8 | |
|
Female | 9 | 4 | |
| Age, years | | | 0.163 |
|
≤65 | 15 | 5 | |
|
>65 | 7 | 7 | |
| Histologic
grade | | | 0.014a |
|
High | 4 | 1 | |
|
Medium | 13 | 2 | |
|
Low | 5 | 9 | |
| TNM stage | | | 0.015a |
|
I | 7 | 0 | |
|
II | 8 | 2 | |
|
III | 4 | 3 | |
|
IV | 3 | 7 | |
| Serum CA19-9,
U/ml | | | 0.556 |
|
≤37 | 3 | 1 | |
|
>37 | 19 | 11 | |
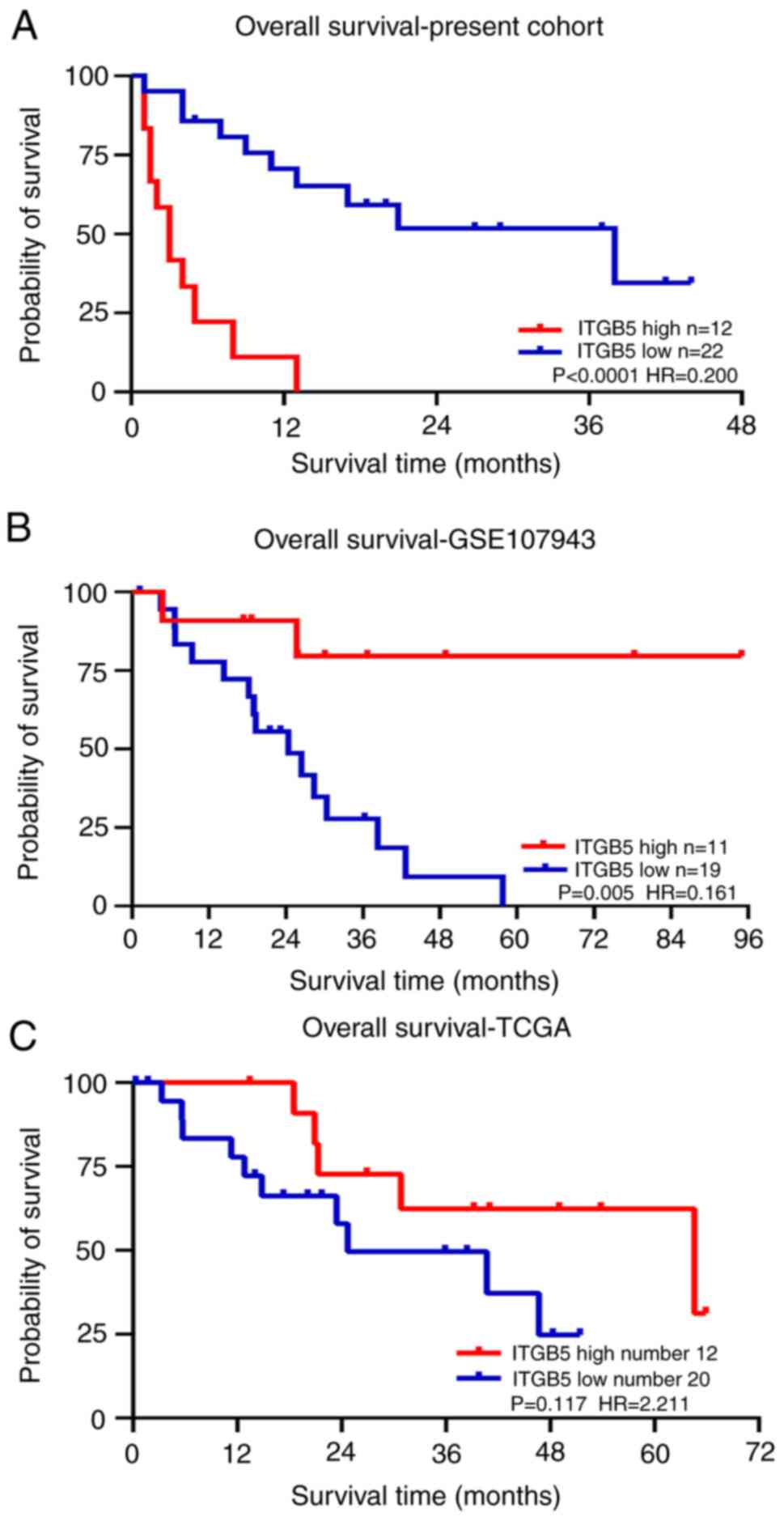
The patients in the high ITGB5 expression level
group had a mean survival of 4.31±1.17 months, which was
significantly reduced compared with that of patients in the low
ITGB5 expression level group (44.23±9.39 months; P<0.001;
Fig. 4A). Survival curves were
produced using the ICC clinical data for the GSE107943 (Fig. 4B) and TCGA (Fig. 4C) datasets. The GSE107943 dataset
indicated that patients in the high ITGB5 expression level group
had a mean OS of 78.89±10.21 months, which was significantly
increased compared with that of patients in the low ITGB5
expression level group (25.90±3.93 months; P=0.005). However, the
results of TCGA showed no significant difference in the mean OS
time between the high and low ITGB5 expression level groups.
Gene enrichment analysis
To elucidate the effect of the screened differential
genes on ICC, gene enrichment analysis was performed using
Metascape, which included GO and KEGG pathway enrichment analyses.
Since the ITGB5 expression level was high, enrichment analysis was
performed on upregulated genes. Through GO enrichment analysis of
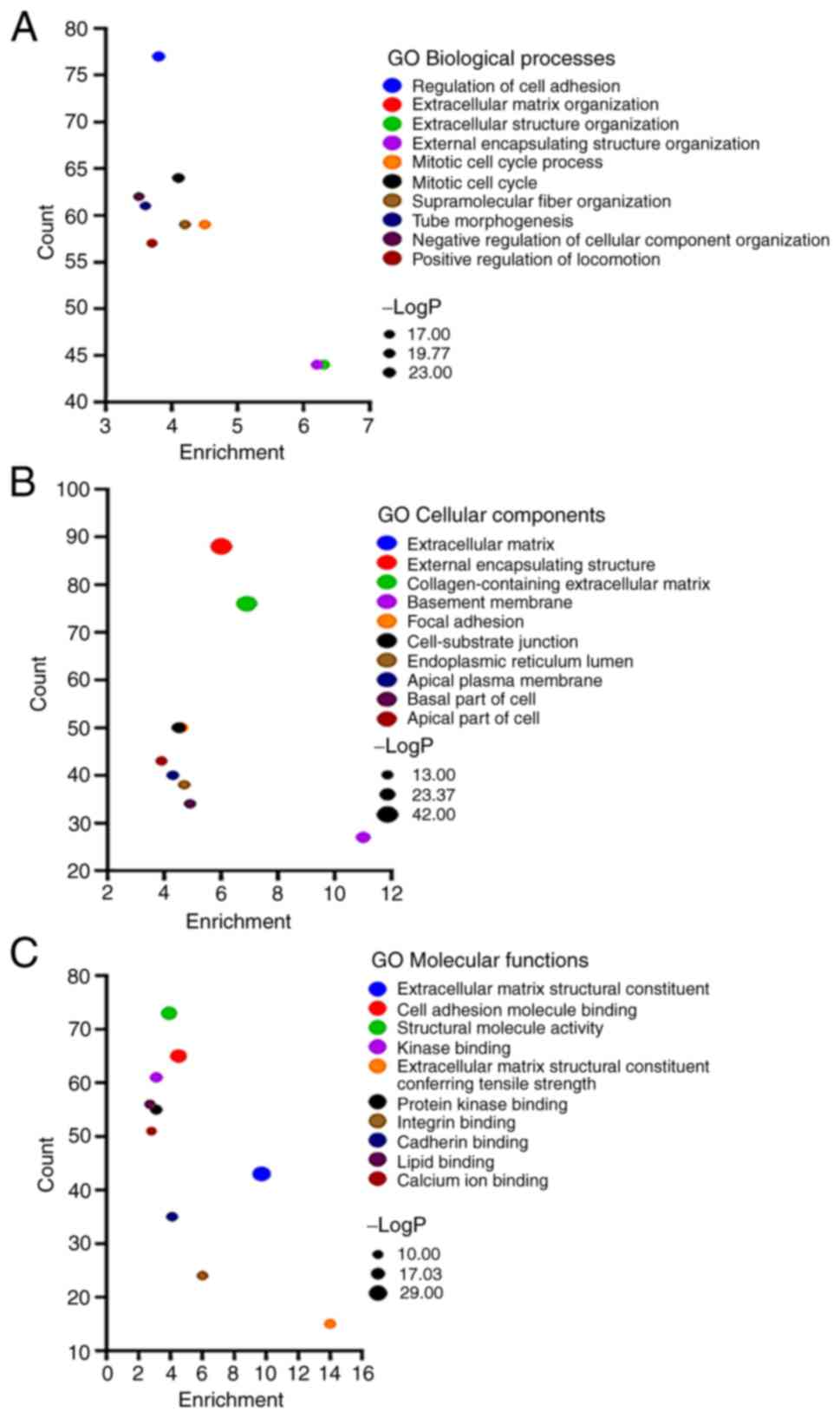
the upregulated genes, numerous enriched gene sets were revealed.
In terms of the biological processes, they were enriched in
‘regulation of cell adhesion’, ‘extracellular matrix organization’,
‘extracellular structure organization’, ‘external encapsulating
structure organization’ and ‘mitotic cell cycle process’ (Fig. 5A). In terms of the cellular
components, they were significantly enriched in ‘extracellular
matrix’, ‘external encapsulating structure’, ‘collagen-containing
extracellular matrix’, ‘basement membrane’ and ‘focal adhesion’
(Fig. 5B). In terms of the
molecular function, they were mainly enriched in ‘extracellular
matrix structural constituent’, ‘cell adhesion molecule binding’,
‘structural molecule activity’, ‘kinase binding’ and ‘extracellular
matrix structural constituent conferring tensile strength’
(Fig. 5C). The functional
significance of differential mRNAs in the development of ICC was
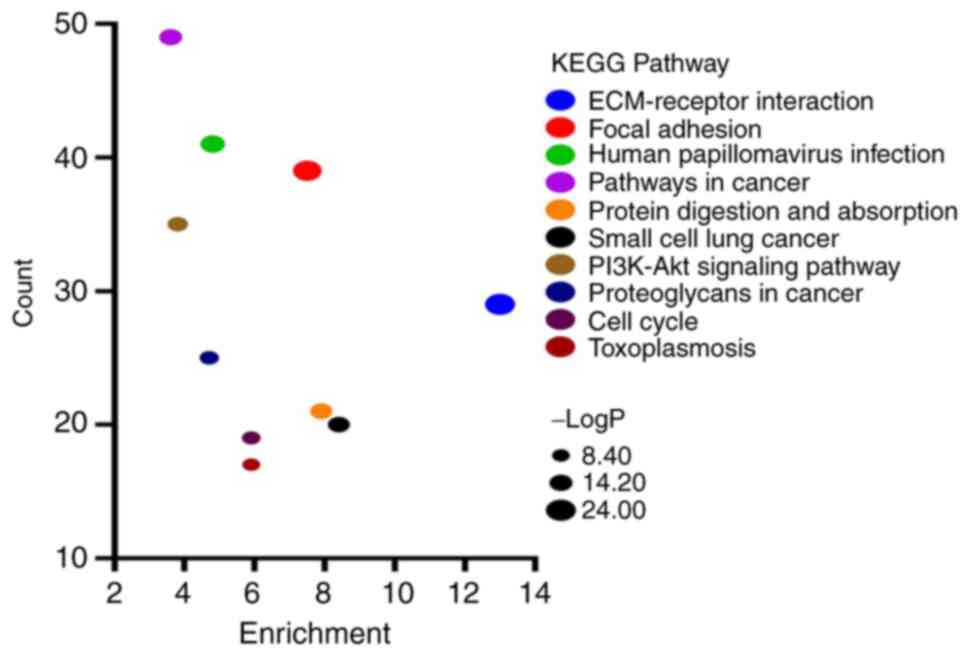
analyzed through KEGG pathway analysis. The results of KEGG
analysis revealed that upregulated genes were significantly
enriched in ‘ECM-receptor interaction’, ‘focal adhesion’, ‘human
papillomavirus infection’, ‘pathways in cancer’ and ‘protein
digestion and absorption’ (Fig.
6).
Prediction of interaction networks of
ITGB5 and enrichment analysis of genes co-expressed with ITGB5
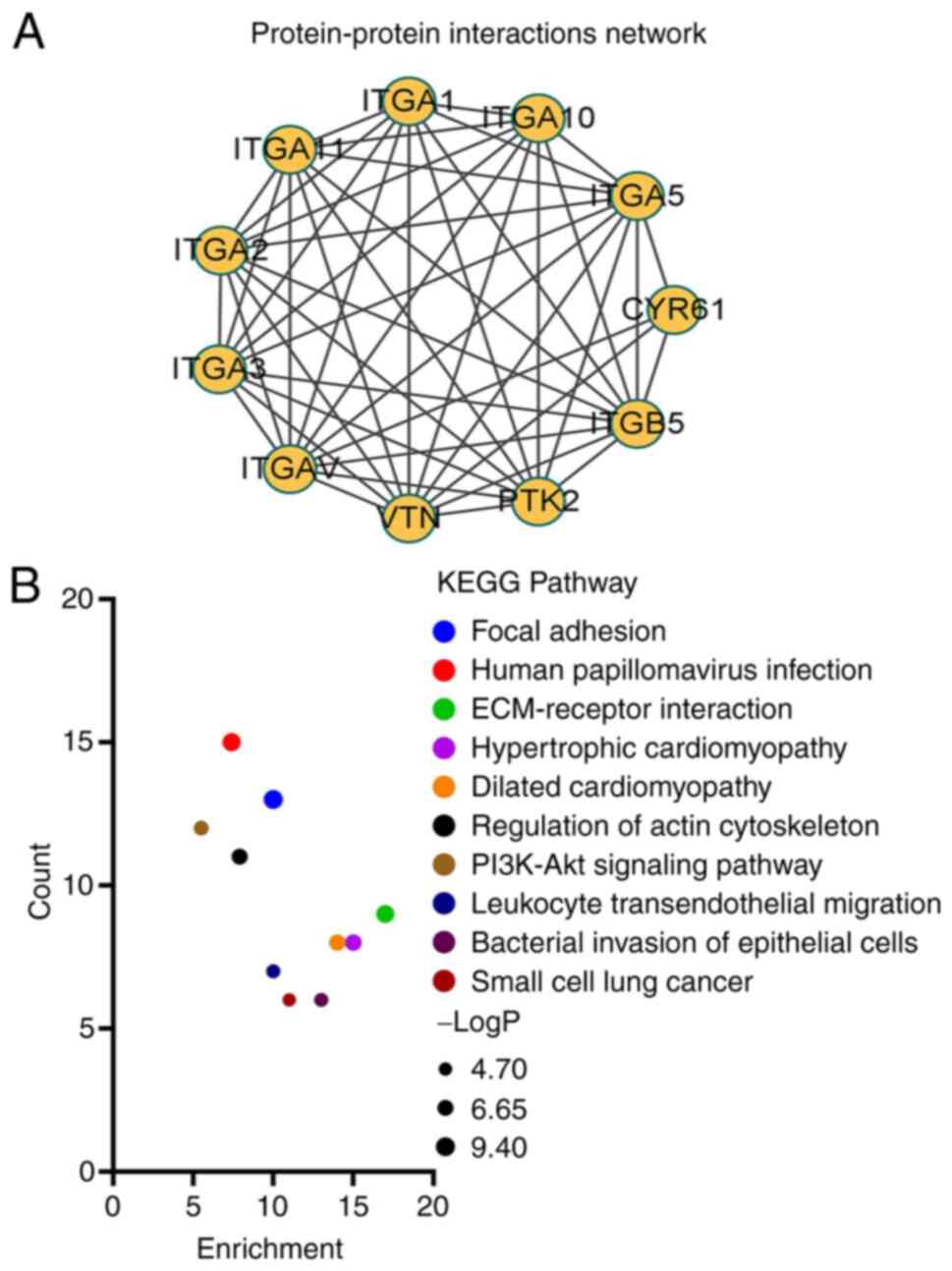
A PPI network for ITGB5 was constructed using the
STRING online database. The node representing ITGB5 was connected
to the nodes of other genes in terms of co-expression and physical
interactions. The PPI network of the top 10 genes was visualized
using Cytoscape (Fig. 7A). For
biological pathway analysis of genes co-expressed with ITGB5, the
top 200 genes strongly correlated with ITGB5 from GEPIA and 10
genes of PPI networks of ITGB5 were all inputted into Metascape for
functional annotations and analyses. The genes associated with
biological pathways were mainly enriched in ‘focal adhesion’,
‘human papillomavirus infection’ and ‘ECM-receptor interaction’
(Fig. 7B). Based on the GSE26566
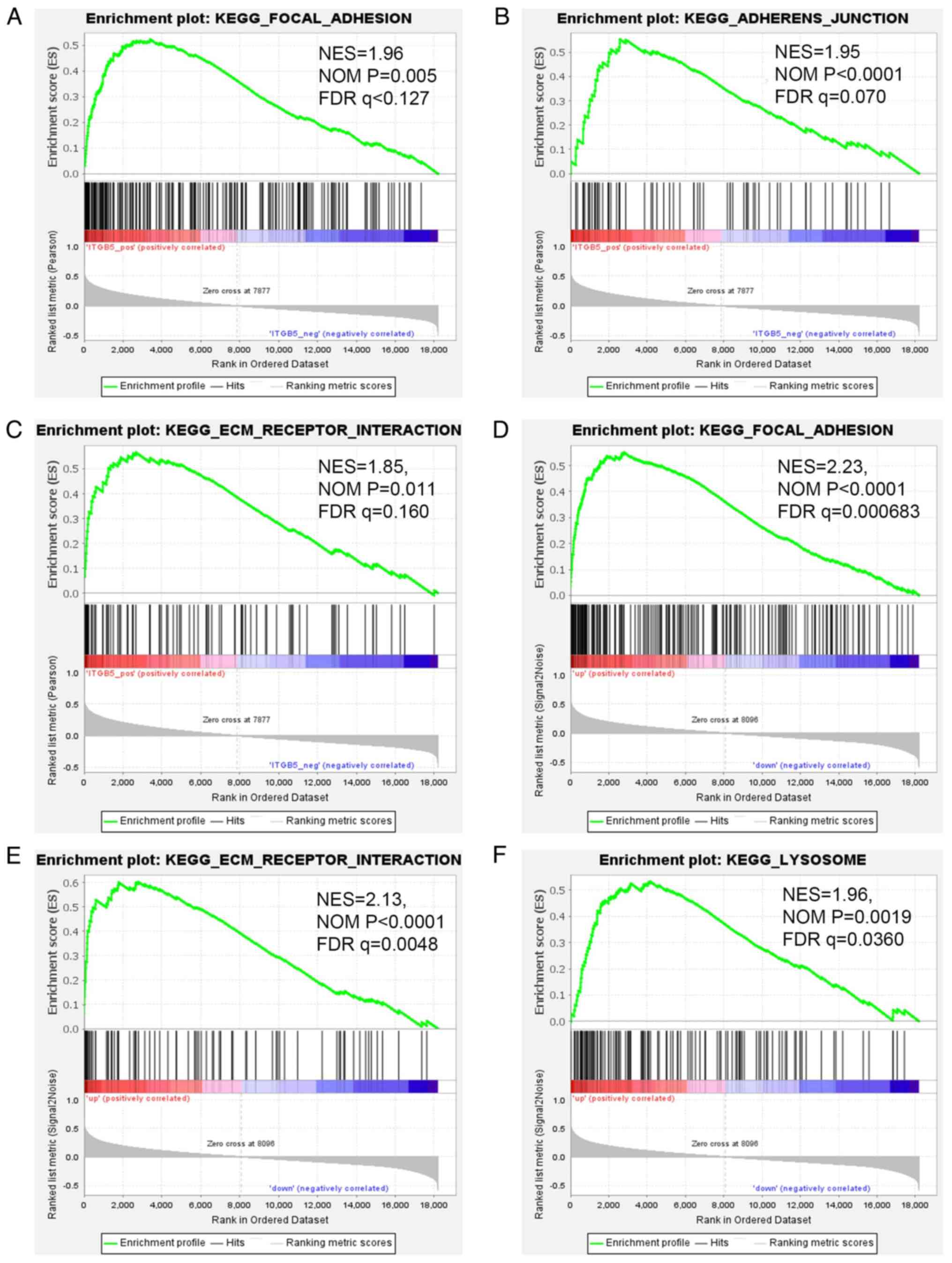
dataset, pathway analysis of genes positively associated to ITGB5
was also performed in GSEA. The genes associated with biological
pathways were mainly enriched in ‘focal adhesion’ (NES=1.96, NOM
P=0.005 and FDR q<0.127), ‘adherens junction’ (NES=1.95, NOM
P<0.0001 and FDR q=0.070) and ‘ECM-receptor interaction’
(NES=1.85, NOM P=0.011 and FDR q=0.160) (Fig. 8A-C). To identify the signaling
pathways activated by the differential upregulation of ITGB5
expression in ICC, GSEA of samples with low and high ITGB5
expression level based on the GSE26566 dataset was performed.
‘Focal adhesion’ (NES=2.23, NOM P<0.0001 and FDR q=0.000683),
‘ECM-receptor interaction’ (NES=2.13, NOM P<0.0001 and FDR
q=0.0048), and ‘lysosome’ (NES=1.96, NOM P=0.0019 and FDR q=0.0360)
were significantly enriched in the high ITGB5 expression level
sample (Fig. 8D-F). The
aforementioned three methods were validated against each other to
determine the most relevant pathway. The results revealed that
ITGB5 may be involved in the progression of ICC by regulating
ECM-receptor interaction and focal adhesion pathways. ECM-receptor
interaction and focal adhesion signaling pathways were also the
most significantly enriched for upregulated DEGs using Metascape
analysis, which is consistent with the aforementioned results.
Knockdown of ITGB5 inhibits the
proliferation and invasion of ICC cells
As ITGB5 was highly expressed in certain patients
with ICC and was significantly correlated with prognosis, the
functional roles of ITGB5 in human ICC cells were investigated.
Western blotting and RT-qPCR were used to test the efficiency of
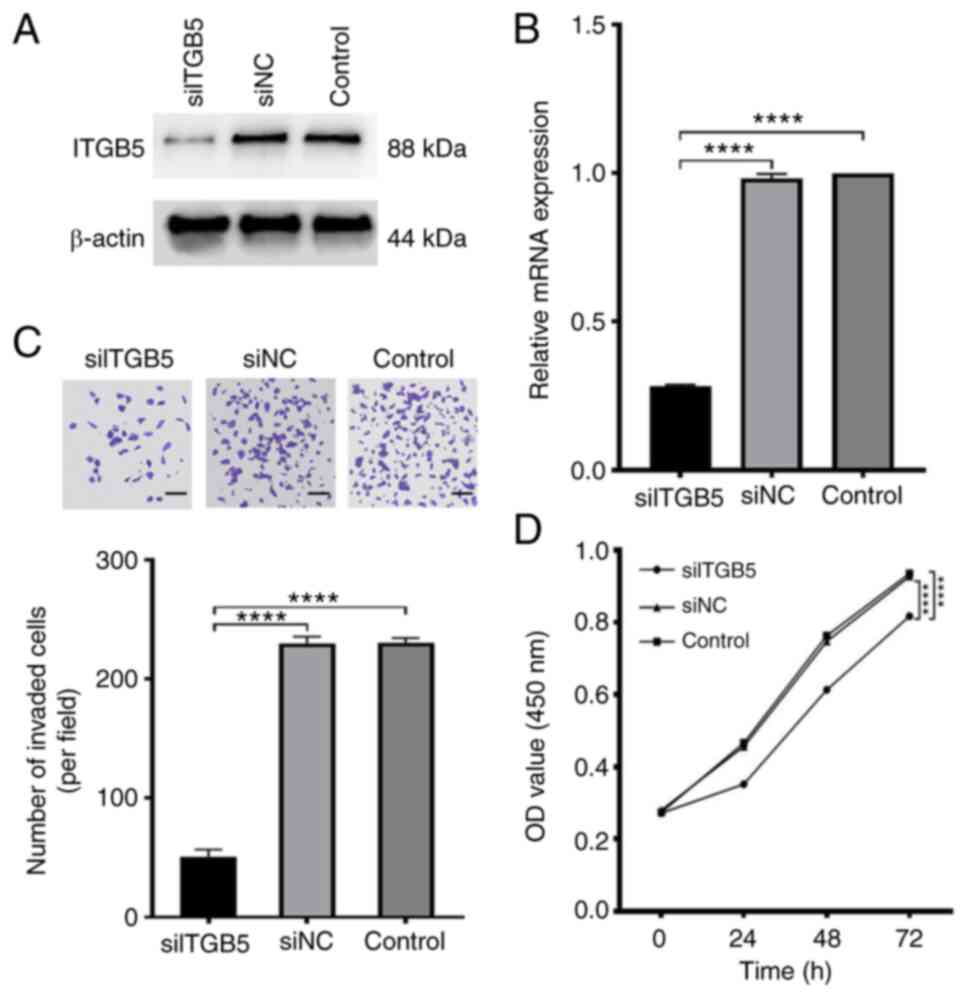
ITGB5 silencing in HuCCT1 cells. The ITGB5 mRNA and protein
expression levels were reduced after transfection with
ITGB5-specific siRNA (Fig. 9A and
B). Transwell invasion assays were
performed in HuCCT1 cells after downregulation of ITGB5. The
invasiveness of ICC cells was significantly reduced by ITGB5
silencing (Fig. 9C). CCK-8 assays
were used to determine the effects of ITGB5 on the viability of ICC
cells. The results demonstrated that ITGB5 silencing in HuCCT1
cells significantly reduced cell viability 72 h after transfection
(Fig. 9D).
Discussion
Given the incidence of ICC increasing from 0.44 to
1.18 cases per 100,000(2) and its
high morbidity, early prediction of prognosis is an arduous and
urgent task. Identifying ICC-specific diagnostic biomarkers has
been a focus among numerous studies, which is associated with
advances in omics technologies. In the past decade, efforts have
been conducted to elucidate the molecular pathogenesis of
cholangiocarcinoma, particularly ICC, through the application of
multi-omics approaches, including genomic, epigenomic,
transcriptomic and metabolomic analyses (43,44).
It has been reported that SMAD4 expression levels are associated
with the prognosis of patients with ICC (45). It has been reported that ITGA6 is
highly expressed in ICC and promotes the proliferation and invasion
of ICC cells (46). ITGB5 is a
potential independent prognostic biomarker and therapeutic target
for patients with HBV-related HCC. Previous studies have
demonstrated that ITGB5 is a prognostic biomarker and new
therapeutic target for human pancreatic (47), breast (48), gastric (49) and ovarian (50) cancer, as well as glioblastoma
(51). The present study revealed
increased ITGB5 mRNA and protein expression levels in ICC tissues,
and the upregulation of ITGB5 was associated with the late TNM
stage and low histological grade. Another novel finding of the
present study was that high ITGB5 levels were independently
correlated with a reduced survival rate in patients with ICC.
Therefore, whether ITGB5 can predict the prognosis of patients with
ICC requires further investigation.
Deregulation of integrin signaling is associated
with carcinogenic effects in a number of malignancies. For example,
in pancreatic cancer, ITGB4 is associated with
epithelial-mesenchymal transition. Overexpression of ITGB4 promotes
pancreatic carcinogenesis and regulates the MEK1-ERK1/2 signaling
pathway (52). ITGB6 promotes the
invasion of various cancer cells, including colorectal and
pancreatic cancer, through the ERK and TGF signaling pathways,
which promote matrix metalloproteinase activation (53,54).
As for ITGB5, Lin et al reported that it was highly
expressed in HCC, and microRNA-185 regulated the expression of
β-catenin in an ITGB5-dependent manner, and affected the
proliferation and migration of HCC cells (24). Tumor cells with knocked out ITGB5
led to a reduced disease burden and a prolonged survival in mice,
demonstrating the contribution of ITGB5 to pancreatic ductal
adenocarcinoma progression (55).
A previous study demonstrated that exosomal ITGB5 regulated liver
tropism, which was associated with liver metastasis in a number of
malignancies, including colorectal, pancreatic and gastric cancer
(56). In the present study, to
investigate the function of ITGB5 in ICC, knockdown experiments
using siRNAs were performed, which demonstrated that HuCCT1 cell
proliferation and invasion were reduced by ITGB5 depletion. For
patients with ICC, TCGA dataset indicated no significant difference
in the prognosis of patients with high or low ITGB5 expression
levels. However, high ITGB5 expression levels reflected an
increased OS rate in the GSE107943 dataset, while high ITGB5
expression levels reflected a reduced OS rate in the data of the
present study. Due to the rarity of ICC, studies on ICC often have
small cohort sizes, which may contribute to the aforementioned
observed difference in OS. Ethnic heterogeneity and differences in
TNM stages might be other factors explaining the differences in
prognosis. The patients with ICC in the cohort of the present study
were of Chinese ethnicity, and the majority exhibited TNM stages
III and IV, while the patients with ICC in the GES107943 dataset
were South Koreans in ethnicity and mainly exhibited TNM stages I
and II.
To investigate the signaling pathways contributing
to ICC progression, the current data were processed through
bioinformatics methods to obtain additional information regarding
ITGB5 and its co-expressed genes. The aforementioned methods were
validated against each other to determine the most relevant
pathway. The results revealed that ITGB5 might be involved in the
progression of ICC by regulating the ECM-receptor interaction and
focal adhesion pathways. The two aforementioned pathways were the
most significantly enriched for upregulated DEGs of ICC using
Metascape analysis, which is consistent with a previous study
(57). These results suggest that
ITGB5 promotes tumor cell proliferation and migration through
ECM-receptor interaction and focal adhesion signaling pathways,
which may lead to the poor survival of patients with ICC. A number
of studies have demonstrated the involvement of ECM-receptor
interaction in the development and formation of metastases in
various tumors, including breast and lung cancer, as well as
glioma, through its regulation of integrin expression levels
(58-61).
The focal adhesion signaling pathway via integrin has an effect on
the regulation of the ECM, cell migration and tumor
microenvironment (62). The focal
adhesion pathway facilitates the interplay between tumors and the
ECM, serving as a crucial link connecting them (63). However, to the best of our
knowledge, although a number of studies have explored the
association between ITGB5 and the ECM-receptor interaction and
focal adhesion signaling pathway in gastric cancer (64,65),
no comprehensive prognostic analysis of ECM-receptor interaction
and focal adhesion-associated genes in ICC has been conducted to
date. Signaling pathways associated to ITGB5 that affect ICC
survival will be the next aim of our future research.
There were a number of limitations in the present
study that should be addressed. Firstly, using the normal bile duct
tissue located next to the ICC tumor as the negative control to
compare the changes in ITGB5 expression levels using IHC would have
improved the present study. However, the present results only
revealed high and low ITGB5 expression levels, which means that the
comparison of ITGB5 in ICC tumor tissue and adjacent normal tissue
is inadequate at the protein level. Secondly, the sample size was
insufficient for the tumor size and macroscopic analysis of
clinical features. Although T stage encompasses more comprehensive
information than tumor size, potential bias may arise in the
results of multivariate Cox regression analysis when considering
tumor size and T stage as independent prognostic factors.
Therefore, it is imperative to assess tumor size as a prognostic
factor through univariate Cox regression analysis. Previous studies
have demonstrated that the macroscopic type affects the prognosis
of patients with ICC (66).
However, the small sample size of the present cohort made it
impossible to evaluate the macroscopic type in the present study.
The inclusion of additional observed variables necessitates a
larger sample size; otherwise, the statistical tests may not meet
the necessary requirements, leading to compromised repeatability
and representativeness. This could potentially result in erroneous
conclusions, including false negatives or false positives.
Considering the purpose of the present study and the small sample
size, the variables presented in Table II were selected. Multi-center
studies on hepatobiliary clinic should be conducted in the future
to examine other factors. Thirdly, no additional cell lines
verified the role of ITGB5 in ICC, and no further experiments
explored the signaling pathways associated with ITGB5 in ICC. In
future studies on ITGB5-related signaling pathways and additional
ICC cell lines (HuH28; RBE; SSP25) should be employed to confirm
the reproducibility of the present findings.
For simplicity and clarity, a flowchart of the
present study has been presented in Fig. 10. In conclusion, ITGB5 may act as
a regulator of ICC development and progression by influencing the
proliferation and invasion of ICC cells. ITGB5 could be a potential
biomarker for a poorer prognosis of ICC in a Chinese population,
and it may be helpful to screen candidates for receiving intensive
therapy. However, future studies with large sample sizes are
required to validate the role of ITGB5 in the prognosis of patients
with ICC.
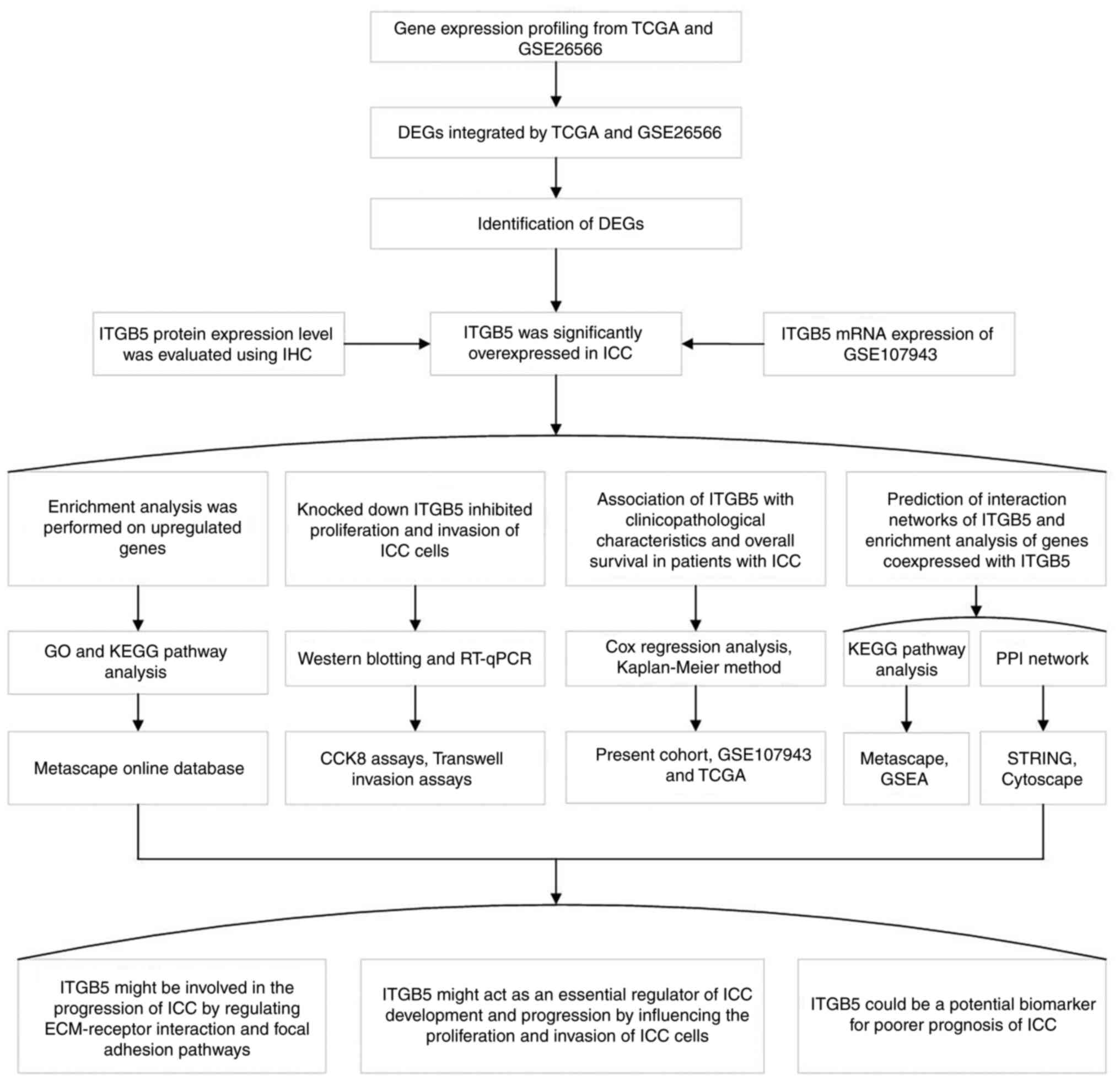 | Figure 10Flowchart of the present study
describing the main methods used and the results obtained. TCGA,
The Cancer Genome Atlas; DEG, differentially expressed gene; ITGB5,
integrin β5; IHC, immunohistochemistry; ICC, intrahepatic
cholangiocarcinoma; GO, Gene Ontology; KEGG, Kyoto Encyclopedia of
Genes and Genomes; RT-qPCR, reverse transcription-quantitative
polymerase chain reaction; PPI, protein-protein interaction; CCK-8,
Cell Counting Kit-8; GSEA, Gene Set Enrichment Analysis; STRING,
Search Tool for the Retrieval of Interacting Genes/Proteins; ECM,
extracellular matrix. |
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
LM and JZ designed the study, performed the
experiments, analyzed the data and wrote the manuscript. LM and KS
collected the data. LM and JZ confirm the authenticity of all the
raw data. All authors agree to be accountable for all aspects of
the research in ensuring that the accuracy or integrity of any part
of the work (including the provided data) are appropriately
investigated and resolved. All authors have read and approved the
final version of the manuscript.
Ethics approval and consent to
participate
Written informed consent was received from each
patient at the time of surgery for the use of their tissues in
research. The present study was conducted in accordance with the
ethical standards defined in the Declaration of Helsinki and was
approved by the Medical Ethics Committee of Taizhou People's
Hospital (approval no. KY 2020-091-01).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Muñoz-Martínez S and Forner A: The
tireless search to improve the prognostic assessment of
intrahepatic cholangiocarcinoma: An urgent need. Liver Int.
41:252–254. 2021.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Saha SK, Zhu AX, Fuchs CS and Brooks GA:
Forty-year trends in cholangiocarcinoma incidence in the U.S.:
Intrahepatic disease on the rise. Oncologist. 21:594–599.
2016.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Rizvi S, Khan SA, Hallemeier CL, Kelley RK
and Gores GJ: Cholangiocarcinoma-evolving concepts and therapeutic
strategies. Nat Rev Clin Oncol. 15:95–111. 2018.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Brunt E, Aishima S, Clavien PA, Fowler K,
Goodman Z, Gores G, Gouw A, Kagen A, Klimstra D, Komuta M, et al:
cHCC-CCA: Consensus terminology for primary liver carcinomas with
both hepatocytic and cholangiocytic differentation. Hepatology.
68:113–126. 2018.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Wu L, Tsilimigras DI, Paredes AZ, Mehta R,
Hyer JM, Merath K, Sahara K, Bagante F, Beal EW, Shen F and Pawlik
TM: Trends in the incidence, treatment and outcomes of patients
with intrahepatic cholangiocarcinoma in the USA: Facility type is
associated with margin status, use of lymphadenectomy and overall
survival. World J Surg. 43:1777–1787. 2019.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Van Dyke AL, Shiels MS, Jones GS, Pfeiffer
RM, Petrick JL, Beebe-Dimmer JL and Koshiol J: Biliary tract cancer
incidence and trends in the United States by demographic group,
1999-2013. Cancer. 125:1489–1498. 2019.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Banales JM, Marin JJG, Lamarca A,
Rodrigues PM, Khan SA, Roberts LR, Cardinale V, Carpino G, Andersen
JB, Braconi C, et al: Cholangiocarcinoma 2020: The next horizon in
mechanisms and management. Nat Rev Gastroenterol Hepatol.
17:557–588. 2020.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Rahnemai-Azar AA, Weisbrod A, Dillhoff M,
Schmidt C and Pawlik TM: Intrahepatic cholangiocarcinoma: Molecular
markers for diagnosis and prognosis. Surg Oncol. 26:125–137.
2017.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Massarweh NN and El-Serag HB: Epidemiology
of hepatocellular carcinoma and intrahepatic cholangiocarcinoma.
Cancer Control. 24(1073274817729245)2017.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Fujita T: Liver transplantation for
intrahepatic cholangiocarcinoma. Lancet. 384(1182)2014.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Khan SA, Davidson BR, Goldin RD, Heaton N,
Karani J, Pereira SP, Rosenberg WM, Tait P, Taylor-Robinson SD,
Thillainayagam AV, et al: Guidelines for the diagnosis and
treatment of cholangiocarcinoma: An update. Gut. 61:1657–1669.
2012.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Lamarca A, Ross P, Wasan HS, Hubner RA,
McNamara MG, Lopes A, Manoharan P, Palmer D, Bridgewater J and
Valle JW: Advanced intrahepatic cholangiocarcinoma: Post hoc
analysis of the ABC-01, -02, and -03 clinical trials. J Natl Cancer
Inst. 112:200–210. 2020.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Ryerson AB, Eheman CR, Altekruse SF, Ward
JW, Jemal A, Sherman RL, Henley SJ, Holtzman D, Lake A, Noone AM,
et al: Annual report to the nation on the status of cancer,
1975-2012, featuring the increasing incidence of liver cancer.
Cancer. 122:1312–1337. 2016.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Hamidi H and Ivaska J: Every step of the
way: Integrins in cancer progression and metastasis. Nat Rev
Cancer. 18:533–548. 2018.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Humphries JD, Byron A and Humphries MJ:
Integrin ligands at a glance. J Cell Sci. 119:3901–3903.
2006.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Arruda Macêdo JK, Fox JW and de Souza
Castro M: Disintegrins from snake venoms and their applications in
cancer research and therapy. Curr Protein Pept Sci. 16:532–548.
2015.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Hussein HA, Walker LR, Abdel-Raouf UM,
Desouky SA, Montasser AK and Akula SM: Beyond RGD: Virus
interactions with integrins. Arch Virol. 160:2669–2681.
2015.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Zhao ZS, Wang YY, Chu YQ, Ye ZY and Tao
HQ: SPARC is associated with gastric cancer progression and poor
survival of patients. Clin Cancer Res. 16:260–268. 2010.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Petricevic B, Vrbanec D, Jakic-Razumovic
J, Brcic I, Rabic D, Badovinac T, Ozimec E and Bali V: Expression
of Toll-like receptor 4 and beta 1 integrin in breast cancer. Med
Oncol. 29:486–494. 2012.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Zhou G, Chiu D, Qin D, Niu L, Cai J, He L,
Tan D and Xu K: Expression of CD44v6 and integrin-β1 for the
prognosis evaluation of pancreatic cancer patients after
cryosurgery. Diagn Pathol. 8(146)2013.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Liu QZ, Gao XH, Chang WJ, Gong HF, Fu CG,
Zhang W and Cao GW: Expression of ITGB1 predicts prognosis in
colorectal cancer: A large prospective study based on tissue
microarray. Int J Clin Exp Pathol. 8:12802–12810. 2015.PubMed/NCBI
|
|
22
|
Takada Y, Ye X and Simon S: The integrins.
Genome Biol. 8(215)2007.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Shi W, He J, Huang Y, Zeng Z, Feng Z, Xu H
and Nie Y: Integrin β5 enhances the malignancy of human colorectal
cancer by increasing the TGF-β signaling. Anticancer Drugs.
32:717–726. 2021.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Lin Z, He R, Luo H, Lu C, Ning Z, Wu Y,
Han C, Tan G and Wang Z: Integrin-β5, a miR-185-targeted gene,
promotes hepatocellular carcinoma tumorigenesis by regulating
β-catenin stability. J Exp Clin Cancer Res. 37(17)2018.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Wortzel I, Dror S, Kenific CM and Lyden D:
Exosome-mediated metastasis: Communication from a distance. Dev
Cell. 49:347–360. 2019.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Shang L, Ye X, Zhu G, Su H, Su Z, Chen B,
Xiao K, Li L, Peng M and Peng T: Prognostic value of integrin
variants and expression in post-operative patients with HBV-related
hepatocellular carcinoma. Oncotarget. 8:76816–76831.
2017.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Andersen JB, Spee B, Blechacz BR, Avital
I, Komuta M, Barbour A, Conner EA, Gillen MC, Roskams T, Roberts
LR, et al: Genomic and genetic characterization of
cholangiocarcinoma identifies therapeutic targets for tyrosine
kinase inhibitors. Gastroenterology. 142:1021–1031.e15.
2012.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Ahn KS, O'Brien D, Kang YN, Mounajjed T,
Kim YH, Kim TS, Kocher JA, Allotey LK, Borad MJ, Roberts LR and
Kang KJ: Prognostic subclass of intrahepatic cholangiocarcinoma by
integrative molecular-clinical analysis and potential targeted
approach. Hepatol Int. 13:490–500. 2019.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Davis S and Meltzer PS: GEOquery: A bridge
between the gene expression omnibus (GEO) and Bioconductor.
Bioinformatics. 23:1846–1847. 2007.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Colaprico A, Silva TC, Olsen C, Garofano
L, Cava C, Garolini D, Sabedot TS, Malta TM, Pagnotta SM,
Castiglioni I, et al: TCGAbiolinks: An R/Bioconductor package for
integrative analysis of TCGA data. Nucleic Acids Res.
44(e71)2016.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Chen C, Chen H, Zhang Y, Thomas HR, Frank
MH, He Y and Xia R: TBtools: An integrative toolkit developed for
interactive analyses of big biological data. Mol Plant.
13:1194–1202. 2020.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Nagtegaal ID, Odze RD, Klimstra D, Paradis
V, Rugge M, Schirmacher P, Washington KM, Carneiro F and Cree IA:
WHO Classification of Tumours Editorial Board. The 2019 WHO
classification of tumours of the digestive system. Histopathology.
76:182–188. 2020.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Chun YS, Pawlik TM and Vauthey JN: 8th
Edition of the AJCC cancer staging manual: Pancreas and
hepatobiliary cancers. Ann Surg Oncol. 25:845–847. 2018.PubMed/NCBI View Article : Google Scholar
|
|
34
|
The Gene Ontology Consortium. The gene
ontology resource: 20 Years and still GOing strong. Nucleic Acids
Res. 47 (D1):D330–D338. 2019.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Kanehisa M and Goto S: KEGG: Kyoto
encyclopedia of genes and genomes. Nucleic Acids Res. 28:27–30.
2000.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Zhou Y, Zhou B, Pache L, Chang M,
Khodabakhshi AH, Tanaseichuk O, Benner C and Chanda SK: Metascape
provides a biologist-oriented resource for the analysis of
systems-level datasets. Nat Commun. 10(1523)2019.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Tang Z, Li C, Kang B, Gao G, Li C and
Zhang Z: GEPIA: A web server for cancer and normal gene expression
profiling and interactive analyses. Nucleic Acids Res. 45
(W1):W98–W102. 2017.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Subramanian A, Tamayo P, Mootha VK,
Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub
TR, Lander ES and Mesirov JP: Gene set enrichment analysis: A
knowledge-based approach for interpreting genome-wide expression
profiles. Proc Natl Acad Sci USA. 102:15545–15550. 2005.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Gao C, Li J, Zeng F, Wang L, Chen K, Chen
D, Hong J and Qu C: MCM6 promotes intrahepatic cholangiocarcinoma
progression by upregulating E2F1 and enhancing
epithelial-mesenchymal transition. Carcinogenesis. 44:279–290.
2023.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Zhang JN, Ding DY, Yang SY, Tao QF, Yang Y
and Zhou WP: The role of tripartite motif containing 59 (TRIM59) in
the proliferation and prognosis of intrahepatic cholangiocarcinoma.
Pathol Res Pract. 236(153989)2022.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Nakamura H, Arai Y, Totoki Y, Shirota T,
Elzawahry A, Kato M, Hama N, Hosoda F, Urushidate T, Ohashi S, et
al: Genomic spectra of biliary tract cancer. Nat Genet.
47:1003–1010. 2015.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Sia D, Hoshida Y, Villanueva A, Roayaie S,
Ferrer J, Tabak B, Peix J, Sole M, Tovar V, Alsinet C, et al:
Integrative molecular analysis of intrahepatic cholangiocarcinoma
reveals 2 classes that have different outcomes. Gastroenterology.
144:829–840. 2013.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Liu J, Ren G, Li K, Liu Z, Wang Y, Chen T,
Mu W, Yang X, Li X, Shi A, et al: The Smad4-MYO18A-PP1A complex
regulates β-catenin phosphorylation and pemigatinib resistance by
inhibiting PAK1 in cholangiocarcinoma. Cell Death Differ.
29:818–831. 2022.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Hozaka Y, Seki N, Tanaka T, Asai S, Moriya
S, Idichi T, Wada M, Tanoue K, Kawasaki Y, Mataki Y, et al:
Molecular pathogenesis and regulation of the miR-29-3p-family:
Involvement of ITGA6 and ITGB1 in intra-hepatic cholangiocarcinoma.
Cancers (Basel). 13(2804)2021.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Ricono JM, Huang M, Barnes LA, Lau SK,
Weis SM, Schlaepfer DD, Hanks SK and Cheresh DA: Specific
cross-talk between epidermal growth factor receptor and integrin
alphavbeta5 promotes carcinoma cell invasion and metastasis. Cancer
Res. 69:1383–1391. 2009.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Bianchi-Smiraglia A, Paesante S and Bakin
AV: Integrin β5 contributes to the tumorigenic potential of breast
cancer cells through the Src-FAK and MEK-ERK signaling pathways.
Oncogene. 32:3049–3058. 2013.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Hung WY, Huang KH, Wu CW, Chi CW, Kao HL,
Li AF, Yin PH and Lee HC: Mitochondrial dysfunction promotes cell
migration via reactive oxygen species-enhanced β5-integrin
expression in human gastric cancer SC-M1 cells. Biochim Biophys
Acta. 1820:1102–1110. 2012.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Tancioni I, Uryu S, Sulzmaier FJ, Shah NR,
Lawson C, Miller NL, Jean C, Chen XL, Ward KK and Schlaepfer DD:
FAK inhibition disrupts a β5 integrin signaling axis controlling
anchorage-independent ovarian carcinoma growth. Mol Cancer Ther.
13:2050–2061. 2014.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Zhang LY, Guo Q, Guan GF, Cheng W, Cheng P
and Wu AH: Integrin beta 5 is a prognostic biomarker and potential
therapeutic target in glioblastoma. Front Oncol.
9(904)2019.PubMed/NCBI View Article : Google Scholar
|
|
52
|
An XZ, Zhao ZG, Luo YX, Zhang R, Tang XQ,
Hao D, Zhao X, Lv X and Liu D: Netrin-1 suppresses the MEK/ERK
pathway and ITGB4 in pancreatic cancer. Oncotarget. 7:24719–24733.
2016.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Yang GY, Guo S, Dong CY, Wang XQ, Hu BY,
Liu YF, Chen YW, Niu J and Dong JH: Integrin αvβ6 sustains and
promotes tumor invasive growth in colon cancer progression. World J
Gastroenterol. 21:7457–7467. 2015.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Reader CS, Vallath S, Steele CW, Haider S,
Brentnall A, Desai A, Moore KM, Jamieson NB, Chang D, Bailey P, et
al: The integrin αvβ6 drives pancreatic cancer through diverse
mechanisms and represents an effective target for therapy. J
Pathol. 249:332–342. 2019.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Hurtado de Mendoza T, Mose ES, Botta GP,
Braun GB, Kotamraju VR, French RP, Suzuki K, Miyamura N, Teesalu T,
Ruoslahti E, et al: Tumor-penetrating therapy for β5 integrin-rich
pancreas cancer. Nat Commun. 12(1541)2021.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Hoshino A, Costa-Silva B, Shen TL,
Rodrigues G, Hashimoto A, Tesic Mark M, Molina H, Kohsaka S, Di
Giannatale A, Ceder S, et al: Tumour exosome integrins determine
organotropic metastasis. Nature. 527:329–335. 2015.PubMed/NCBI View Article : Google Scholar
|
|
57
|
Li H, Long J, Xie F, Kang K, Shi Y, Xu W,
Wu X, Lin J, Xu H, Du S, et al: Transcriptomic analysis and
identification of prognostic biomarkers in cholangiocarcinoma.
Oncol Rep. 42:1833–1842. 2019.PubMed/NCBI View Article : Google Scholar
|
|
58
|
Wu D, Sun J, Wang H and Ma C: LncRNA
SOCS2-AS1 promotes the progression of glioma via regulating ITGB1
expression. Neurosci Lett. 765(136248)2021.PubMed/NCBI View Article : Google Scholar
|
|
59
|
Que ZJ, Yang Y, Liu HT, Shang-Guan WJ, Yu
P, Zhu LH, Li HG, Liu HM and Tian JH: Jinfukang regulates
integrin/Src pathway and anoikis mediating circulating lung cancer
cells migration. J Ethnopharmacol. 267(113473)2021.PubMed/NCBI View Article : Google Scholar
|
|
60
|
Bergamaschi A, Tagliabue E, Sørlie T,
Naume B, Triulzi T, Orlandi R, Russnes HG, Nesland JM, Tammi R,
Auvinen P, et al: Extracellular matrix signature identifies breast
cancer subgroups with different clinical outcome. J Pathol.
214:357–367. 2008.PubMed/NCBI View Article : Google Scholar
|
|
61
|
Yang J, Hou Y, Zhou M, Wen S, Zhou J, Xu
L, Tang X, Du YE, Hu P and Liu M: Twist induces
epithelial-mesenchymal transition and cell motility in breast
cancer via ITGB1-FAK/ILK signaling axis and its associated
downstream network. Int J Biochem Cell Biol. 71:62–71.
2016.PubMed/NCBI View Article : Google Scholar
|
|
62
|
Eke I and Cordes N: Focal adhesion
signaling and therapy resistance in cancer. Semin Cancer Biol.
31:65–75. 2015.PubMed/NCBI View Article : Google Scholar
|
|
63
|
Murphy JM, Rodriguez YAR, Jeong K, Ahn EE
and Lim SS: Targeting focal adhesion kinase in cancer cells and the
tumor microenvironment. Exp Mol Med. 52:877–886. 2020.PubMed/NCBI View Article : Google Scholar
|
|
64
|
Liu D, Liu S, Fang Y, Liu L and Hu K:
Comprehensive analysis of the expression and prognosis for ITGBs:
Identification of ITGB5 as a biomarker of poor prognosis and
correlated with immune infiltrates in gastric cancer. Front Cell
Dev Biol. 9(816230)2022.PubMed/NCBI View Article : Google Scholar
|
|
65
|
Yang X, Chen L, Mao Y, Hu Z and He M:
Progressive and prognostic performance of an extracellular
matrix-receptor interaction signature in gastric cancer. Dis
Markers. 2020(8816070)2020.PubMed/NCBI View Article : Google Scholar
|
|
66
|
Shimizu S, Okumura T, Oshiro Y, Fukumitsu
N, Fukuda K, Ishige K, Hasegawa N, Numajiri H, Murofushi K, Ohnishi
K, et al: Clinical outcomes of previously untreated patients with
unresectable intrahepatic cholangiocarcinoma following proton beam
therapy. Radiat Oncol. 14(241)2019.PubMed/NCBI View Article : Google Scholar
|